Development and Validation of a Quantitative Polymerase Chain Reaction Assay for the Detection of Red Sea Bream Iridovirus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Primers, Hydrolysis Probe, and the Real-Time PCR Assay
2.2. Fish Populations
2.3. Experimental Infection
2.4. Nucleic Acid Extraction
2.5. Real-Time PCR Assay Validation: Analytical Evaluation
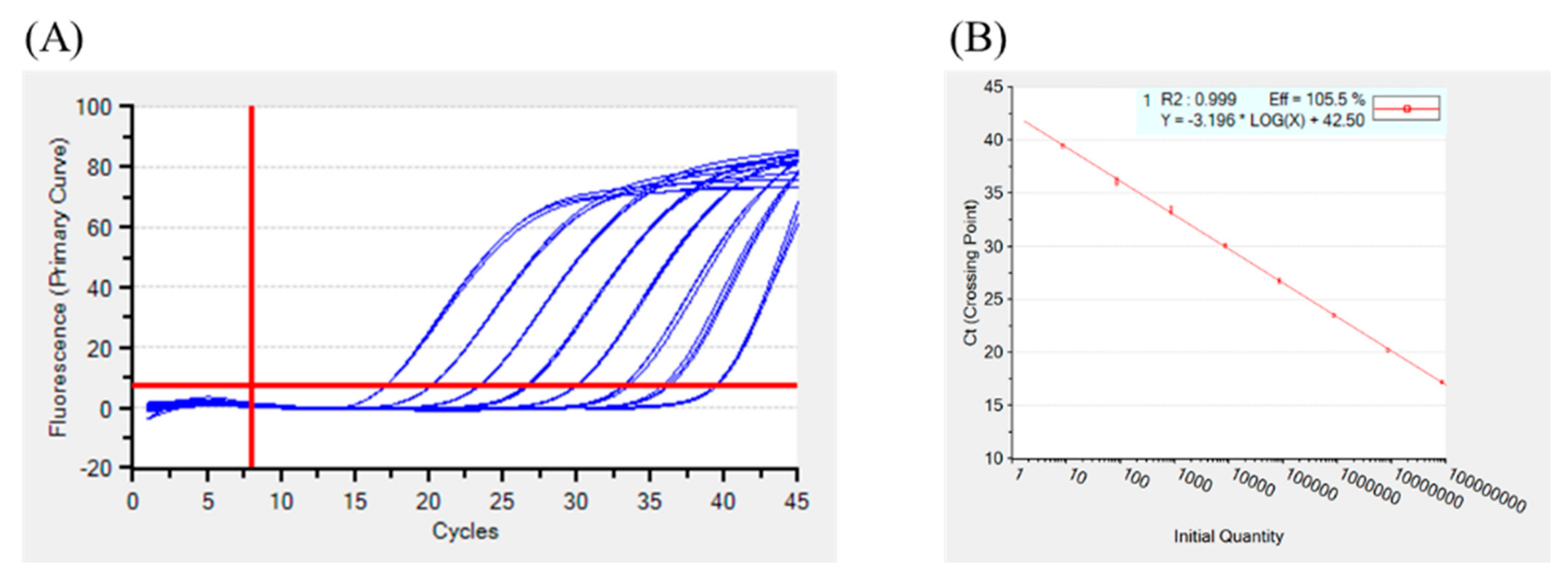
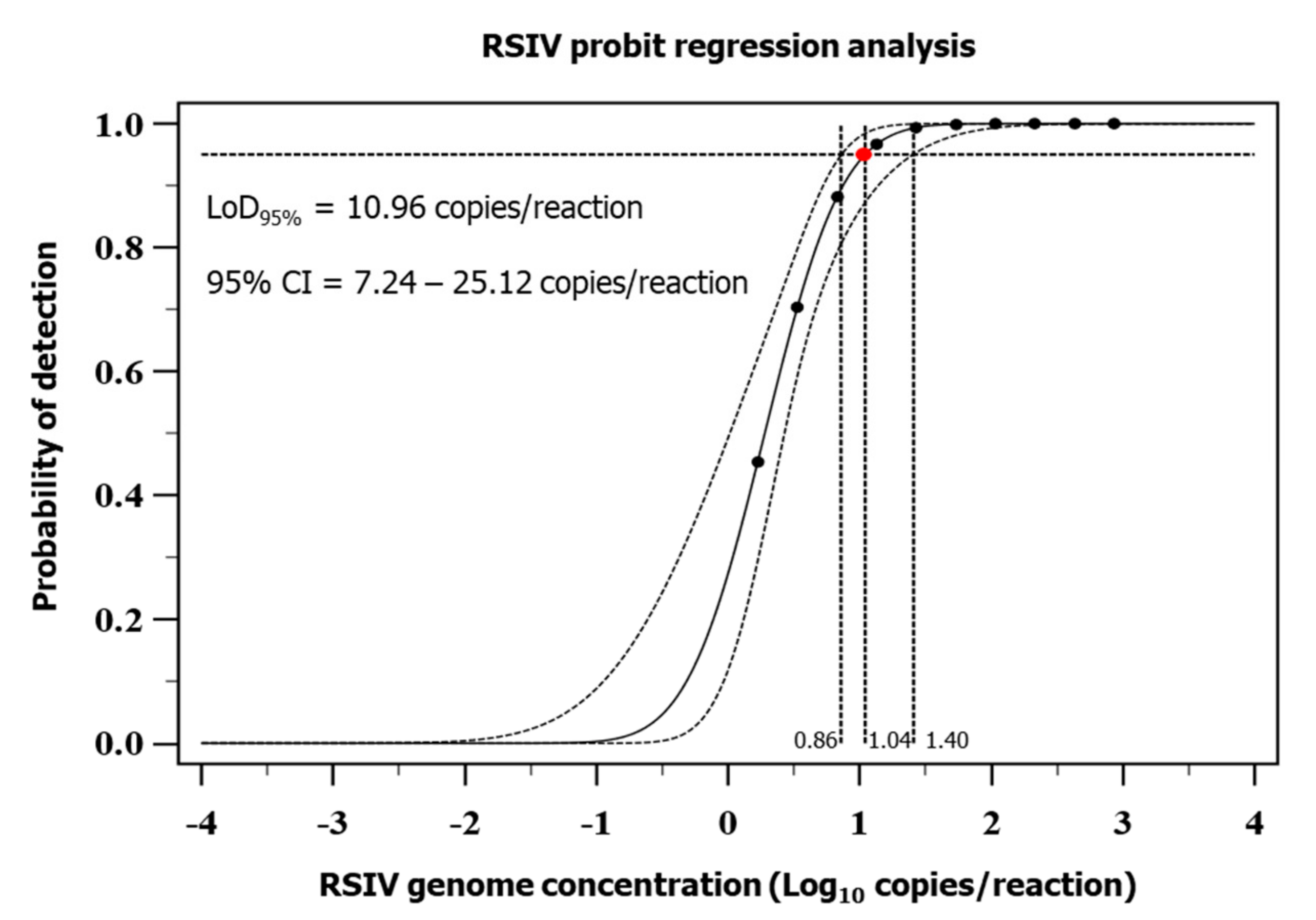
2.5.1. Analytical Sensitivity (ASe)
