Abstract
Transforming growth factor-β (TGF-β)-activated kinase 1 (TAK1), TAK1-binding protein 1 (TAB1) and TAB2 are components of the mitogen-activated protein kinase (MAPK) pathway. In this study, TAK1, TAB1 and TAB2 were characterized from golden pompano (Trachinotus ovatus), a marine fish of great economic value, and named as trTAK1, trTAB1 and trTAB2, respectively. The lengths of the cDNA sequences of the three genes were 2429 bp, 2068 bp and 4229 bp and encoded 575, 506 and 759 amino acids, respectively. The trTAK1, trTAB1 and trTAB2 genes shared high sequence identities and were well clustered with their counterparts from other fish species. Real-time qPCR analysis showed that the three genes were constitutively expressed in all the selected tissues of healthy pompano, and the expression levels of the three genes were significantly up-regulated in head kidney and spleen following Vibrio alginolyticus, lipolysaccharide (LPS) and polyinosinic polycytidylic acid (poly I:C) challenge, indicating their roles in the immune response against pathogens in golden pompano. Our results provide a basis for further study of the functions of these genes in golden pompano.
1. Introduction
Transforming growth factor-β (TGF-β) activated kinase 1 (TAK1, also known as MPKKK7), a member of the mitogen-activated protein kinase (MAPK) family, plays a key role in the “upstream of MAPK” pathway [1,2]. TAK1 can be activated by cytokines (such as: TGF-β, TNF-α and IL-1β), bone morph proteins (BMP), toll-like receptors (TLRs), immune-related B-cell receptors (BCR) and T-cell receptors (TCR) [1,3,4]. In vitro over-expression studies reveal that TAK1 is involved in the IKK, JNK and p38/MAPK signaling pathways mediated by TNFR1 and IL-1R/TLR, confirming its special role in inflammatory signaling pathways [5,6]. In the innate immune response of Drosophila, TAK1 is essential for the expression of many antimicrobial peptides and participates in the activation of JNK and NF-κB during resistance to Gram-negative bacteria [7,8]. Moreover, the IMD signaling pathway is severely impaired in TAK1-deficient Drosophila [9,10]. Additionally, TAK1-deficient mice showed a significant reduction in bone marrow Ig-positive B-cell numbers and the expression of survival factor Bcl-2 [11]. These results indicate that TAK1 plays an important role in the immune response against pathogens.
The activation of TAK1 requires TAK1-binding protein 1 (TAB1), TAB2 and TAB3 [12]. TAB1 binds to the N-terminus of TAK1 to promote the autophosphorylation of two threonine residues (Thr-184 and Thr-187) and one serine residue (Ser-192) of TAK1, which is essential for TAK1 kinase activity [13]. In addition, TAB1 can interact with P38α, leading to the phosphorylation and activation of P38α. The activated P38α then phosphorylates TAB1 at Ser-423, Thr-431 and Ser-438, and phosphorylated TAB1 reduces TAK1 activity, thereby establishing a feedback control mechanism to terminate TAK1 activation [14,15]. Therefore, TAB1 can selectively activate or inhibit TAK1 activation under the stimulation of different immune signals. Unlike TAB1, TAB2 binds to the C-terminus of TAK1 and promotes TAK1 activation through TNF receptor-associated factor (TRAF) [16]. TAB2 and TAB3 both contain a C-terminal NZF ubiquitin-binding domain, which can link to the lysine-63 (K-63) multiubiquitin chain of tumor necrosis factor receptor-associated factor 6 (TRAF6). Thus, when TAB2 is deficient, TAB3 can replace TAB2 to fulfil its functions [17]. Additionally, mammalian TAK1, TAB1, TAB2 and TAB3 trigger a signal cascade through the formation of a TAK1–TABs complex, leading to the activation of NF-κB, thereby promoting the secretion of a variety of immune factors, including pro-inflammatory cytokines and chemokines [18]. It has been found that the TAK1–TABs complex mediates a wide range of biological functions, including immunity, inflammatory responses, proliferation and differentiation, angiogenesis and myocardial homeostasis [19,20,21].
Thus far, TAK1 and/or TAB1 and/or TAB2 have been identified in several marine species, such as large yellow croaker (Larimichthys crocea) [22,23], grass carp (Ctenopharyngodon idellus) [24,25], orange-spotted grouper (Epinephelus coioides) [26,27], black carp (Mylopharyngodon piceus) [28,29], Japanese flounder (Paralichthys olivaceus) [30] and rainbow trout (Oncorhynchus mykiss) [31], providing valuable information about the roles of TAK1, TAB1 and TAB2 in fish innate immunity.
Golden pompano, Trachinotus ovatus, which is a species of percoid fish of high economic value, is widely distributed in tropical and subtropical oceans [32]. The annual production of golden pompano has reached approximately 120,000 tons in South China [33]. However, the frequent outbreaks and spread of diseases caused by bacteria [34], viruses [35] and parasites [36,37] limit the expansion of pompano farming and production. Understanding the roles of TAK1, TAB1 and TAB2 may contribute to preventing the diseases of golden pompano, as most diseases are accompanied by inflammatory responses. In the present study, the cDNA of trTAK1, trTAB1 and trTAB2 were cloned by RT-PCR and RACE-PCR methods. The sequence features of these three genes were analyzed by bioinformatics methods, and their transcription levels in normal tissues and in Vibrio alginolyticus, LPS and poly I:C infections were detected using real-time quantitative PCR (qPCR). Our study will contribute to further study of the functions of these genes in golden pompano.
2. Materials and Methods
2.1. Fish and Tissue Samples
Eighty healthy golden pompano weighing about 400 g were purchased from a local farm in Qinzhou (Guangxi, China), maintained at 26 ± 2 °C in a tank with air-pumped seawater in our breeding base for at least 2 weeks before experiments. The gill, brain, heart, head kidney, muscle, liver, spleen and intestine tissues were randomly isolated from four pompano for basal gene expression analysis.
The rest of the fish were randomly divided into the LPS-injected group, poly I:C-injected group, V. alginolyticus-injected group and control group. Fish in the LPS-injected group were intraperitoneally (i.p.) injected with LPS (dissolved in PBS, 2 μg/μL). Fish in the poly I:C-injected group were i.p. injected with poly I:C (dissolved in PBS, 2 μg/μL). Fish in the V. alginolyticus-injected group were i.p. injected with V. alginolyticus (dissolved in PBS, 5 × 108 CFU/mL) and fish in the control group were i.p. injected with PBS. The injection volume was 100 μL per fish. Additionally, at 0, 6, 12, 24, 48 h post-injection (hpi), head kidney (HK) and spleen tissues were collected from three fish per group. All tissue samples were immediately frozen in liquid nitrogen and stored at −80 °C until use.
2.2. Total RNA Extraction and cDNA Synthesis
A TRIzol Kit (TaKaRa, Japan) was used to extract the total RNA from the spleen tissue of four pompano following the manufacturer’s instructions. Spectrophotometry (Eppendorf, Germany) and 1% agarose gel electrophoresis were used to determine the quality and quantity of total RNA. A RevertAid First Strand cDNA Synthesis Kit (Thermo, USA) was used for transcribing the total RNA into first strand cDNA, and 3′ RACE cDNA and 5′ RACE cDNA were produced using a SMART RACE cDNA Amplification Kit (TaKaRa, Japan) according to the manufacturer’s instructions.
