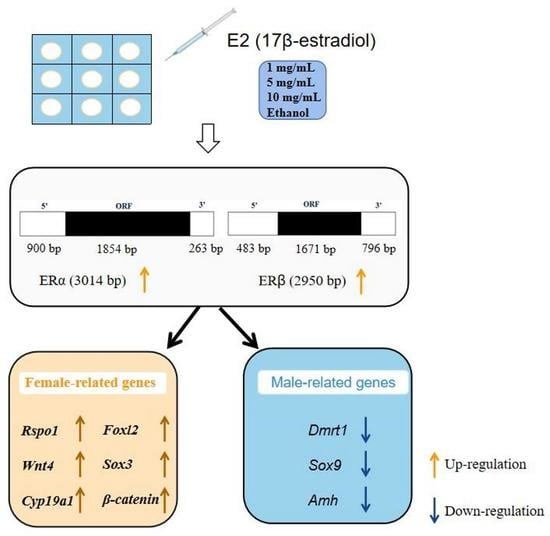
Effect of Estradiol on Estrogen Nuclear Receptors Genes Expression on Embryonic Development Stages in Chinese Soft-Shelled Turtle (Pelodiscus sinensis)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Maintenance of the Chinese Soft-Shelled Turtles
2.2. Estradiol Treatment
2.3. Samples Collection
2.4. RNA Extraction
2.5. Full-Length cDNA Cloning of ERs
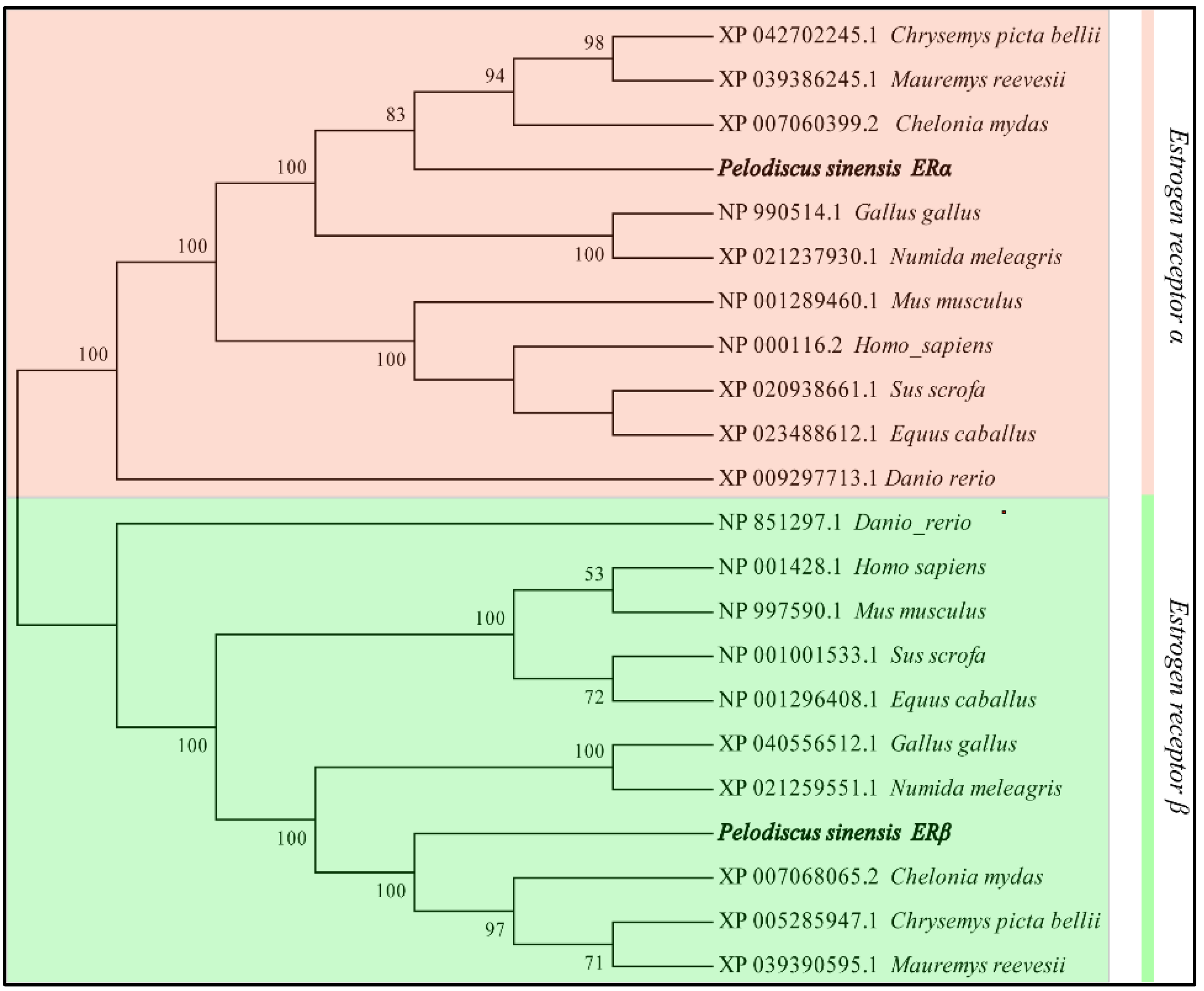
2.6. Sequence and Phylogenetic Analysis
2.7. Gene Expression Analysis by Quantitative Real-Time Reverse Transcription-PCR
2.8. Statistical Analysis
3. Results
3.1. Sequence Analysis of P. sinensis ERs Gene
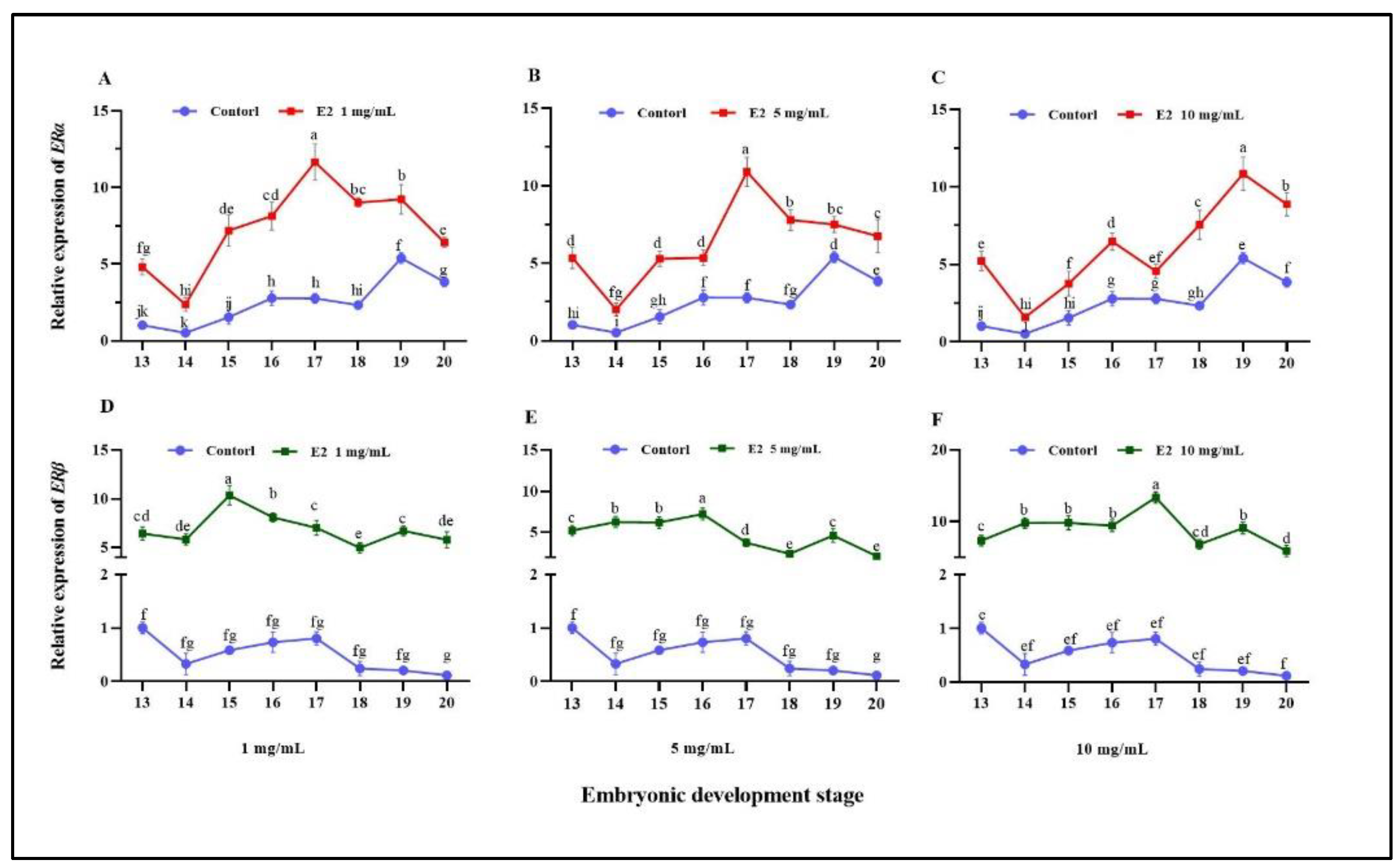
3.2. Tissue and Embryonic Development Stage Distribution of ERs
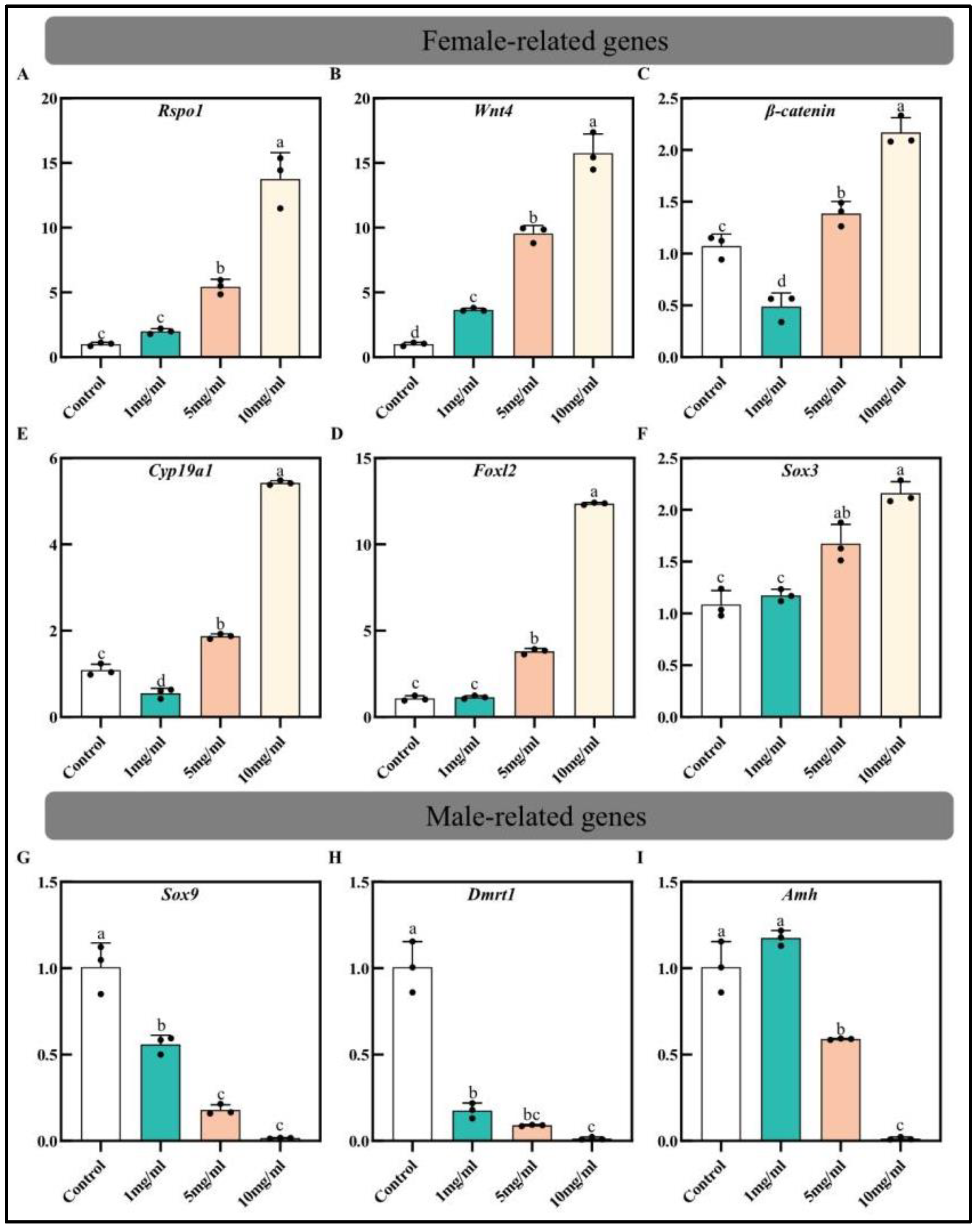
3.3. Effect of Estradiol on ERs Expression and Sex-Related Genes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Gong, S.P.; Vamberger, M.; Auer, M.; Praschag, P.; Fritz, U. Millennium-old farm breeding of Chinese softshell turtles (Pelodiscus sinensis) results in massive erosion of biodiversity. Sci. Nat. 2018, 105, 34. [Google Scholar] [CrossRef]
- Liang, H.W.; Wang, L.H.; Sha, H.; Zou, G.W. Development and Validation of Sex-Specific Markers in Pelodiscus Sinensis Using Restriction Site-Associated DNA Sequencing. Genes 2019, 10, 302. [Google Scholar] [CrossRef]
- Cornejo-Paramo, P.; Lira-Noriega, A.; Ramirez-Suastegui, C.; Mendez-de-la-Cruz, F.R.; Szekely, T.; Urrutia, A.O.; Cortez, D. Sex determination systems in reptiles are related to ambient temperature but not to the level of climatic fluctuation. BMC Evol. Biol. 2020, 20, 103. [Google Scholar] [CrossRef] [PubMed]
- Mu, Y.; Zhao, B.; Tang, W.Q.; Sun, B.J.; Zeng, Z.G.; Valenzuela, N.; Du, W.G. Temperature-Dependent Sex Determination Ruled Out in the Chinese Soft-Shelled Turtle (Pelodiscus sinensis) via Molecular Cytogenetics and Incubation Experiments across Populations. Sex. Dev. 2015, 9, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.S.; Kettlewell, J.R.; Anderson, R.C.; Bardwell, V.J.; Zarkower, D. Sexually dimorphic expression of multiple doublesex-related genes in the embryonic mouse gonad. Gene Expr. Patterns 2003, 3, 77–82. [Google Scholar] [CrossRef]