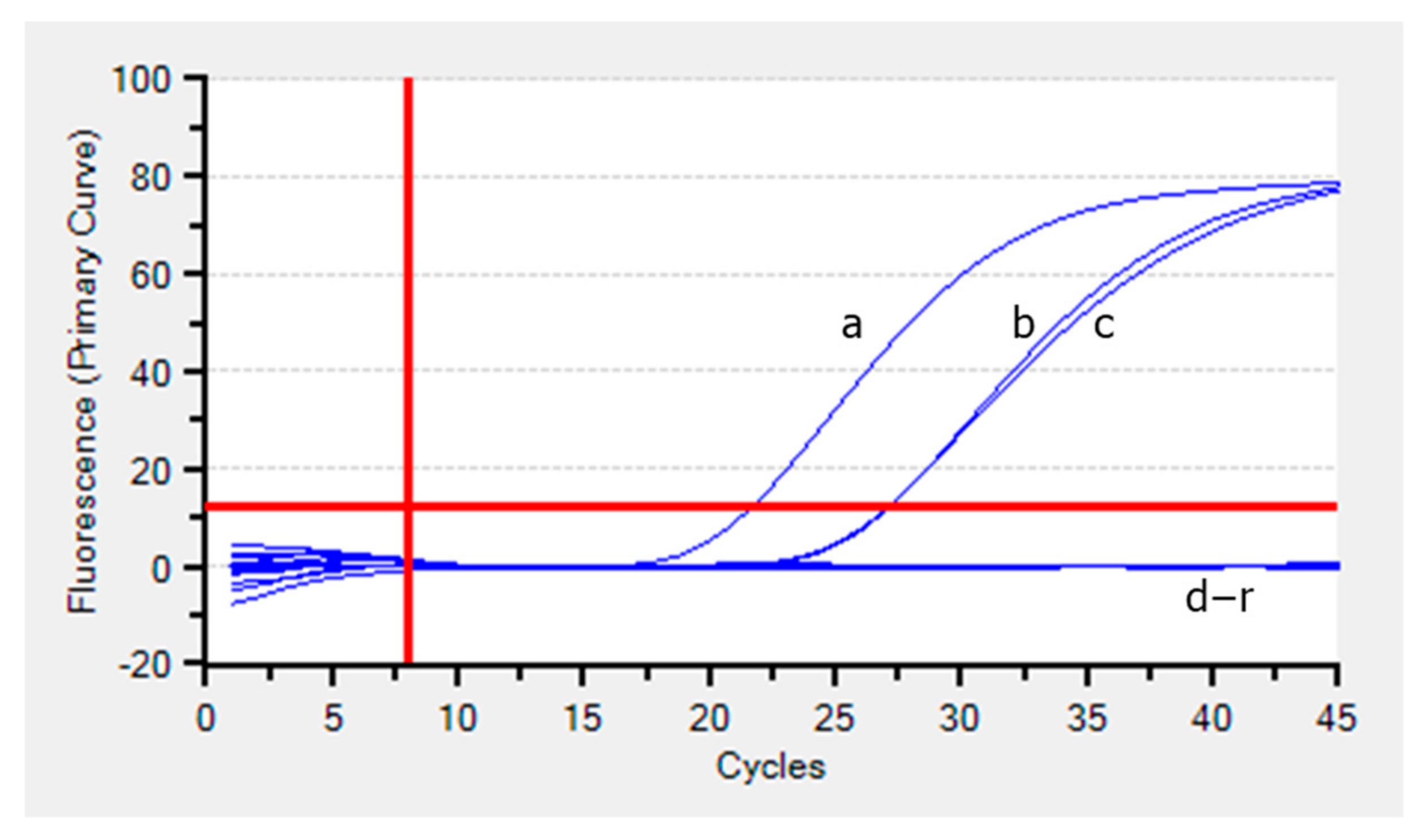
2.5.2. Analytical Specificity (ASp)
2.5.3. Effect of Interfering Substances
2.5.4. Analytical Repeatability
2.6. Real-Time PCR Assay Validation: Diagnostic Performance Evaluation
2.6.1. Diagnostic Sensitivity (DSe) and Diagnostic Specificity (DSp)
2.6.2. Comparison of the Reproducibility of the Real-Time PCR Assay between Technicians
3. Results
3.1. Analytical Sensitivity of the Real-Time PCR Assay