2.3. Molecular Cloning of trTAK1, trTAB1 and trTAB2 cDNA
Degenerate primers (Table 1) were designed based on the TAK1, TAB1 and TAB2 conserved sequences of large yellow croaker (GenBank accession numbers: MG210566.1, MG210567.1 and XM_019253325.1) and orange-spotted grouper (GenBanck accession number:s JX856141.1, KM669146.1 and KM669147.1) available in the GenBank, separately. Nested PCR (Bio-Rad, California, USA) was used to amplify partial cDNA fragments. The PCR amplification reaction system was 25 μL, including 1 μL first strand cDNA, 1 μL upstream and downstream primers (10 μM), 9.5 μL ddH2O and 12.5 μL 2 × EasyTaq® PCR SuperMix (TaKaRa, Osaka, Japan), and the PCR program was as follows: 94 °C for 4 min, 30 cycles of 94 °C for 30 s, 66 °C for 30 s, and 72 °C for 2 min, and 72 °C for 10 min. The 5′- and 3′-ends were obtained by nested 5′ and 3′ RACE PCR with intragenic primers and universal primers (UPM and NUP) (Table 1). The nested RACE PCR program was 94 °C for 4 min, followed by 30 cycles of 94 °C for 30 s, 66 °C for 30 s, 72 °C for 2 min, and a final extension at 72 °C for 10 min. All the PCR products were ligated into the pMD18-T vector (TaKaRa, Japan) and sequenced (Sangon, Shanghai, China). The full-length cDNA sequences of trTAK1, trTAB1 and trTAB2 were assembled by DNAstar software 7.0, separately.

Table 1.
Primers used in this study.
2.4. Sequence Analysis
The online website BLASTX (http://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 2 June 2020) was used to annotate the three gene sequences of golden pompano. The ExPASy website (http://web.expasy.org/translate/, accessed on 12 June 2021) was used for deducing the corresponding amino acids, theoretical molecular weight (MW) and isoelectric point (pI). The MegAlign software of the DNAstar 7 software package (http://www.dnastar.com/, accessed on 7 July 2021) was used to calculate the TAK1, TAB1 and TAB2 protein identities between golden pompano and other vertebrates. The simple modular architecture research tool (SMART) (http://smart.emblheidelberg.de/, accessed on 21 July 2021) was used to predict the protein domain. Phylogenetic analysis was constructed by MEGA 5.0 software using the neighbor-joining method with bootstrap settings at 10,000 replicates.
2.5. RT-qPCR
Real-time PCR for the target genes was performed using a SYBR® Green PreMix Ex TaqTM II (TaKaRa, Japan) and quantified on the LightCycler480 (Roche Applied Science, Basel Switzerland). The PCR program comprised denaturation at 94 °C for 3 min followed by 40 cycles of 94 °C for 30 s, 60 °C for 30 s, 72 °C for 20 s. The reaction volume was 20 μL, including 10 μL SYBR® Green PreMix Ex TaqTM ii (TaKaRa, Japan), 0.5 μL per primer (0.2 μM), 2 μL cDNA template, and 7 μL ddH2O. Each reaction was performed in triplicate. The relative expression levels of trTAK1, trTAB1 and trTAB2 in healthy fish tissues were normalized as β-actin expression levels. The transcript changes of trTAK1, trTAB1 and trTAB2 in the head kidney and spleen of pompano after bacterial, PBS and poly I:C stimulated were analyzed by the 2△△CT method, and the control group was simulated with PBS at the same sampling time point and calculated by fold change [38]. The primers for qPCR are listed in Table 1.
2.6. Statistical Analysis
The qPCR data were calculated in GraphPad Prism 7.0 software and expressed as the mean ± standard error (SE). All assumptions were met prior to data analysis, and the results were analyzed by one-way analysis of variance (ANOVA) using SPSS 25.0 software. p values of less than 0.05 were regarded as statistically significant; p values of less than 0.01 were considered to represent an extremely significant difference.
3. Results
3.1. Sequence Features of trTAK1
The full cDNA sequence of trTAK1 (GenBank accession number: MN539080) was 2429 bp, containing a 111 bp 5′-untranslated region (5′-UTR), a 590 bp 3′-UTR with a typical poly(A) structure and a 1728 bp open reading frame (ORF) encoding 575 amino acids (aa) (Figure 1A). The predicted molecular weight and the theoretical pI of trTAK1 were 64.28 kDa and 5.99, respectively. Multiple sequence alignment showed that trTAK1 had a 95.1–97.9% identity with its counterparts in other fish species (Table 2), among which trTAK1 had the highest identity with greater amberjack (Seriola dumerili) TAK1 (97.9%), followed by large yellow croaker TAK1 (97%), and the lowest identity with yellowtail kingfish (Seriola lalandi) TAK1 (95.1%). Additionally, it had higher identities with TAK1s of human (80.5%) and mouse (81.1%) than trTAB1 and trTAB2 with TAB1s and TAB2s of human and mouse. Protein domain analysis revealed that trTAK1 had a S_TKc domain at its N-terminus and a CCR domain at its C-terminus (Figure 1B). These domains, which were conserved, exist in the large yellow croaker, human, emperor penguin and western painted turtle (Figure 2).

Figure 1.
Nucleotide and deduced amino acid sequence of the trTAK1 gene (A) and its predicted domains (B). The start codon (atg) and stop codon (tga) are marked in cyan. The CCR domain is marked in green, and the S_TKc domain is marked in red.

Table 2.
The identities (%) between the three genes’ amino acid sequence and other vertebrates.

Figure 2.
Multiple amino acid sequence alignment between trTAK1 and other vertebrates’ TAK1. The S_TKc and CCR domains of trTAK1 are boxed. Asterisk is regarded as the number of amino acids, and there will be one asterisk for every 10 amino acids. The species are golden pompano (T. ovatus), large yellow croaker (L. croea), human (H. sapiens), emperor penguin (A. forsteri) and western painted turtle (C. picta), and their GenBank accession numbers are listed in phylogenetic tree.
3.2. Sequence Features of trTAB1
The full-length trTAB1cDNA sequence (GenBank accession number: MN539081) was 2068 bp, containing a 19 bp 5′-untranslated region (5′-UTR), a 528 bp 3′-UTR with a typical poly(A) structure and a 1521 bp open reading frame (ORF) encoding 506 amino acids (aa) (Figure 3A). The predicted molecular weight and the theoretical pI of trTAB1 were 54.69 kDa and 5.67, respectively. Multiple sequence alignment showed that pompano TAB1 had a 92.1–96.4% identity (Table 2) with those in other fish species, among which trTAB1 had the highest identity with greater amberjack TAB1 and yellowtail kingfish TAB1 (96.4%), followed by orange-spotted grouper TAB1 (96%), and the lowest identity with half smooth tongue sole TAB1 (92.1%); in addition, it had lower identities with that of human (65.0%) and mouse (64.8%). Protein domain analysis revealed that trTAB1 contained a sigma factor PP2C-like phosphatase (PP2C_SIG) domain (Figure 3B). Multiple sequence analysis indicated that the TAB1s of other vertebrates also contained typical PP2C domains at their amino termini (Figure 4).