- Wang, Y.; Luo, X.; Qu, C.; Xu, T.; Zou, G.; Liang, H. The Important Role of Sex-Related Sox Family Genes in the Sex Reversal of the Chinese Soft-Shelled Turtle (Pelodiscus sinensis). Biology 2022, 11, 83. [Google Scholar] [CrossRef]
- Liang, H.; Meng, Y.; Cao, L.; Li, X.; Zou, G. Effect of exogenous hormones on R-spondin 1 (RSPO1) gene expression and embryo development in Pelodiscus sinensis. Reprod. Fertil. Dev. 2019, 31, 1425–1433. [Google Scholar] [CrossRef]
- Cheng, S.B.; Dong, J.; Pang, Y.F.; LaRocca, J.; Hixon, M.; Thomas, P.; Filardo, E.J. Anatomical location and redistribution of G protein-coupled estrogen receptor-1 during the estrus cycle in mouse kidney and specific binding to estrogens but not aldosterone. Mol. Cell. Endocrinol. 2014, 382, 950–959. [Google Scholar] [CrossRef]
- Zhou, L.Y.; Charkraborty, T.; Yu, X.G.; Wu, L.M.; Liu, G.; Mohapatra, S.; Wang, D.S.; Nagahama, Y. R-spondins are involved in the ovarian differentiation in a teleost, medaka (Oryzias latipes). BMC Dev. Biol. 2012, 12, 36. [Google Scholar] [CrossRef]
- Parma, P.; Radi, O.; Vidal, V.; Chaboissier, M.C.; Dellambra, E.; Valentini, S.; Guerra, L.; Schedl, A.; Camerino, G. R-spondin1 is essential in sex determination, skin differentiation and malignancy. Nat. Genet. 2006, 38, 1304–1309. [Google Scholar] [CrossRef]
- Maatouk, D.M.; DiNapoli, L.; Alvers, A.; Parker, K.L.; Taketo, M.M.; Capel, B. Stabilization of beta-catenin in XY gonads causes male-to-female sex-reversal. Hum. Mol. Genet. 2008, 17, 2949–2955. [Google Scholar] [CrossRef] [PubMed]
- Stewart, M.K.; Mattiske, D.M.; Pask, A.J. Estrogen suppresses SOX9 and activates markers of female development in a human testis-derived cell line. BMC Mol. Cell Biol. 2020, 21, 66. [Google Scholar] [CrossRef] [PubMed]
- Iwamatsu, T.; Kobayashi, H. Sex reversal in the medaka treated in vitro 17 alpha-methyldihydrotestosterone during oocyte maturation. Zool. Sci. 2005, 22, 1505. [Google Scholar]