3.2. Analytical Specificity of the Real-Time PCR Assay
3.3. Interfering Substances
3.4. Analytical Repeatability
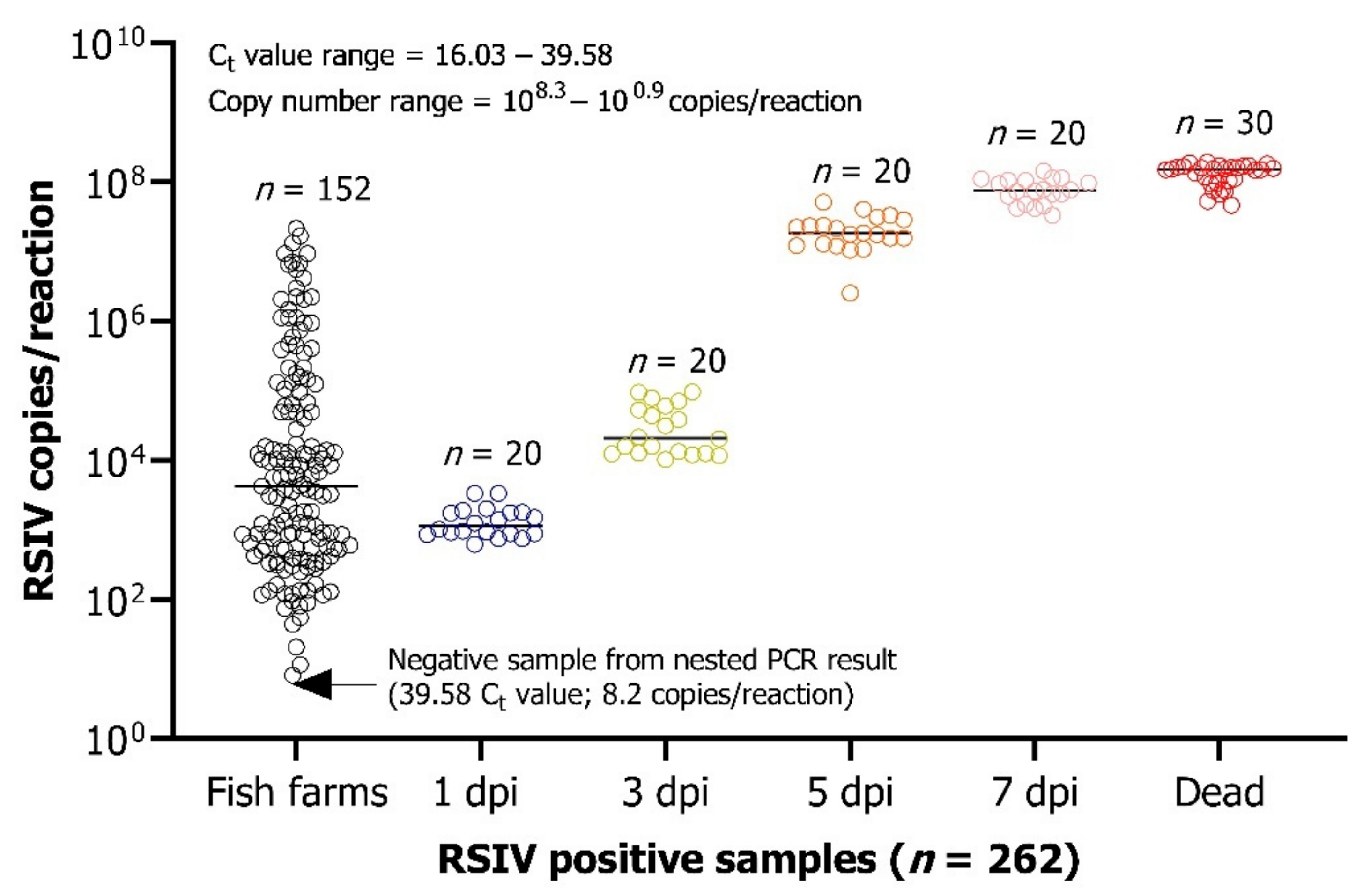
3.5. Diagnostic Performance of the Real-Time PCR Assay on Fish Samples
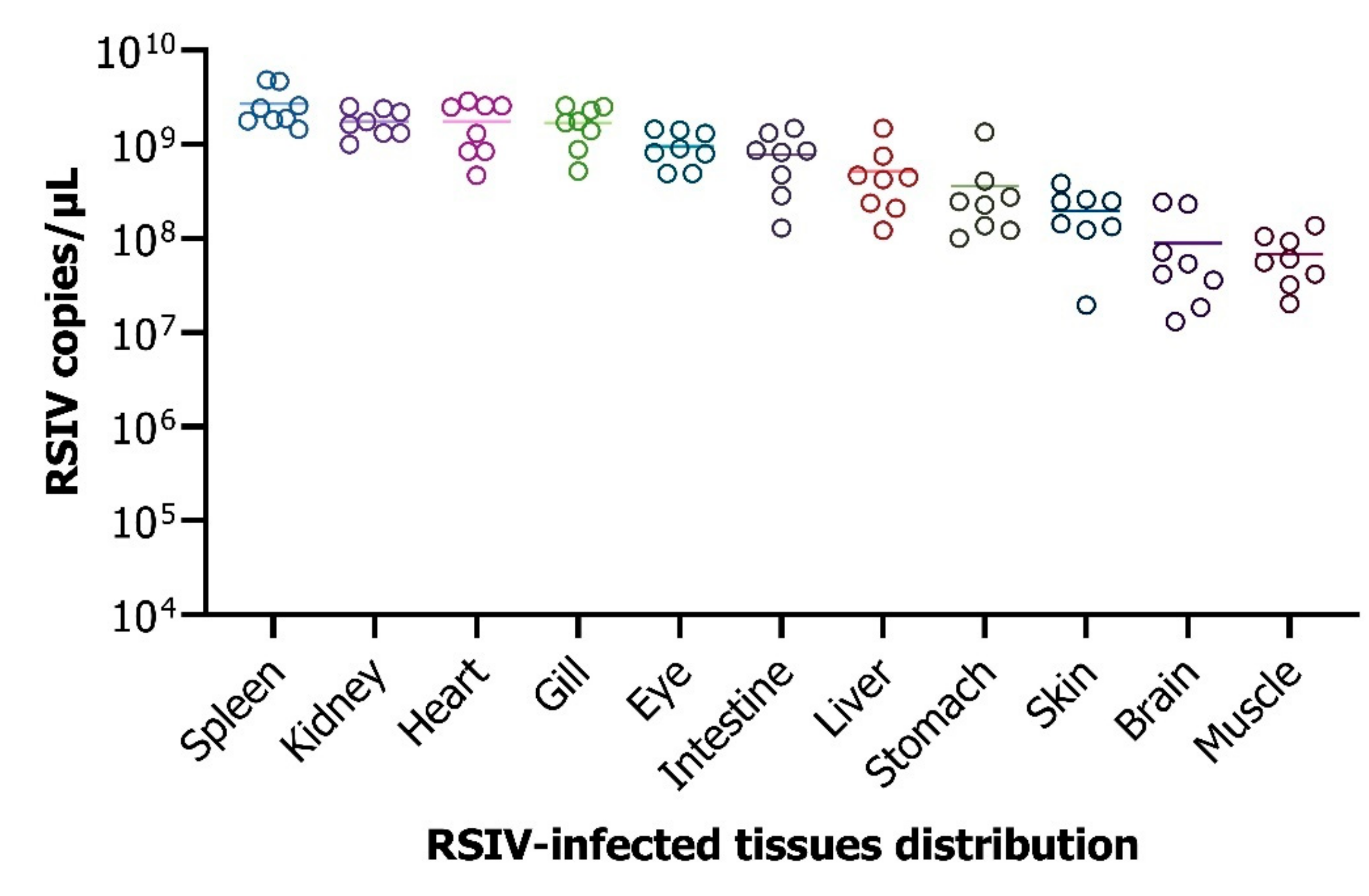
3.6. Analysis of the Viral Load in the Different Tissues Infected by the Virus
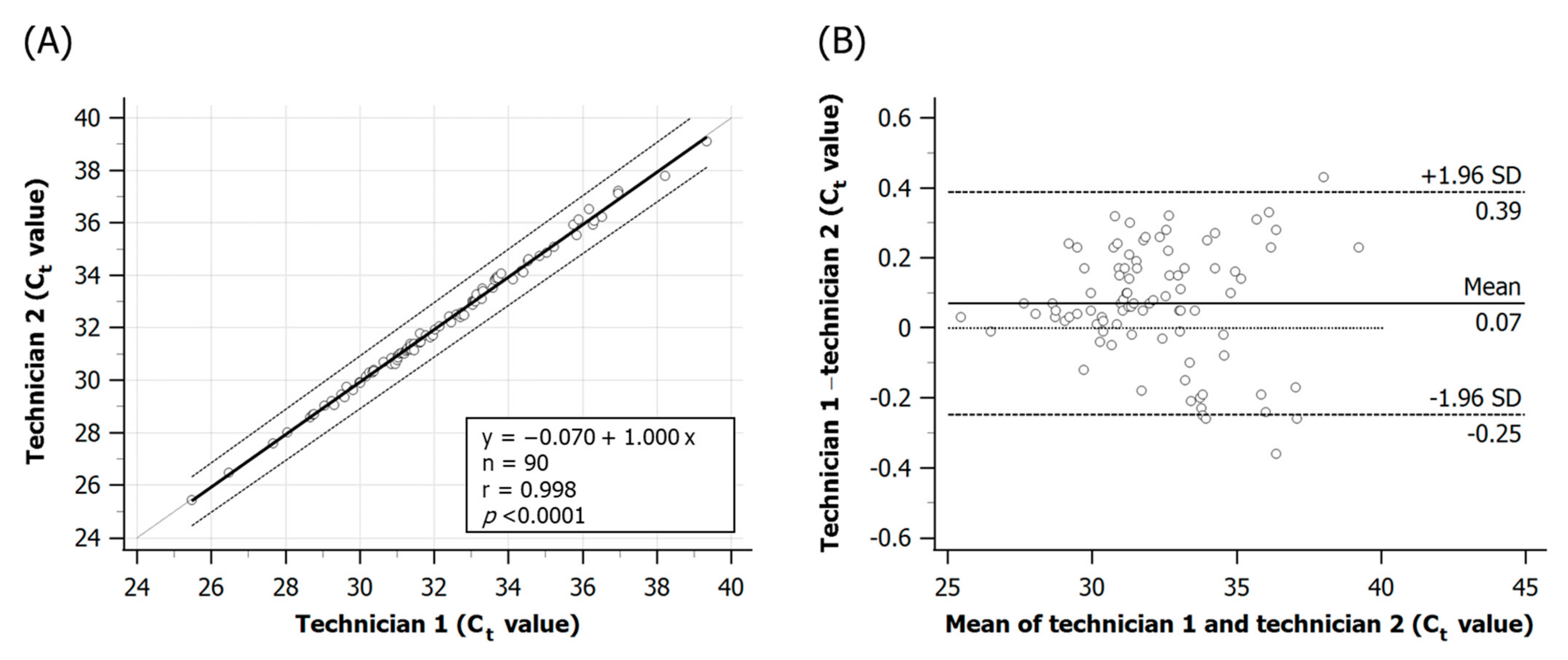
3.7. Within-Laboratory Reproducibility of the Real-Time PCR Analysis of Fish Samples