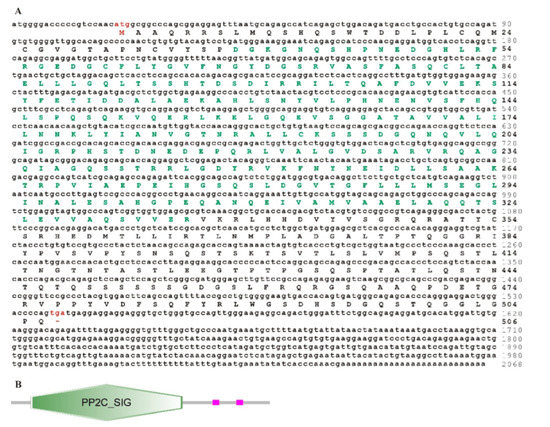
Figure 3.
Nucleotide and deduced amino acid sequence of the trTAB1 gene (A) and its predicted domains (B). The start codon (atg) and stop codon (tga) are marked in red. The PP2C_SIGdomain is marked in green.
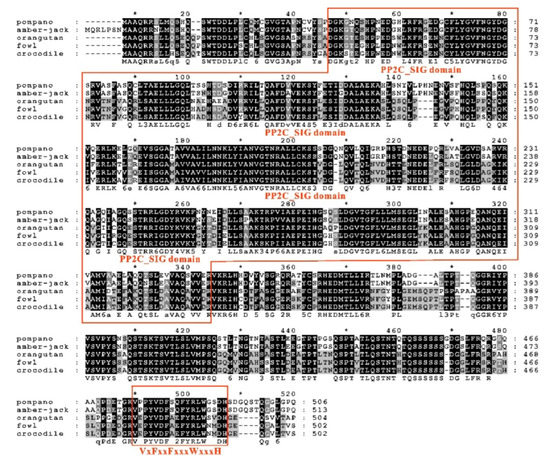
Figure 4.
Multiple amino acid sequence alignment between trTAB1 and other vertebrates’ TAB1. The PP2C_SIG domains and ‘VxFxxFxxxWxxxH’ sequence of trTAB1 are boxed. Asterisk is regarded as the number of amino acids, and there will be one asterisk for every 10 amino acids. The species are golden pompano (T. ovatus), greater amber jack (S. dumerili), Sumatran orangutan (P. abelii), red jungle fowl (G. gallus) and etuarine crocodile (C. porosus), and their GenBank accession numbers are listed in phylogenetic tree.
3.3. Sequence Features of trTAB2
The complete cDNA sequence of trTAB2 (GenBank accession number: MN539082) was 4229 bp, containing a 363 bp 5′-untranslated region (5′-UTR), a 1586 bp 3′-UTR with a typical poly(A) structure and a 2280 bp open reading frame (ORF) encoding 759 amino acids (aa) (Figure 5A). The predicted molecular weight and the theoretical pI of trTAB1 were 54.69 kDa and 5.67, respectively. Multiple sequence alignment showed that the identities between trTAB2 and TAB2 of other fish species ranged from 86.3% to 98.1% (Table 2). trTAB2 had the highest identity with yellowtail kingfish TAB2 (98.1%), followed by greater amberjack TAB2 (97.9%), and the lowest identity with half smooth tongue sole TAB2 (86.3%). Domain prediction showed that trTAB2 contains an amino-terminating CUE domain, a CCR domain and a carboxy-terminating ZnF domain (Figure 5B), similar to TAB2 of other vertebrates (Figure 6).

Figure 5.
Nucleotide and deduced amino acid sequence of the trTAB2 gene (A) and its predicted domains (B). The start codon (atg) and stop codon (tga) are marked in pink. The CCR domain is marked in green, the ZnF domain is marked in cyan and the CUE domain is marked in red.
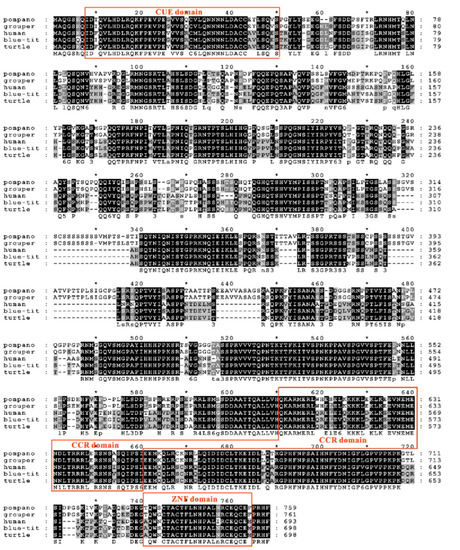
Figure 6.
Multiple amino acid sequence alignment between trTAB2 and other vertebrates’ TAB2. The CUE, CCR and ZNF domains of trTAB2 are boxed. Asterisk is regarded as the number of amino acids, and there will be one asterisk for every 10 amino acids. The species are golden pompano (T. ovatus), orange-spotted grouper (E. coioides), human (H. sapiens), Eurasian blue tit (C. caeruleus) and western painted turtle (C. picta), and their GenBank accession numbers are listed in phylogenetic tree.
3.4. Phylogenetic Analysis of trTAK1, trTAB1 and trTAB2
Phylogenetic trees were constructed with the TAK1, TAB1 and TAB2 protein sequences of various species to investigate the evolutionary relationship of trTAK1, trTAB1 and trTAB2. The results showed that TAK1 (Figure 7A), TAB1 and TAB2 (Figure 7B) homologues could be divided into four groups: birds, reptiles, mammals and fishes, in agreement with previous studies [23]. Moreover, trTAK1 was clustered closely with Japanese flounder TAK1; trTAB1 shared the closest evolutionary relationship with greater amberjack TAB1 and yellowtail kingfish TAB1. Similarly, trTAB2 was grouped closely with the TAB2s of great amberjack and yellowtail kingfish, which was in line with the results of the sequence comparison (Table 2). Additionally, the phylogenetic analysis of trTAK1, trTAB1 and trTAB2 showed a highly correlated evolutionary relationship with other piscine TAK1, TAB1 and TAB2.
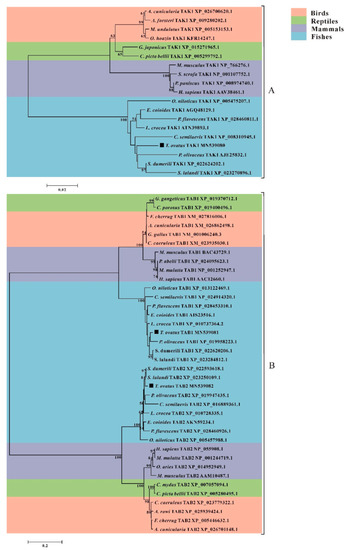
Figure 7.
Evolutionary relationships of the TAK1 (A), TAB1 and TAB2 (B) of vertebrates. The tree was inferred using the neighbor-joining method with bootstrap settings of 10,000 replicates. The trTAK1, trTAB1 and trTAB2 analyzed in the study are marked by black boxes. The evolutionary distances were computed using the JTT matrix-based method. The analysis of A and B involved 19 and 38 amino acid sequences, separately. The sequences used for phylogenetic analysis of TAK1 and/or TAB1 and/or TAB2 in different species, including T. ovatus, S. dumerili, E. coioides, L. crocea, P. olivaceus, C. semilaevis, P. flavescens, S, lalandi, O. niloticus, P. paniscus, H. sapiens, M. musculus, A. cunicularia, A. forsteri, M. undulatus, O. hoazin, G. japonicus, C. picta bellii, S. scrofa, M. mulatta, P. abelii, C. caeruleus, F. cherrug, G. gallus, G. gangeticus, C. porosus, O. aries, A. rowi, C. mydas, and the accession numbers for each sequence are given after the gene name.