- Rose, E.; Flanagan, S.P.; Jones, A.G. The Effects of Synthetic Estrogen Exposure on the Sexually Dimorphic Liver Transcriptome of the Sex-Role-Reversed Gulf Pipefish. PLoS ONE 2015, 10, e0139401. [Google Scholar] [CrossRef]
- Eick, G.N.; Thornton, J.W. Evolution of steroid receptors from an estrogen-sensitive ancestral receptor. Mol. Cell. Endocrinol. 2011, 334, 31–38. [Google Scholar] [CrossRef]
- Jensen, E.V.; Jacobson, H.I.; Walf, A.A.; Frye, C.A. Estrogen action: A historic perspective on the implications of considering alternative approaches. Physiol. Behav. 2010, 99, 151–162. [Google Scholar] [CrossRef]
- Kowalski, A.A.; Graddy, L.G.; Vale-Cruz, D.S.; Choi, I.; Katzenellenbogen, B.S.; Simmen, F.A.; Simmen, R.C.M. Molecular cloning of porcine estrogen receptor-beta complementary DNAs and developmental expression in periimplantation embryos. Biol. Reprod. 2002, 66, 760–769. [Google Scholar] [CrossRef]
- Edwards, D.P. Regulation of signal transduction pathways by estrogen and progesterone. Annu. Rev. Physiol. 2005, 67, 335–376. [Google Scholar] [CrossRef]
- Hewitt, S.C.; Korach, K.S. Estrogen Receptors: New Directions in the New Millennium. Endocr. Rev. 2018, 39, 664–675. [Google Scholar] [CrossRef]
- Qin, X.; Xiao, Y.; Ye, C.; Jia, J.; Liu, X.; Liang, H.; Zou, G.; Hu, G. Pituitary Action of E2 in Prepubertal Grass Carp: Receptor Specificity and Signal Transduction for Luteinizing Hormone and Follicle-Stimulating Hormone Regulation. Front. Endocrinol. 2018, 9, 308. [Google Scholar] [CrossRef]
- Marino, M.; Galluzzo, P.; Ascenzi, P. Estrogen signaling multiple pathways to impact gene transcription. Curr. Genom. 2006, 7, 497–508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pietras, R.J.; Marquez-Garban, D.C. Membrane-associated estrogen receptor signaling pathways in human cancers. Clin. Cancer Res. 2007, 13, 4672–4676. [Google Scholar] [CrossRef] [PubMed]
- Barton, M.; Filardo, E.J.; Lolait, S.J.; Thomas, P.; Maggiolini, M.; Prossnitz, E.R. Twenty years of the G protein-coupled estrogen receptor GPER: Historical and personal perspectives. J. Steroid Biochem. Mol. Biol. 2018, 176, 4–15. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Sha, H.; Chen, M.; Chen, G.; Zou, G.; Liang, H. MicroRNAs May Play an Important Role in Sexual Reversal Process of Chinese Soft-Shelled Turtle, Pelodiscus sinensis. Genes 2021, 12, 1696. [Google Scholar] [CrossRef]