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Jancovich, J.K.; Chinchar, V.G.; Hyatt, A.; Miyazaki, T.; Williams, T.; Zhang, Q.Y. Iridoviridae. In Virus Taxonomy: 9th Report of the International Committee on Taxonomy of Viruses; King, A.M.Q., Adams, M.J., Carstens, E.B., Lefkowitz, E.J., Eds.; Academic Press: London, UK, 2012; pp. 207–210. [Google Scholar]
- Kurita, J.; Nakajima, K. Megalocytiviruses. Viruses 2012, 4, 521–538. [Google Scholar] [CrossRef] [PubMed]
- Inouye, K.; Yamano, K.; Maeno, Y.; Nakajima, K.; Matsuoka, M.; Wada, Y.; Sorimachi, M. Iridovirus infection of cultured red sea bream, Pagrus major. Fish Pathol. 1992, 27, 19–27. [Google Scholar] [CrossRef]
- OIE (World Organisation for Animal Health). Red sea bream iridoviral disease. In Manual of Diagnostic Tests for Aquatic Animals; OIE: Paris, France, 2012; Chapter 2.3.7; Available online: https://www.oie.int/fileadmin/Home/eng/Health_standards/aahm/current/2.3.07_RSIVD.pdf (accessed on 29 March 2022).