3.5. Basal Tissue Expression of trTAK1, trTAB1 and trTAB2
The expression of trTAK1, trTAB1 and trTAB2 in tissues of healthy golden pompano was detected using qPCR. The results showed that the transcription of trTAK1 and trTAB1 was constitutive in all tested healthy pompano tissues (Figure 8). Furthermore, the mRNA of trTAK1 was highly expressed in the liver, intestine and brain, but weakly expressed in the heart and muscle, and the expression of trTAB1 was in agreement with that of grass carp TAB1 and large yellow croaker TAB1 [22,25], whereas the highest expression of trTAB2 was observed in the intestine, followed by the liver, brain, gill, spleen, head kidney, heart and muscle.
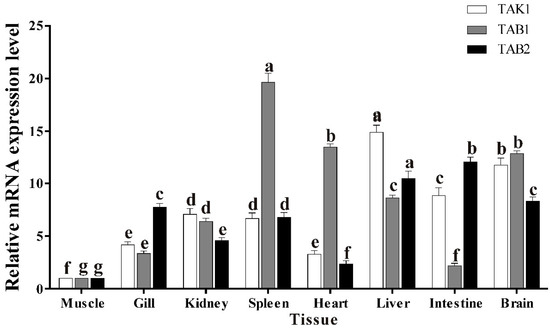
Figure 8.
Expression patterns of trTAK1, trTAB1 and trTAB2 in tissues of healthy golden pompano. The expression values were normalized to β-actin, and the data were expressed as mean ± SE. The expression level in muscle was set to 1.0 as control. Bars that do not share a letter represent a significant difference (p < 0.05) of the same gene among different tissues.
3.6. Transcripts Changes of trTAK1, trTAB1 and trTAB2 in Response to LPS, Poly I:C and V. alginolyticus Challenges
To investigate the roles of trTAK1, trTAB1 and trTAB2 during pathogen invasion, the fish were stimulated with LPS, poly I:C and V. alginolyticus, and the expression of these three genes was analyzed. The results are shown in Figure 9A–F. Following LPS stimulation, the expression level of trTAK1 reached the highest level in the HK and spleen at 12 h, then gradually returned to the normal value, and remained with no significant change at 48 h. In contrast, trTAB1 reached peaks in the HK and spleen at 12 h and 24 h post LPS stimulation, respectively, and then decreased to the normal level. The expression of trTAB2 in the HK and spleen increased from 24 h to 48 h post LPS stimulation, and peaked at 24 h.
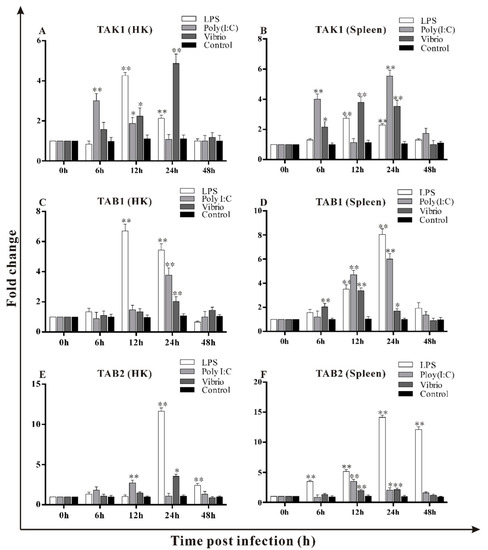
Figure 9.
Expression patterns of trTAK1, trTAB1 and trTAB2 in spleen and head kidney (HK) following the Vibrio alginolyticus, LPS and poly I:C challenge (A–F). The expression changes of the three genes were analyzed using the 2△△CT method. The relative expressions of target genes in each tissue were normalized to that of β-actin and fold-change relative to the expression levels at time 0 (set as 1). Results are expressed as mean ± SE from three independent triplicated experiments. Asterisks and double stars indicate that the same experimental group is significantly different from 0 h (* p < 0.05, ** p < 0.01).
After poly I:C stimulation, the expression of trTAK1 in the HK and spleen peaked at 6 h, and then began to decline. At 24 h, the expression of trTAK1 in the spleen peaked again, but remained unchanged in HK. The expression of trTAB1 in the HK and spleen reached its highest at 24 h, and decreased to the normal level at 48 h. The expression of trTAB2 in the HK and spleen peaked at 12 h, and then decreased.
After V. alginolyticus challenge, the expression level of trTAK1 in the HK and spleen increased significantly from 12 h to 24 h, and returned to the normal level at 48 h. The expression level of trTAB1 peaked at 24 h and 12 h in the HK and spleen, respectively. The expression of trTAB2 in the HK and spleen peaked at 24 h, then began to decline; there were no changes at 48 h.
4. Discussion
TAK1, which can be activated by many pro-inflammatory signals, plays a critical role in inflammation and regulates innate and adaptive immune responses [39]. In mammals, TABs activate TAK1 by forming a kinase complex (TAK–TABs), which is required for the activation of NF-κB [40]. However, the molecular characteristics of TAK1 and TABs in teleost fish, as well as their immune function, remain largely unknown—although they have been reported in grass carp [25], black carp [28] and large yellow croaker [22]. To understand the characteristics and roles of trTAK1, trTAB1 and trTAB2, their gene and amino acid sequences and expression patterns before and after LPS, poly I:C and V. alginolyticus infection were investigated.
In this study, trTAK1 possessed a S_TKc domain at its N-terminus and a CCR domain at its C-terminus (Figure 1B). These domains are conserved in the large yellow croaker, human, emperor penguin and western painted turtle (Figure 2). Similar results were also observed in TAK1s of grass carp [24], Japanese flounder [30], blunt snout bream (Megalobrama amblycephala) [41] and mammals [42,43]. The S_TKc domain plays an important role in the interaction between TAK1 and TAB1 and is essential for the activation of IKK and MAPKs [44,45]. Meanwhile, the CCR domain plays key roles in the interaction between TAK1 and TAB2, and mediating the combination of TAK1 and TRAF6 [13,46]. The finding of these two domains in pompano TAK1 indicates the conserved functions of TAK1 in fishes and mammals. trTAB1 contained a sigma factor PP2C-like phosphatase (PP2C_SIG) domain (Figure 3B), in agreement with those of TAB1s in black carp [29], grass carp [24], orange-spotted grouper and large yellow croaker [22]. PP2C is a protein phosphatase involved in the regulation of the MAPK signaling pathway [47]. Although TAB1 contains a PP2C-like domain, it lacks key residues required for binding divalent cations and catalytic activity, and therefore has no phosphatase activity, so TAB1 is considered a pseudophosphatase [48]. Previous studies have shown that TAB1 Phe-484, located in the conserved motif (PYVDXA/TXF) that was at the α-helix of the carboxyl terminus region of TAB1s in large yellow croaker [22,23], orange-spotted grouper [27], grass carp and human [25], was crucial for TAK1 binding and activation [18,49]. Interestingly, a motif ‘PYVDFSQFYRLWGSDH’ (478–493aa) and a conserved Phe-482 residue were also found in the carboxy-terminal region of trTAB1, suggesting that TAB1s are highly conserved in fish species and the Phe residue within the conserved motif of TAB1 proteins might be involved in TAK1 activation and interaction [22]. In contrast, trTAB2 was composed of an amino-terminating CUE domain, a CCR domain and a carboxy-terminating ZnF domain (Figure 5B), similar to the TAB2s of other vertebrates (Figure 6) and fishes [25,26]. CUE is a conserved ubiquitin-binding domain and is essential for TAB2-dependent cell responses [50,51]; the CCR domain plays an important role in the interaction between TAB2 and TAK1 [46], and ZnF domain has a significant role in the binding between TAB2 and TRAF6 [52]. These results suggest that trTAB2 may also have similar functions to mammalian TAB2 in the innate immune response. In addition, trTAK1, trTAB1 and trTAB2 showed a highly correlated evolutionary relationship (Figure 7) and identities (Table 2) with other piscine TAK1s, TAB1s and TAB2s, which indicates that trTAK1, trTAB1 and trTAB2 might have a similar function with those of other fish species [31].