- Sun, J.-J.; Sun, Z.-H.; Wei, J.-L.; Ding, J.; Song, J.; Chang, Y.-Q. Identification and functional analysis of foxl2 and nodal in sea cucumber, Apostichopus japonicus. Gene. Expr. Patterns GEP 2022, 44, 119245. [Google Scholar] [CrossRef]
- Liang, H.W.; Meng, Y.; Cao, L.H.; Li, X.; Zou, G.W. Expression and characterization of the cyp19a gene and its responses to estradiol/letrozole exposure in Chinese soft-shelled turtle (Pelodiscus sinensis). Mol. Reprod. Dev. 2019, 86, 480–490. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Cai, H.; Zhang, G.; Zhang, H.; Bao, H.; Wang, L.; Ye, J.; Qian, G.; Ge, C. Dmrt1 is required for primary male sexual differentiation in Chinese soft-shelled turtle Pelodiscus sinensis. Sci. Rep. 2018, 8, 4433. [Google Scholar] [CrossRef] [PubMed]
- Dean, R.; Mank, J.E. The role of sex chromosomes in sexual dimorphism: Discordance between molecular and phenotypic data. J. Evol. Biol. 2014, 27, 1443–1453. [Google Scholar] [CrossRef]
- Luo, A.; Zhang, X. ERRF is essential for Estrogen-Estrogen Receptor alpha signaling pathway in ER positive breast cancer cells. Biochem. Biophys. Res. Commun. 2016, 474, 400–405. [Google Scholar] [CrossRef]
- Misra, S.; Pandey, A.K.; Gupta, S.; Kumar, A.; Khanna, P.; Shankar, J.; Ram, K.R. Estrogen related receptor is required for the testicular development and for the normal sperm axoneme/mitochondrial derivatives in Drosophila males. Sci. Rep. 2017, 7, 40372. [Google Scholar] [CrossRef]
- Li, P.; Guo, Y.; Jin, L.; Liang, X.; Chen, G.A.; Sun, W.; Xiao, L.; Qian, G.Y.; Ge, C.T. ESR1 mediates estrogen-induced feminization of genetic male Chinese soft-shelled turtle. Biol. Reprod. 2022, ioac088. [Google Scholar] [CrossRef] [PubMed]
- Toyota, K.; Masuda, S.; Sugita, S.; Miyaoku, K.; Yamagishi, G.; Akashi, H.; Miyagawa, S. Estrogen Receptor 1 (ESR1) Agonist Induces Ovarian Differentiation and Aberrant Mullerian Duct Development in the Chinese Soft-shelled Turtle, Pelodiscus sinensi. Zool. Stud. 2020, 59, e54. [Google Scholar] [PubMed]