- Jun, L.J.; Jeong, J.B.; Kim, J.H.; Nam, J.H.; Shin, K.W.; Kim, J.K.; Kang, J.C.; Jeong, H.D. Influence of temperature shifts on the onset and development of red sea bream iridoviral disease in rock bream Oplegnathus fasciatus. Dis. Aquat. Org. 2009, 84, 201–208. [Google Scholar] [CrossRef]
- Jung, M.H.; Jung, S.J.; Vinay, T.N.; Nikapitiya, C.; Kim, J.O.; Lee, J.H.; Lee, J.; Oh, M.J. Effects of water temperature on mortality in Megalocytivirus-infected rock bream Oplegnathus fasciatus (Temminck et Schlegel) and development of protective immunity. J. Fish Dis. 2015, 38, 729–737. [Google Scholar] [CrossRef] [PubMed]
- Jung, M.H.; Nikapitiya, C.; Vinay, T.N.; Lee, J.H.; Jung, S.J. Rock bream iridovirus (RBIV) replication in rock bream (Oplegnathus fasciatus) exposed for different time periods to susceptible water temperatures. Fish Shellfish Immunol. 2017, 70, 731–735. [Google Scholar] [CrossRef]
- Kawato, Y.; Yamashita, H.; Yuasa, K.; Miwa, S.; Nakajima, K. Development of a highly permissive cell line from spotted knifejaw (Oplegnathus punctatus) for red sea bream iridovirus. Aquaculture 2017, 473, 291–298. [Google Scholar] [CrossRef]
- Nakajima, K.; Maeno, Y.; Fukudome, M.; Fukuda, Y.; Tanaka, S.; Matsuoka, S.; Sorimachi, M. Immunofluorescence test for the rapid diagnosis of red sea bream iridovirus infection using monoclonal antibody. Fish Pathol. 1995, 30, 115–119. [Google Scholar] [CrossRef]
- Caipang, C.M.; Haraguchi, I.; Ohira, T.; Hirono, I.; Aoki, T. Rapid detection of a fish iridovirus using loop-mediated isothermal amplification (LAMP). J. Virol. Methods 2004, 121, 155–161. [Google Scholar] [CrossRef]
- He, J.G.; Zeng, K.; Weng, S.P.; Chan, S.M. Systemic disease caused by an iridovirus-like agent in cultured mandarinfish, Siniperca chuatsi (Basilewsky), in China. J. Fish. Dis. 2000, 23, 219–222. [Google Scholar] [CrossRef]
- Kurita, J.; Nakajima, K.; Hirono, I.; Aoki, T. Polymerase chain reaction (PCR) amplification of DNA of red sea bream iridovirus (RSIV). Fish Pathol. 1998, 33, 17–23. [Google Scholar] [CrossRef] [Green Version]
- Oshima, S.; Hata, J.; Hirasawa, N.; Ohtaka, T.; Hirono, I.; Aoki, T.; Yamashita, S. Rapid diagnosis of red sea bream iridovirus infection using the polymerase chain reaction. Dis. Aquat. Org. 1998, 32, 87–90. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.K.; Kwon, S.R.; Nam, Y.K.; Kim, S.K.; Kim, K.H. Organ distribution of red sea bream iridovirus (RSIV) DNA in asymptomatic yearling and fingerling rock bream (Oplegnathus fasciatus) and effects of water temperature on transition of RSIV into acute phase. Aquaculture 2006, 256, 23–26. [Google Scholar] [CrossRef]
- Caipang, C.M.; Hirono, I.; Aoki, T. Development of a Real-time PCR Assay for the Detection and Quantification of Red Seabream Iridovirus (RSIV). Fish Pathol. 2003, 38, 1–7. [Google Scholar] [CrossRef]
- Mohr, P.G.; Moody, N.J.; Williams, L.M.; Hoad, J.; Cummins, D.M.; Davies, K.R.; StJ Crane, M. Molecular confirmation of infectious spleen and kidney necrosis virus (ISKNV) in farmed and imported ornamental fish in Australia. Dis. Aquat. Org. 2015, 116, 103–110. [Google Scholar] [CrossRef]
- Lee, E.S.; Cho, M.; Min, E.Y.; Jung, S.H.; Kim, K.I. Novel peptide nucleic acid-based real-time PCR assay for detection and genotyping of Megalocytivirus. Aquaculture 2020, 518, 734818. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Choi, K.M.; Joo, M.S.; Kang, G.; Woo, W.S.; Kim, K.H.; Jeong, S.H.; Son, M.Y.; Kim, D.H.; Park, C.I. First report of eosinophil peroxidase in starry flounder (Platichthys stellatus): Gene identification and gene expression profiling. Fish Shellfish Immunol. 2021, 118, 155–159. [Google Scholar] [CrossRef]
- Kim, K.I.; Hwang, S.D.; Cho, M.Y.; Jung, S.H.; Kim, Y.C.; Jeong, H.D. A natural infection by the red sea bream iridovirus-type Megalocytivirus in the golden mandarin fish Siniperca scherzeri. J. Fish. Dis. 2018, 41, 1229–1233. [Google Scholar] [CrossRef]