RT-qPCR results showed that trTAK1, trTAB1 and trTAB2 were constitutively distributed in all of the tested healthy pompano tissues and their expression levels were distinct in different tissues (Figure 8), consistent with their expression in human [53], rainbow trout [31], large yellow croaker and grass carp [23,54], suggesting that these genes may perform similar biological functions in different species. The different expressions of trTAK1, trTAB1 and trTAB2 in different tissues indicate these three genes might be tissue-specific [55]. Furthermore, trTAK1 was the highest in the liver, followed by the brain, intestine, head kidney, spleen, gill, heart and muscle, similar to that of grass carp TAK1 and large yellow croaker TAK1 [22,24], implying that they might have a close relationship in many physiological events. However, different tissue expression profiles of TAK1s were observed in Japanese flounder and grass crab [30,31], which might be explained by differences in the fish species and feeding conditions [56]. The spleen is an important systemic lymphoid organ in fish. The highest expression of trTAB1 was observed in the spleen and heart, indicating that TAB1 actively participates in the fish immune response and plays an important role in the heart [57]. Meanwhile, trTAB2 was most highly expressed in the liver. Additionally, the high expression of TAB2 in the liver was observed for that of large yellow croaker [23] and grass carp [25]. Besides, a TAB2-deficient mouse resulted in embryonic lethality due to liver degeneration and apoptosis [58]. These results indicate that TAB2 plays a crucial role in the liver [59], and fish TAB2 might play a similar role in the function of the liver [60].
TAK1 plays an important role in both innate and adaptive immune responses in mediating inflammation [28], and performs different biological roles in many signaling pathways, such as TGF-β, WNT and NF-κB [7]. In mammals, TAK1 and its binding proteins (TAB1, TAB2 and TAB3), which are important signal transduction proteins in MyD88-dependent TLR signaling pathways, can activate the host immune response [61,62,63,64]. In addition, previous studies have shown that TAK1 is involved in the innate immune response and inducing the activity of NF-κB by forming a complex with TAB1 and TAB2 in teleost fishes [22,23,25,41]. In the present study, following LPS stimulation, the expression level of trTAK1 reached its highest level in the HK and spleen at 12 h. In contrast, trTAB1 reached its highest level in the HK and spleen at 12 h and 24 h, respectively, whereas the expression of trTAB2 in the HK and spleen peaked at 24 h. The changes in the expression patterns of these three pompano genes were consistent with those observed in previous studies of other fish. For example, amphioxus TAK1 was significantly increased after LPS stimulation [65,66]; Japanese flounder TAK1 expression showed significant upregulation after PAMPs challenge [30]; large yellow croaker TAK1 and TAB1 were induced in LCK cells after LPS and poly I:C stimulation, and the interaction of TAK1 and TAB1 enhanced LPS-induced cytokine release by modulating NF-κB activation [22]. In addition, it also showed that large yellow croaker TAK1, TAB1 and TAB2 were increased in the spleen and HK by V. parahemolyticus stimulation [23]. These results indicate that TAK1, TAB1 and TAB2 are involved in the immune response against bacterial infection in golden pompano. After poly I:C stimulation, the expression of trTAK1, trTAB1 and trTAB2 in the HK and spleen were up-regulated. Similarly, high expressions of TAK1, TAB1 and TAB2 following poly I:C stimulation have also been reported in other fish species. The large yellow croaker TAK1 and TAB1 were induced in the spleen after poly I:C stimulation [67]. Black carp TAK1 was induced in MPAK cells after LPS and poly I:C stimulation [28]. Additionally, the interaction between TAK1 and TAB1 can boost the IFN signaling mediated by IRF7 during black carp innate immune activation against GCRV and SVCV [29]. In addition, it has been reported that the knockdown of TAB2 significantly reduced the production of the inflammatory cytokines TNF-α, IL-8 and IL-1β in miiuy croaker (Miichthys miiuy) macrophages [68]. These results suggest that the trTAK1, trTAB1 and trTAB2 genes are involved in the antiviral process of golden pompano. After V. alginolyticus challenge, the expression level of trTAK1 in the HK and spleen increased significantly from 12 h to 24 h, and returned to the normal level at 48 h. The expression level of trTAB1 peaked at 24 h and 12 h in the HK and spleen, respectively. The expression of trTAB2 in the HK and spleen peaked at 24 h, then began to decline, and there were no changes at 48 h. Similar results were observed in other fish species. For example, rainbow trout TAK1 was strongly induced in RTH-149 cells following E. tarda and LPS stimulation [31]. Soiny mullet (Liza haematocheila) TAK1 was up-regulated in the spleen and HK after S. dysgalactiae infection [56]. Western mosquitofish (Gambusia affinis) TAK1 was significantly increased post A. hydrophila injection [69]. In addition, the expressions of pro-inflammatory cytokines (IL-1β, IL-8 and TNF-α) were induced in grass carp TAK1/TAB1 co-overexpressed cells after the potential vaccine antigen of V. mimicus stimulation. These results suggest that TAK1, TAB1 and TAB2 play an important role in the fish immune defense against microbial pathogens.
5. Conclusions
In the present study, TAK1 and its binding proteins (TAB1 and TAB2) were characterized from golden pompano and their expression levels in normal tissues, and HK and spleen following LPS, poly I:C and V. alginolyticus stimulation, were analyzed. The studied trTAK1, trTAB1 and trTAB2 shared similar characteristics with their counterparts in other vertebrates. High expression levels of these three genes were observed in the spleen and liver. Following LPS, poly I:C and V. alginolyticus infection, these three genes were up-regulated in the HK and spleen, indicating their roles in the immune response against pathogens.
Author Contributions
Conceptualization, Y.W.; methodology, Y.W. and K.L.; software, Y.W.; validation, Y.X. and J.H.; formal analysis, K.L. and Y.X.; investigation, K.L. and Y.X.; resources, Y.W.; data curation, K.L., Y.X. and J.H.; writing—original draft preparation, Y.X. and K.L.; writ-ing—review and editing, Y.W., J.H. and Y.X.; visualization, Y.X., K.L. and J.H.; supervision, Y.W.; project administration, Y.W. and K.L.; funding acquisition, Y.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China, grant number 31860736.