- Ouyang, H.; Han, C.; Zhu, Q.; Xu, L.; Huang, J.; Li, S.; Li, G.; Lin, H.; Zhang, Y. Molecular cloning and characterization of estrogen and androgen receptors in Mandarin fish, Siniperca chuatsi. Aquac. Rep. 2021, 21, 100834. [Google Scholar] [CrossRef]
- Pelletier, G.; Labrie, C.; Labrie, F. Localization of oestrogen receptor alpha, oestrogen receptor beta and androgen receptors in the rat reproductive organs. J. Endocrinol. 2000, 165, 359–370. [Google Scholar] [CrossRef] [PubMed]
- Schoen, J.; Neumann, S.; Wildt, D.E.; Pukazhenthi, B.S.; Jewgenow, K. Localization of Oestrogen Receptors in the Epididymis During Sexual Maturation of the Domestic Cat. Reprod. Domest. Anim. 2009, 44, 294–301. [Google Scholar] [CrossRef]
- Blazquez, M.; Gonzalez, A.; Papadaki, M.; Mylonas, C.; Piferrer, F. Sex-related changes in estrogen receptors and aromatase gene expression and enzymatic activity during early development and sex differentiation in the European sea bass (Dicentrarchus labrax). Gen. Comp. Endocrinol. 2008, 158, 95–101. [Google Scholar] [CrossRef]
- Wojnarowski, K.; Cholewinska, P.; Palic, D.; Bednarska, M.; Jarosz, M.; Wisniewska, I. Estrogen Receptors Mediated Negative Effects of Estrogens and Xenoestrogens in Teleost Fishes-Review. Int. J. Mol. Sci. 2022, 23, 2605. [Google Scholar] [CrossRef]
- Yan, L.; Feng, H.; Wang, F.; Lu, B.; Liu, X.; Sun, L.; Wang, D. Establishment of three estrogen receptors (esr1, esr2a, esr2b) knockout lines for functional study in Nile tilapia. J. Steroid Biochem. Mol. Biol. 2019, 191, 105379. [Google Scholar]
- Saito, K.; Cui, H. Emerging Roles of Estrogen-Related Receptors in the Brain: Potential Interactions with Estrogen Signaling. Int. J. Mol. Sci. 2018, 19, 1091. [Google Scholar] [CrossRef]
- O’Brien, M.H.; Pitot, H.C.; Chung, S.-H.; Lambert, P.F.; Drinkwater, N.R.; Bilger, A. Estrogen Receptor-alpha Suppresses Liver Carcinogenesis and Establishes Sex-Specific Gene Expression. Cancers 2021, 13, 2355. [Google Scholar] [CrossRef]