- Godornes, C.; Leader, B.T.; Molini, B.J.; Centurion-Lara, A.; Lukehart, S.A. Quantitation of rabbit cytokine mRNA by real-time RT-PCR. Cytokine 2007, 38, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Kralik, P.; Ricchi, M. A Basic Guide to Real Time PCR in Microbial Diagnostics: Definitions, Parameters, and Everything. Front. Microbiol. 2017, 8, 108. [Google Scholar] [CrossRef] [PubMed]
- Caraguel, C.G.; Stryhn, H.; Gagné, N.; Dohoo, I.R.; Hammell, K.L. Selection of a cutoff value for real-time polymerase chain reaction results to fit a diagnostic purpose: Analytical and epidemiologic approaches. J. Vet. Diagn. Investig. 2011, 23, 2–15. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.B.; Kim, H.Y.; Jun, L.J.; Lyu, J.H.; Park, N.G.; Kim, J.K.; Jeong, H.D. Outbreaks and risks of infectious spleen and kidney necrosis virus disease in freshwater ornamental fishes. Dis. Aquat. Org. 2008, 78, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.I.; Jin, J.W.; Kim, Y.C.; Jeong, H.D. Detection and genetic differentiation of megalocytiviruses in shellfish, via high-resolution melting (HRM) analysis. Korean J. Fish Aquat. Sci. 2014, 47, 241–246. [Google Scholar]
- Burd, E.M. Validation of Laboratory-Developed Molecular Assays for Infectious Diseases. Clin. Microbiol. Rev. 2010, 23, 550–576. [Google Scholar] [CrossRef]
- Schrader, C.; Schielke, A.; Ellerbroek, L.; Johne, R. PCR inhibitors–occurrence, properties and removal. J. Appl. Microbiol. 2012, 113, 1014–1026. [Google Scholar] [CrossRef]
- Bilić-Zulle, L. Comparison of methods: Passing and Bablok regression. Biochem. Med. 2011, 21, 49–52. [Google Scholar] [CrossRef]
- Kirsch, K.; Detilleux, J.; Serteyn, D.; Sandersen, C. Comparison of two portable clinical analyzers to one stationary analyzer for the determination of blood gas partial pressures and blood electrolyte concentrations in horses. PLoS ONE 2019, 14, e0211104. [Google Scholar] [CrossRef]
- OIE (World Organisation for Animal Health). Principles and methods of validation of diagnostic assays for infectious diseases. In Manual of Diagnostic Tests and Vaccines for Terrestrial Animals; OIE: Paris, France, 2013; Chapter 1.1.6; Available online: https://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/1.01.06_VALIDATION.pdf (accessed on 24 November 2021).
- Tidona, C.A.; Schnitzler, P.; Kehm, R.; Darai, G. Is the major capsid protein of iridoviruses a suitable target for study of viral Evolution? Virus Genes 1998, 16, 59–66. [Google Scholar] [CrossRef]
- Eaton, H.E.; Metcalf, J.; Penny, E.; Tcherepanov, V.; Upton, C.; Brunetti, C.R. Comparative genomic analysis of the family Iridoviridae: Re-annotating and defining the core set of iridovirus genes. Virol. J. 2007, 4, 11. [Google Scholar] [CrossRef]
- Fu, X.; Li, N.; Lai, Y.; Liu, L.; Lin, Q.; Shi, C.; Huang, Z.; Wu, S. Protective immunity against iridovirus disease in mandarin fish induced by recombinant major capsid protein of infectious spleen and kidney necrosis virus. Fish Shellfish Immunol. 2012, 33, 880–885. [Google Scholar] [CrossRef] [PubMed]
- Hirotsu, Y.; Mochizuki, H.; Omata, M. Double-quencher probes improve detection sensitivity toward Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) in a reverse-transcription polymerase chain reaction (RT-PCR) assay. J. Virol. Methods 2020, 284, 113926. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.; Gravelsina, S.; Öhrmalm, C.; Ottoson, J.; Blomberg, J. Development of single-tube nested real-time PCR assays with long internally quenched probes for detection of norovirus genogroup II. Biotechniques 2016, 60, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Holland, P.M.; Abramson, R.D.; Watson, R.; Gelfand, D.H. Detection of specific polymerase chain reaction product by utilizing the 5′–3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc. Natl. Acad. Sci. USA 1991, 88, 7276–7280. [Google Scholar] [CrossRef]
- Wilhelm, J.; Pingoud, A. Real-time polymerase chain reaction. ChemBioChem 2003, 4, 1120–1128. [Google Scholar] [CrossRef]
- Rimmer, A.E.; Becker, J.A.; Tweedie, A.; Whittington, R.J. Development of a quantitative polymerase chain reaction (qPCR) assay for the detection of dwarf gourami iridovirus (DGIV) and other megalocytiviruses and comparison with the Office International des Epizooties (OIE) reference PCR protocol. Aquaculture 2012, 358, 155–163. [Google Scholar] [CrossRef]