Institutional Review Board Statement
This study was approved by the Animal Care and Welfare Committee of Guangxi University (Gxu-2022-281).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ajibade, A.A.; Wang, H.Y.; Wang, R.-F. Cell type-specific function of TAK1 in innate immune signaling. Trends. Immunol. 2013, 34, 307–316. [Google Scholar] [CrossRef] [PubMed]
- Hirose, T.; Fujimoto, W.; Tamaai, T.; Kim, K.H.; Matsuura, H.; Jetten, A.M. TAK1: Molecular cloning and characterization of a new member of the nuclear receptor superfamily. Mol. Endocrinol. 1994, 8, 1667–1680. [Google Scholar] [PubMed] [Green Version]
- Dai, L.; Thu, C.A.; Liu, X.-Y.; Xi, J.; Cheung, P.C.F. TAK1, more than just innate immunity. Iubmb Life 2012, 64, 825–834. [Google Scholar] [CrossRef]
- Shirakabe, K.; Yamaguchi, K.; Shibuya, H.; Irie, K.; Matsuda, S.; Moriguchi, T.; Gotoh, Y.; Matsumoto, K.; Nishida, E. TAK1 mediates the ceramide signaling to stress-activated protein kinase c-Jun N-terminal kinase. J. Biol. Chem. 1997, 272, 8141–8144. [Google Scholar] [CrossRef] [Green Version]
- Takaesu, G.; Surabhi, R.M.; Park, K.J.; Ninomiya-Tsuji, J.; Matsumoto, K.; Gaynor, R.B. TAK1 is critical for I kappa B kinase-mediated activation of the NF-kappa B pathway. J. Mol. Biol. 2003, 326, 105–115. [Google Scholar] [CrossRef]
- Shim, J.H.; Xiao, C.C.; Paschal, A.E.; Bailey, S.T.; Rao, P.; Hayden, M.S.; Lee, K.Y.; Bussey, C.; Steckel, M.; Tanaka, N.; et al. TAK1, but not TAB1 or TAB2, plays an essential role in multiple signaling pathways in vivo. Genes Dev. 2005, 19, 2668–2681. [Google Scholar] [CrossRef] [Green Version]
- Delaney, J.R.; Mlodzik, M. TGF-beta activated kinase-1—New insights into the diverse roles of TAK1 in development and immunity. Cell Cycle 2006, 5, 2852–2855. [Google Scholar] [CrossRef] [Green Version]
- Paquette, N.; Conlon, J.; Sweet, C.; Rus, F.; Wilson, L.; Pereira, A.; Rosadini, C.V.; Goutagny, N.; Weber, A.N.R.; Lane, W.S.; et al. Serine/threonine acetylation of TGF beta-activated kinase (TAK1) by Yersinia pestis YopJ inhibits innate immune signaling. Proc. Natl. Acad. Sci. USA 2012, 109, 12710–12715. [Google Scholar] [CrossRef] [Green Version]
- Boutros, M.; Agaisse, H.; Perrimon, N. Sequential activation of signaling pathways during innate immune responses in Drosophila. Dev. Cell 2002, 3, 711–722. [Google Scholar] [CrossRef] [Green Version]
- Park, J.M.; Brady, H.; Ruocco, M.G.; Sun, H.Y.; Williams, D.; Lee, S.J.; Kato, T.; Richards, N.; Chan, K.; Mercurio, F.; et al. Targeting of TAK1 by the NF-kappa B protein Relish regulates the JNK-mediated immune response in Drosophila. Genes Dev. 2004, 18, 584–594. [Google Scholar] [CrossRef] [Green Version]
- Shinohara, H.; Nagashima, T.; Cascalho, M.I.; Kurosaki, T. TAK1 maintains the survival of immunoglobulin -chain-positive B cells. Genes Cells 2016, 21, 1233–1243. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.-R.; Lei, C.-Q. TAK1-TABs Complex: A Central Signalosome in Inflammatory Responses. Front. Immunol. 2021, 11, 608976. [Google Scholar] [CrossRef] [PubMed]
- Shibuya, H.; Yamaguchi, K.; Shirakabe, K.; Tonegawa, A.; Gotoh, Y.; Ueno, N.; Irie, K.; Nishida, E.; Matsumoto, K. TAB1: An activator of the TAK1 MAPKKK in TGF-beta signal transduction. Science 1996, 272, 1179–1182. [Google Scholar] [CrossRef]
- Ge, B.X.; Gram, H.; Di Padova, F.; Huang, B.; New, L.; Ulevitch, R.J.; Luo, Y.; Han, J.H. MAPKK-independent activation of p38 alpha mediated by TAB1-dependent autophosphorylation of p38 alpha. Science 2002, 295, 1291–1294. [Google Scholar] [CrossRef] [PubMed]
- Shin, M.-S.; Shinghirunnusorn, P.; Sugishima, Y.; Nishimura, M.; Suzuki, S.; Koizumi, K.; Saiki, I.; Sakurai, H. Cross interference with TNF-alpha-induced TAK1 activation via EGFR-mediated p38 phosphorylation of TAK1-binding protein 1. BBA-Mol. Cell. Res. 2009, 1793, 1156–1164. [Google Scholar]
- Mihaly, S.R.; Ninomiya-Tsuji, J.; Morioka, S. TAK1 control of cell death. Cell Death Differ. 2014, 21, 1667–1676. [Google Scholar] [CrossRef] [Green Version]
- Aashaq, S.; Batool, A.; Andrabi, K.I. TAK1 mediates convergence of cellular signals for death and survival. Apoptosis 2019, 24, 3–20. [Google Scholar] [CrossRef]
- Ono, K.; Ohtomo, T.; Sato, S.; Sugamata, Y.; Suzuki, M.; Hisamoto, N.; Ninomiya-Tsuji, J.; Tsuchiya, M.; Matsumoto, K. An evolutionarily conserved motif in the TAB1 C-terminal region is necessary for interaction with and activation of TAK1 MAPKKK. J. Biol. Chem. 2001, 276, 24396–24400. [Google Scholar] [CrossRef] [Green Version]
- Landstrom, M. The TAK1-TRAF6 signalling pathway. Int. J. Biochem. Cell. 2010, 42, 585–589. [Google Scholar] [CrossRef]
- Koitabashi, N.; Danner, T.; Zaiman, A.L.; Pinto, Y.M.; Rowell, J.; Mankowski, J.; Zhang, D.; Nakamura, T.; Takimoto, E.; Kass, D.A. Pivotal role of cardiomyocyte TGF-beta signaling in the murine pathological response to sustained pressure overload. J. Clin. Investig. 2011, 121, 2301–2312. [Google Scholar] [CrossRef]
- Morioka, S.; Inagaki, M.; Komatsu, Y.; Mishina, Y.; Matsumoto, K.; Ninomiya-Tsuji, J. TAK1 kinase signaling regulates embryonic angiogenesis by modulating endothelial cell survival and migration. Blood 2012, 120, 3846–3857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bao, S.-Y.; Sun, Q.-X.; Yao, C.-L. The interaction of TAK1 and TAB1 enhances LPS-induced cytokine release via modulating NF-kappa B activation (Larimichthys crocea). Fish Shellfish. Immunol. 2018, 74, 450–458. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Jiang, L.; Liu, G.; Wu, C.; Liu, B.; Liu, L.; Lv, Z.; Gong, L.; Song, X. Molecular characterization and expression of TAK-binding proteins (TAB1-3) in Larimichthys crocea infected by Vibrio parahemolyticus and LPS. Dev. Comp. Immunol. 2019, 98, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Li, Y.-W.; Pan, H.-J.; Wu, S.-Q.; Shi, C.-B.; Luo, X.-C.; Li, A.-X. Grass carp (Ctenopharyngodon idella) TRAF6 and TAK1: Molecular cloning and expression analysis after Ichthyophthirius multifiliis infection. Fish Shellfish. Immunol. 2013, 34, 1514–1523. [Google Scholar] [CrossRef]