- Hwang, W.J.; Lee, T.Y.; Kim, N.S.; Kwon, J.S. The Role of Estrogen Receptors and Their Signaling across Psychiatric Disorders. Int. J. Mol. Sci. 2021, 22, 373. [Google Scholar] [CrossRef]
- Guillette, L.J.; Iguchi, T.; Doheny, B.M.; Bryan, T.A.; Zhu, J.; Katsu, Y.; Bernhard, M.C.; Kohno, S. Estrogen Receptor 1 (ESR1; ERα), not ESR2 (ERβ), Modulates Estrogen-Induced Sex Reversal in the American Alligator, a Species with Temperature-Dependent Sex Determination. Endocrinology 2015, 156, 1887–1899. [Google Scholar]
- Gustafsson, J.-A. Historical overview of nuclear receptors. J. Steroid Biochem. Mol. Biol. 2016, 157, 3–6. [Google Scholar] [CrossRef] [PubMed]
- Lindberg, M.K.; Moverare, S.; Skrtic, S.; Gao, H.; Dahlman-Wright, K.; Gustafsson, J.A.; Ohlsson, C. Estrogen receptor (ER)-beta reduces ER alpha-regulated gene transcription, supporting a “Ying Yang” relationship between ER alpha and ER beta in mice. Mol. Endocrinol. 2003, 17, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.Q.; Weng, Y.Z.; Ye, R.Z.; Liu, L.L. Immunolocalization of aromatase, estrogen and estrogen receptor alpha and beta in the epithelium of digestive tract and enteric neurons of amphioxus (Branchiostoma belcheri). Prog. Nat. Sci.-Mater. Int. 2005, 15, 157–161. [Google Scholar]
- Wang, L.; Cen, S.; Shi, X.; Zhang, H.; Wu, L.; Tian, X.; Ma, W.; Li, X.; Ma, X. Molecular characterization and functional analysis of Esr1 and Esr2 in gonads of Chinese soft-shelled turtle (Pelodiscus sinensis). J. Steroid Biochem. Mol. Biol. 2022, 222, 106147. [Google Scholar] [CrossRef] [PubMed]
- O’Lone, R.; Frith, M.C.; Karlsson, E.K.; Hansen, U. Genomic targets of nuclear estrogen receptors. Mol. Endocrinol. 2004, 18, 1859–1875. [Google Scholar] [CrossRef]
- Agrawal, R.; Sahoo, B.K.; Saini, D.K. Cross-talk and specificity in two-component signal transduction pathways. Future Microbiol. 2016, 11, 685–697. [Google Scholar] [CrossRef]
- Chen, D.; Li, Q.; Chen, H.; Huang, Q.; Zeng, M. Estrogen receptor regulates immune defense by suppressing NF-kappa B signaling in the Crassostrea hongkongensis. Fish Shellfish. Immunol. 2020, 106, 796–803. [Google Scholar] [CrossRef]