- Hoffmann, B.; Beer, M.; Reid, S.M.; Mertens, P.; Oura, C.A.; van Rijn, P.A.; Slomka, M.J.; Banks, J.; Brown, I.H.; Alexander, D.J.; et al. A review of RT-PCR technologies used in veterinary virology and disease control: Sensitive and specific diagnosis of five livestock diseases notifiable to the World Organisation for Animal Health. Vet. Microbiol. 2009, 139, 1–23. [Google Scholar] [CrossRef]
- Sidstedt, M.; Rådström, P.; Hedman, J. PCR inhibition in qPCR, dPCR and MPS-mechanisms and solutions. Anal. Bioanal. Chem. 2020, 412, 2009–2023. [Google Scholar] [CrossRef] [Green Version]
| Materials | Stock Concentration | Mean Ct | SD | CV (%) | ΔCt | t-Test | ||
|---|---|---|---|---|---|---|---|---|
| Only Positive Sample | Positive Sample + Interfering Substance | Negative Sample + Interfering Substance | ||||||
| L-Ascorbic acid | 10 mM | 32.77 | 32.99 | N.D. | 0.11 | 0.32 | −0.22 | p = 0.054 |
| Fucoidan | 50 μg/mL | 32.77 | 32.84 | N.D. | 0.08 | 0.25 | −0.07 | p = 0.4 |
| Beta-glucan | 50 mg/mL | 32.77 | 32.72 | N.D. | 0.15 | 0.46 | 0.05 | p = 0.701 |
| Enrofloxacin | 5 mg/mL | 32.77 | 32.71 | N.D. | 0.06 | 0.20 | 0.06 | p = 0.481 |
| Ampicillin | 10 mg/mL | 32.77 | 32.64 | N.D. | 0.20 | 0.62 | 0.13 | p = 0.386 |
| Kanamycin | 10 mg/mL | 32.77 | 32.72 | N.D. | 0.15 | 0.47 | 0.05 | p = 0.702 |
| Amoxicillin | 10 mg/mL | 32.77 | 32.57 | N.D. | 0.15 | 0.45 | 0.2 | p = 0.129 |
| Florfenicol | 10 mg/mL | 32.77 | 32.80 | N.D. | 0.10 | 0.30 | −0.03 | p = 0.704 |
| Trimethoprim | 10 mg/mL | 32.77 | 32.71 | N.D. | 0.10 | 0.29 | 0.06 | p = 0.519 |
| Concentrations (Copies/Reaction) | Intra-Assay Repeatability | Inter-Assay Repeatability | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Technician 1 | Technician 2 | Technician 3 | Technicians 1–3 | |||||||||
| Mean Ct (n = 33) | SD | CV (%) | Mean Ct (n = 33) | SD | CV (%) | Mean Ct (n = 33) | SD | CV (%) | Mean Ct (n = 99) | SD | CV (%) | |
| 8.60 × 105 | 23.27 | 0.07 | 0.3 | 23.38 | 0.17 | 0.73 | 23.37 | 0.18 | 0.78 | 23.34 | 0.16 | 0.67 |
| 8.60 × 103 | 29.88 | 0.08 | 0.28 | 29.98 | 0.22 | 0.72 | 29.92 | 0.36 | 1.21 | 29.93 | 0.25 | 0.83 |
| 8.60 × 101 | 36.39 | 0.39 | 1.08 | 36.55 | 0.49 | 1.34 | 36.39 | 0.39 | 1.08 | 36.46 | 0.44 | 1.2 |
| No. of Samples with the Following Result in RSIV Nested PCR Assay | ||||
|---|---|---|---|---|
| Known Positive | Known Negative | Total | ||
| TaqMan qPCR assay (Ct cut-off value 39.75) | Positive | 261 | 1 | 262 |
| Negative | 0 | 248 | 248 | |
| Total | 261 | 249 | ||
| Performance parameter | Value | 95% CI | ||
| Sensitivity (%) | 100 | 98.60 to 100 | ||
| Specificity (%) | 99.6 | 97.78 to 99.99 | ||
| Positive predictive value (%) | 99.62 | 97.36 to 99.95 | ||
| Negative predictive value (%) | 100 | |||
| Accuracy (%) | 99.8 | 98.91 to 100 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, K.-H.; Choi, K.-M.; Kang, G.; Woo, W.-S.; Sohn, M.-Y.; Son, H.-J.; Yun, D.; Kim, D.-H.; Park, C.-I. Development and Validation of a Quantitative Polymerase Chain Reaction Assay for the Detection of Red Sea Bream Iridovirus. Fishes 2022, 7, 236. https://doi.org/10.3390/fishes7050236
Kim K-H, Choi K-M, Kang G, Woo W-S, Sohn M-Y, Son H-J, Yun D, Kim D-H, Park C-I. Development and Validation of a Quantitative Polymerase Chain Reaction Assay for the Detection of Red Sea Bream Iridovirus. Fishes. 2022; 7(5):236. https://doi.org/10.3390/fishes7050236
Chicago/Turabian StyleKim, Kyung-Ho, Kwang-Min Choi, Gyoungsik Kang, Won-Sik Woo, Min-Young Sohn, Ha-Jeong Son, Dongbin Yun, Do-Hyung Kim, and Chan-Il Park. 2022. "Development and Validation of a Quantitative Polymerase Chain Reaction Assay for the Detection of Red Sea Bream Iridovirus" Fishes 7, no. 5: 236. https://doi.org/10.3390/fishes7050236
APA StyleKim, K.-H., Choi, K.-M., Kang, G., Woo, W.-S., Sohn, M.-Y., Son, H.-J., Yun, D., Kim, D.-H., & Park, C.-I. (2022). Development and Validation of a Quantitative Polymerase Chain Reaction Assay for the Detection of Red Sea Bream Iridovirus. Fishes, 7(5), 236. https://doi.org/10.3390/fishes7050236