- Zhao, F.; Li, Y.-W.; Pan, H.-J.; Shi, C.-B.; Luo, X.-C.; Li, A.-X.; Wu, S.-Q. TAK1-binding proteins (TAB1 and TAB2) in grass carp (Ctenopharyngodon idella): Identification, characterization, and expression analysis after infection with Ichthyophthirius multifiliis. Fish Shellfish. Immunol. 2014, 38, 389–399. [Google Scholar] [CrossRef]
- Li, Y.W.; Li, X.; Wang, Z.; Mo, Z.Q.; Dan, X.M.; Luo, X.C.; Li, A.X. Orange-spotted grouper Epinephelus coioides Tak1: Molecular identification, expression analysis and functional study. J. Fish. Biol. 2015, 86, 417–430. [Google Scholar] [CrossRef]
- Hu, Y.-Z.; Li, X.; Han, R.; Jiang, B.; Li, Y.-W.; Dan, X.-M.; Li, A.-X. Molecular identification and expression analysis of TAB1 from orange-spotted grouper (Epinephelus coioides). Dev. Comp. Immunol. 2019, 90, 152–156. [Google Scholar] [CrossRef]
- Wang, C.; Peng, J.; Zhou, M.; Liao, G.; Yang, X.; Wu, H.; Xiao, J.; Feng, H. TAK1 of black carp positively regulates IRF7-mediated antiviral signaling in innate immune activation. Fish Shellfish. Immunol. 2019, 84, 83–90. [Google Scholar] [CrossRef]
- Zou, Z.; Xie, X.; Li, W.; Song, X.; Tan, Y.; Wu, H.; Xiao, J.; Feng, H. Black carp TAB1 up-regulates TAK1/IRF7/IFN signaling during the antiviral innate immune activation. Fish Shellfish. Immunol. 2019, 89, 736–744. [Google Scholar] [CrossRef]
- Zhao, H.; Chen, Y.; Wang, Z.; Zhang, Q.; Yu, H. Molecular Characterization, Expression Profiles, and Immunostimulation Responses of TRAF6 and TAK1 in Japanese Flounder (Paralichthys olivaceus). J. Ocean Univ. China 2019, 18, 165–176. [Google Scholar] [CrossRef]
- Jang, J.H.; Kim, H.; Cho, J.H. Molecular cloning and functional characterization of TRAF6 and TAK1 in rainbow trout, Oncorhynchus mykiss. Fish Shellfish Immunol. 2019, 84, 927–936. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Zhu, K.-C.; Guo, L.; Liu, B.-S.; Guo, H.-Y.; Zhang, N.; Yang, J.-W.; Zhang, D.-C. Molecular characterization of GRP94 and HSP90 alpha from Trachinotus ovatus, Linnaeus 1758 and their expression responses to various levels of stocking density stress and Cryptocaryon irritans infection. Aquaculture 2020, 529, 739601. [Google Scholar] [CrossRef]
- Ning, L.; Gao, L.; Zhou, W.; Liu, S.; Chen, X.; Pan, Q. Beneficial effects of dietary mulberry leaf along with multi-enzyme premix on the growth, immune response and disease resistance of golden pompano Trachinotus Ovatus. Aquaculture 2021, 535, 736396. [Google Scholar] [CrossRef]
- Sun, Y.; Ding, S.; He, M.; Liu, A.; Long, H.; Guo, W.; Cao, Z.; Xie, Z.; Zhou, Y. Construction and analysis of the immune effect of Vibrio harveyi subunit vaccine and DNA vaccine encoding TssJ antigen. Fish Shellfish. Immunol. 2020, 98, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Yu, Q.; Li, F.; Qin, X.; Dong, D.; Chen, B.; Qin, Q. First identification of the nervous necrosis virus isolated from cultured golden pompano (Trachinotus ovatus) in Guangxi, China. J. Fish. Dis. 2018, 41, 1177–1180. [Google Scholar] [CrossRef] [PubMed]
- Hamdi, I.; Hermida, M.; Kamanli, S.A.; Benmansour, B.; Ozak, A.A.; Boxshall, G.A. Caligus madeirensissp. nov. (Copepoda: Caligidae) Parasitic on Pompano, Trachinotus ovatus (Linnaeus, 1758), from Eastern Atlantic Waters, Surrounding the Madeira Archipelago, Portugal. Acta Parasitol. 2021, 66, 361–376. [Google Scholar] [CrossRef]
- Qiao, Y.; Shao, Y.; Pengsakul, T.; Chen, C.; Zheng, S.; Wu, W.; Hardjo, T.B. Morphological and molecular characterization of Ceratomyxa batam n. sp. (Myxozoa: Ceratomyxidae) infecting the gallbladder of the cultured Trachinotus ovatus (Perciformes: Carangidae) in Batam Island, Indonesia. Parasitol. Res. 2019, 118, 1647–1651. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T) (-Delta Delta C) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Oelze, B.; Elger, K.; Schadzek, P.; Burmeister, L.; Hamm, A.; Laggies, S.; Seiffart, V.; Gross, G.; Hoffmann, A. The inflammatory signalling mediator TAK1 mediates lymphocyte recruitment to lipopolysaccharide-activated murine mesenchymal stem cells through interleukin-6. Mol. Cell. Biochem. 2021, 476, 3655–3670. [Google Scholar] [CrossRef]
- Fechtner, S.; Fox, D.A.; Ahmed, S. Transforming growth factor beta activated kinase 1: A potential therapeutic target for rheumatic diseases. Rheumatology 2017, 56, 1060–1068. [Google Scholar]
- Xu, Y.D.; Zhu, B.; Zhang, R.; Tang, J.Z.; Liu, Y.; Wang, W.J.; Wang, Z.Z.; Mao, Y.; Zeng, G.Q.; Yan, J.P. TAK1 of blunt snout bream promotes NF-kappa B activation via interaction with TAB1 in response to pathogenic bacteria. Fish Shellfish Immunol. 2022, 120, 481–496. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, K.; Shirakabe, T.; Shibuya, H.; Irie, K.; Oishi, I.; Ueno, N.; Taniguchi, T.; Nishida, E.; Matsumoto, K. Identification of a member of the MAPKKK family as a potential mediator of TGF-beta signal-transduction. Science 1995, 270, 2008–2011. [Google Scholar] [CrossRef] [PubMed]
- Dempsey, C.E.; Sakurai, H.; Sugita, T.; Guesdon, F. Alternative splicing and gene structure of the transforming growth factor beta-activated kinase 1. BBA-Gene Struct. Expr. 2000, 1517, 46–52. [Google Scholar] [CrossRef]
- Scholz, R.; Sidler, C.L.; Thali, R.F.; Winssinger, N.; Cheung, P.C.F.; Neumann, D. Autoactivation of Transforming Growth Factor beta-activated Kinase 1 Is a Sequential Bimolecular Process. J. Biol. Chem. 2010, 285, 25753–25766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singhirunnusorn, P.; Suzuki, S.; Kawasaki, N.; Saiki, I.; Sakurai, H. Critical roles of threonine 187 phosphorylation in cellular stress-induced rapid and transient activation of transforming growth factor-beta-activated kinase 1 (TAK1) in a signaling complex containing TAK1-binding protein TAB1 and TAB2. J. Biol. Chem. 2005, 280, 7359–7368. [Google Scholar] [CrossRef] [Green Version]