- Hollingshead, B.D.; Beischlag, T.V.; DiNatale, B.C.; Ramadoss, P.; Perdew, G.H. Inflammatory signaling and aryl hydrocarbon receptor mediate synergistic induction of interleukin 6 in MCF-7 cells. Cancer Res. 2008, 68, 3609–3617. [Google Scholar] [CrossRef]
- Wissink, S.; van der Burg, B.; Katzenellenbogen, B.S.; van der Saag, P.T. Synergistic activation of the serotonin-1A receptor by nuclear factor-kappa B and estrogen. Mol. Endocrinol. 2001, 15, 543–552. [Google Scholar] [PubMed] [Green Version]


| Primer Name | Primer Sequence (5′–3′) | Application | Related References |
|---|---|---|---|
| ERα-F | GTTGATCCCTCCGCTGACAGT | CDS amplification | |
| ERα-R | CTCGCAAGACCAGACTCCATAAT | ||
| ERβ-F | TGACGTTACTACAGCCAGCATCAC | ||
| ERβ-R | CGACCTCCACATCAGACCCATC | ||
| ERα-GSP5-1 | ATCTGGTGGAGCATGGCAACTC | 5′ RACE | |
| ERα-GSP5-2 | GAATCTGGTGGAGCATGGCAAC | ||
| ERβ-GSP5-1 | TAATCAAAGCTCGTGGAGTGGC | ||
| ERβ-GSP5-2 | GCGTACGTGTATTTGTCGGTCA | ||
| ERα-GSP3-1 | GCCAGTTAACAACTGCATCAACTT | 3′ RACE | |
| ERα-GSP3-2 | AAGCAGGGAAGATGAGAATTTGC | ||
| ERβ-GSP3-1 | ATGCTAGATGCTCACCGATTGC | ||
| ERβ-GSP3-2 | CAGGCACATGAGCAATAAAGGG | ||
| UPM short | CTAATACGACTCACTATAGGGC | 5′ and 3′ RACE | |
| UPM long | CTAATACGACTCACTATAGGGCAAGCA | ||
| ERα-F | CCGACTGCGAAAGTGCTATGA | qPCR | |
| ERα-R | ACGCTGGACTGTTCTTCTTGCTA | ||
| ERβ-F | GCAACAGACAACTCGCATGG | ||
| ERβ-R | GTGTGTGCATTCAGCATCTCC | ||
| Rspo1-F | CCTGCTGGAGAGGAATGACA | [7] | |
| Rspo1-R | CCCACTCGCTCATTTCACA | ||
| Wnt4-F | GAGGTGATGGACTCGGTGCG | ||
| Wnt4-R | CCCGTTCTTGAGGTCGTGGTC | ||
| β-catenin-F | GCTTTGGGACTCCACCTTACAG | ||
| β-catenin-R | ATCACCAGCCCGAAGAACAGT | ||
| Foxl2-F | ATCTGTTTTTATTAGCACGGTT | [25] | |
| Foxl2-R | CCTTCTCAGGAGGAGTTTCGT | ||
| Cyp19a1-F | TCGTGGCTGTACAAGAAATACGAA | [26] | |
| Cyp19a1-R | CCAGTCATATCTCCACGGCTCT | ||
| Sox3-F | GAGTGTAGAGGTGGAATGGAAACG | ||
| Sox3-R | AAACCCTCAAGCAGGATACGG | ||
| Sox9-F | TACGACTACACCGACCACCA | ||
| Sox9-R | GTAGTGTCTGCAATGGGCGT | ||
| Dmrt1-F | CCGCCTCGGGAAAGAAGTC | [27] | |
| Dmrt1-R | TGCTGGATGCCGTAGTTGC | ||
| Amh-F | CGGCTACTCCTCCCACACG | ||
| Amh-R | CCTGGCTGGAGTATTTGACGG | ||
| Ps4085-F | GTTTGAAGTGCTGCTGGGAAG | Sex identification | [2] |
| Ps4085-R | TTCCCCGTATAAAGCCAGGG | ||
| COI-F | CAACCAACCACAAAGACATTGGCAC | ||
| COI-R | ACCTCAGGGTGTCCGAAAATCAAA | ||
| Gapdh-F | AGAACATCATTCCAGCATCCA | Internal control | |
| Gapdh-R | CTTCATCACCTTCTTAATGTCGTC |
| ERα | ERβ | |||
|---|---|---|---|---|
| Species | Accession Number | Identity (%) | Accession Number | Identity (%) |
| Mauremys mutica | XP 039386245.1 | 88.33 | XP 039390595.1 | 93.17 |
| Chelonia mydas | XP 007060399.2 | 82.80 | XP 007068065.2 | 86.35 |
| Chrysemys picta bellii | XP 042702245.1 | 79.77 | XP 005285947.1 | 87.54 |
| Numida meleagris | XP 021237930.1 | 76.01 | XP 021259551.1 | 80.03 |
| Gallus | NP 990514.1 | 74.28 | XP 040556512.1 | 80.38 |
| Homo sapiens | NP 000116.2 | 67.49 | NP 001428.1 | 69.11 |
| Sus scrofa | XP 020938661.1 | 67.49 | NP 001001533.1 | 70.31 |
| Mus musculus | NP 001289460.1 | 66.47 | NP 997590.1 | 72.01 |
| Equus caballus | XP 023488612.1 | 64.88 | NP 001296408.1 | 70.31 |
| Danio rerio | XP 009297713.1 | 41.18 | NP 851297.1 | 49.66 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, G.; Zhou, T.; Chen, M.; Zou, G.; Liang, H. Effect of Estradiol on Estrogen Nuclear Receptors Genes Expression on Embryonic Development Stages in Chinese Soft-Shelled Turtle (Pelodiscus sinensis). Fishes 2022, 7, 223. https://doi.org/10.3390/fishes7050223
Chen G, Zhou T, Chen M, Zou G, Liang H. Effect of Estradiol on Estrogen Nuclear Receptors Genes Expression on Embryonic Development Stages in Chinese Soft-Shelled Turtle (Pelodiscus sinensis). Fishes. 2022; 7(5):223. https://doi.org/10.3390/fishes7050223
Chicago/Turabian StyleChen, Guobin, Tong Zhou, Meng Chen, Guiwei Zou, and Hongwei Liang. 2022. "Effect of Estradiol on Estrogen Nuclear Receptors Genes Expression on Embryonic Development Stages in Chinese Soft-Shelled Turtle (Pelodiscus sinensis)" Fishes 7, no. 5: 223. https://doi.org/10.3390/fishes7050223
APA StyleChen, G., Zhou, T., Chen, M., Zou, G., & Liang, H. (2022). Effect of Estradiol on Estrogen Nuclear Receptors Genes Expression on Embryonic Development Stages in Chinese Soft-Shelled Turtle (Pelodiscus sinensis). Fishes, 7(5), 223. https://doi.org/10.3390/fishes7050223