- Takaesu, G.; Kishida, S.; Hiyama, A.; Yamaguchi, K.; Shibuya, H.; Irie, K.; Ninomiya-Tsuji, J.; Matsumoto, K. TAB2, a novel adaptor protein, mediates activation of TAK1 MAPKKK by linking TAK1 to TRAF6 in the IL-1 signal transduction pathway. Mol. Cell 2000, 5, 649–658. [Google Scholar] [CrossRef]
- Fjeld, C.C.; Denu, J.M. Kinetic analysis of human serine threonine protein phosphatase 2C alpha. J. Biol. Chem. 1999, 274, 20336–20343. [Google Scholar] [CrossRef] [Green Version]
- Conner, S.H.; Kular, G.; Peggie, M.; Shepherd, S.; Schuttelkopf, A.W.; Cohen, P.; Van Aalten, D.M.F. TAK1-binding protein 1 is a pseudophosphatase. Biochem. J. 2006, 399, 427–434. [Google Scholar] [CrossRef]
- Brown, K.; Vial, S.C.M.; Dedi, N.; Long, J.M.; Dunster, N.J.; Cheetham, G.M.T. Structural basis for the interaction of TAK1 kinase with its activating protein TAB1. J. Mol. Biol. 2005, 354, 1013–1020. [Google Scholar] [CrossRef]
- Kanayama, A.; Seth, R.B.; Sun, L.J.; Ea, C.K.; Hong, M.; Shaito, A.; Chiu, Y.H.; Deng, L.; Chen, Z.J. TAB2 and TAB3 activate the NF-kappa B pathway through binding to polyubiquitin chains. Mol. Cell 2004, 15, 535–548. [Google Scholar] [CrossRef]
- Kishida, S.; Sanjo, H.; Akira, S.; Matsumoto, K.; Ninomiya-Tsuji, J. TAK1-binding protein 2 facilitates ubiquitination of TRAF6 and assembly of TRAF6 with IKK in the IL-1 signaling pathway. Genes Cells 2005, 10, 447–454. [Google Scholar] [CrossRef] [PubMed]
- Kulathu, Y.; Akutsu, M.; Bremm, A.; Hofmann, K.; Komander, D. Two-sided ubiquitin binding explains specificity of the TAB2 NZF domain. Nat. Struct. Mol. Biol. 2009, 16, 1328–1330. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Cao, M.; Ni, Y.; Han, L.; Dai, A.; Chen, R.; Ning, X.; Liu, X.; Ke, K. Up-regulation of TAB3 is involved in neuronal apoptosis after intracerebral hemorrhage. Cell. Mol. Neurobiol. 2017, 37, 607–617. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.F.; Cao, S.L.; Liu, X.Y.; Zhu, X.C.; Zhou, X.H.; Li, J.N. Grass carp (Ctenopharyngodon idella) TAK1 promotes NF-kappa B activation via interaction with TAB1 in response to potential vaccine antigen of Vibrio mimicus. Aquaculture 2020, 529, 735712. [Google Scholar] [CrossRef]
- Chen, Z.J.; Bhoj, V.; Seth, R.B. Ubiquitin, TAK1 and IKK: Is there a connection? Cell Death Differ. 2006, 13, 687–692. [Google Scholar] [CrossRef]
- Qi, Z.; Xu, Y.; Wang, X.; Wang, S.; Zhang, Q.; Wang, Z.; Gao, Q. TLR13, TLR22, TRAF6, and TAK1 in the soiny mullet (Liza haematocheila): Molecular characterization and expression profiling analysis. Dev. Comp. Immunol. 2020, 112, 103774. [Google Scholar] [CrossRef]
- Komatsu, Y.; Shibuya, H.; Takeda, N.; Ninomiya-Tsuji, J.; Yasui, T.; Miyado, K.; Sekimoto, T.; Ueno, N.; Matsumoto, K.; Yamada, G. Targeted disruption of the Tab1 gene causes embryonic lethality and defects in cardiovascular and lung morphogenesis. Mech. Develop. 2002, 119, 239–249. [Google Scholar] [CrossRef]
- Sanjo, H.; Takeda, K.; Tsujimura, T.; Ninomiya-Tsuji, J.; Matsumoto, K.; Akira, S. TAB2 is essential for prevention of apoptosis in fetal liver but not for interleukin-1 signaling. Mol. Cell. Biol. 2003, 23, 1231–1238. [Google Scholar] [CrossRef] [Green Version]
- Ikeda, Y.; Morioka, S.; Matsumoto, K.; Ninomiya-Tsuji, J. TAK1 binding protein 2 is essential for liver protection from stressors. PLoS ONE 2014, 9, e88037. [Google Scholar] [CrossRef]
- Deng, F.; Xu, G.; Cheng, Z.; Huang, Y.; Ma, C.; Luo, C.; Yu, C.; Wang, J.; Xu, X.; Liu, S.; et al. Hepatitis B surface antigen suppresses the activation of nuclear factor kappa B pathway via interaction with the TAK1-TAB2 complex. Front. Immunol. 2021, 12, 618196. [Google Scholar] [CrossRef]
- Egan, C.E.; Sukhumavasi, W.; Butcher, B.A.; Denkers, E.Y. Functional aspects of Toll-like receptor/MyD88 signalling during protozoan infection: Focus on Toxoplasma gondii. Clin. Exp. Immunol. 2009, 156, 17–24. [Google Scholar] [CrossRef] [PubMed]
- McDonald, V.; Korbel, D.S.; Barakat, F.M.; Choudhry, N.; Petry, F. Innate immune responses against Cryptosporidium parvum infection. Parasite Immunol. 2013, 35, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Sasai, M.; Yamamoto, M. Pathogen recognition receptors: Ligands and signaling pathways by Toll-Like receptors. Int. Rev. Immunol. 2013, 32, 116–133. [Google Scholar] [CrossRef] [PubMed]
- Yarovinsky, F. Innate immunity to Toxoplasma gondii infection. Nat. Rev. Immunol. 2014, 14, 109–121. [Google Scholar] [CrossRef]
- Yin, D.; Li, W.; Fu, M.; Chen, L.; Ma, F.; Jin, P. Identification and characterization of a TAB1 gene involved in innate immunity of amphioxus (Branchiostoma belcheri). Gene 2016, 575, 294–302. [Google Scholar] [CrossRef]
- Cao, Y.; Jin, N.; Fan, M.; Lv, C.; Song, X.; Jin, P.; Ma, F. Phylogenetically conserved TAK1 participates in Branchiostoma belcheri innate immune response to LPS stimulus. Fish Shellfish Immunol. 2019, 94, 264–270. [Google Scholar] [CrossRef]
- Mu, Y.; Li, M.; Ding, F.; Ding, Y.; Ao, J. De Novo Characterization of the spleen transcriptome of the large yellow croaker (Pseudosciaena crocea) and analysis of the immune relevant genes and pathways involved in the antiviral response. PLoS ONE 2014, 9, e101069. [Google Scholar] [CrossRef] [Green Version]
- Ren, X.; Cui, J.; Xu, T.; Sun, Y. microRNA-128 inhibits the inflammatory responses by targeting TAB2 in miiuy croaker, Miichthys miiuy. Dev. Comp. Immunol. 2021, 117, 103976. [Google Scholar] [CrossRef]
- Liu, S.L.; Zhou, A.G.; Xie, S.L.; Sun, D.; Zhang, Y.; Sun, Z.L.; Chen, Y.F.; Zou, J.X. Immune-related genes expression analysis of Western mosquitofish (Gambusia affinis) challenged with Aeromonas hydrophila. Fish Shellfish Immunol. 2020, 102, 92–100. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).