Abstract
The quality of sea urchin gonad is important to consumers with high standards for nutrition and taste. However, few studies have been conductedon the molecular mechanisms that determine the quality of male and female sea urchins. In this study, our goal was to understand the differences and characteristics of gonad quality between sea urchin (Strongylocentrotus intermedius) males and females. The transcriptomes of males and females were obtained, with totals of 43,797,146 and 56,222,782 raw reads, respectively, comprising 128,979 transcripts and 85,745 unigenes. After comparative transcriptome analysis, a total of 6736 differentially expressed genes (DEGs) between the males and females were identified, of which 2950 genes were up-regulated and 3786 genes were down-regulated in males. We compared the expression of twelve DEGs with significant differences their expression levels and functional annotations to confirm the reliability of the RNA-Seq data. Five DEGs related to gonadal quality were found through enrichment analysis of KEGG pathways: 17β-HSD8, PGDH, FAXDC2, C4MO, and PNPLA7. Our study analyzes genes related to the taste and flavor of sea urchin gonads among the sexes and provides reference sequences and fundamental information concerning the nutrition and taste of S. intermedius gonads.
1. Introduction
Echinoderms, the highest class of invertebrates among deuterostome phyla, have always attracted the focus of researchers. The extant echinoderms include Echinoidea, Holothuroidea, Crinoidea, Asteroidea, and Ophiuroidea [1]. Sea urchins are all marine organisms, with approximately 850 species worldwide [2,3,4]. There are only six edible species in China: Glyptocidari screnularis, Hemicentrotus pulcherrimus, Tripneustes gratilla, S. intermedius, S. purpuratus, and S. nudus [5]. S. intermedius was introduced to China by Dalian Ocean University [6], having originally been distributed in Japan’s island of Hokkaido and Russia. Since S. intermedius was introduced into China, it has become the dominant species that is widely cultivated on the coasts of both Liaoning and Shandong Provinces [7,8].
Sea urchin gonads are the only edible part; they are delicious and rich in protein, amino acids, sugars, various fatty acids, and other active nutrients. They also have potential medicinal value [8,9,10]. In addition, sea urchin gonads are considered a delicacy in many parts of the world, especially in China and Japan, due to their distinctive taste and high level of nutrition. The economic value of sea urchins is increasing, as they are being consumed by more people [9,10,11]. Sea urchin gonad quality has an important influence on market value, with the most important characteristics being flavor and nutritional value. The flavor refers to the proportions of variousamino acids, and the nutritional value is a function of the type and content of fatty acids [8,12]. There are five critical amino acids in the gonads of sea urchins, as different amino acids have different tastes. Among these, the free amino acids, umamiamino acids such as glutamic acid (Glu), threonine (Thr), glycine (Gly), and alanine (Ala) are sweet, and valine (Val), lysine (Lys), and arginine (Arg) are bitter; lysine (Lys), arginine (Arg), tyrosine (Tyr), and glutamic (Glu) acid are the most abundant [13,14,15,16]. Osako et al. [14] studied the types of free amino acids in the gonads and the effects of feed on the composition of free amino acids in the cultured sea urchin Anthocidaris crassispina. Ayyagariet al. [16] investigated nutritional composition and characteristics, including moisture, ash, proteins, lipids, and carbohydrates in S. variolaris gonads. S. intermedius, the main breeding species in the northern China, contains proteins, lipids, and amino acids in the gonads; the lipids are primarily phospholipids but also include cholesterol, free fatty acids, and triglycerides [17]. A growing number of consumers hold the view that the male gonads are more palatable and nutritious than females in S. intermedius. Previous studies have found that sea urchin gonads differ in taste and flavor by sex due to the composition and content of amino acids, especially the umami amino acids; however, there are also studies reporting that females have a higher fatty acid content and better color than males, which affects market price. This is also likely due to variation among species and developmental stages [18,19]. Some researchers, such as Murata et al. [9], have also studied the differences between male and female sea urchin gonads, and they have identified sources ofbitterness in the green sea urchin Strongylocentrotus droebachiensis, particularly in the mature ovaries. Zhao et al. [19] reported that S. intermedius had differences in flavor and gonad characteristics between the sexes. Li et al. [17] also studied the contents of various amino acids among the sexes of S. intermedius. All previous studies have concentrated on macroscopic aspects such as appearance, composition, hardness, particle size, amino acid content, and the influence of these aspects [20]. There are few studies on the molecular mechanisms related to these characteristics. Therefore, we performed transcriptome analyses of S. intermedius females and males by RNA-Seq. Comparing the differences related to gonad quality would enhance our understanding of the variation in the nutritional value of sea urchins by sex and provide new information with which to understand the gonad differences between male and female in Strongylocentrotus intermedius.
2. Materials and Methods
2.1. Ethics Statement
The procedures involving animals in this study were conducted in accordance with the Guidelines for Experimental Animals established by the Ministry of Science and Technology (Beijing, China).All individuals were anesthetized and then sampled. All experimental procedures in the present study were approved by the Animal Care and Use Committee (ACUC) of Dalian Ocean University, China. The Ethics Approval Code was DLOU-2019-036.
2.2. Sample, RNA Preparation, and cDNA Library Construction
Experimental sea urchins (S. intermedius) were collected from an aquaculture company in Dalian, China. The female sea urchin Gonadosomatic index (GSI) was 0.21 ± 0.06, and the male sea urchin GSI was 0.20 ± 0.05. A total of two mixed samples (five female and five male sea urchin gonads in each mixed sample) were collected in1.5 mL RNase-free centrifuge tubes. Then, samples were preserved according to the method of Konget al. [21], and total RNA was extracted from each sample according to the experimental method of Cui et al. [22]. The preliminary detection of RNA used 1% agarose gels, an Agilent Bioanalyzer 2100 (Agilent Technologies, Palo Alto, CA, USA), and a Nano Photometer® spectrophotometer (IMPLEN, LA CA, USA) were used to check RNA purity and concentration. High-quality RNA samples were sent to Novogene Biological Information Technology Corporation (Beijing, China) for the construction of the cDNA libraries and Illumina HiSeq sequencing.
2.3. De Novo Transcriptome Assembly, Mapping and Annotation
The produced raw reads were filtered to obtain clean reads, Trinity software [23] was used to assemble the clean reads into transcripts, and the longest transcripts were selected as unigenes. Then, all unigenes were annotated via six major public databases for bioinformatic analysis; these were DIAMOND (with an e-value ≤ 1 × 10−5), NR, COG, Swiss-Prot, BLAST2GO annotated to GO, HMMER [24] (e-value ≤ 0.01), and KOBAS [25] (e-value ≤ 1 × 10−10).The sequences were then annotated to the Pfam protein family and the KEGG database. After the annotation was complete, the results described the functions of the unigenes, and the best-matching annotation was selected to predict the functions of each gene.
2.4. Gene Expression Quantification and Differentially Expressed Gene Enrichment Analysis
FPKM (fragments per kb per million fragments) is the number of reads of the length of a gene per kilobase per million reads (a commonly used method to estimate gene expression levels) [4,26]. The read count data for all genes were standardized by HMM [27], and we used DESeq [28,29] software to analyze the differentially expressed genes (DEGs) between groups. The default parameters were p-adjust < 0.001 and|log2FC| ≥ 4. We annotated all DEGs to the COG, GO, and KEGG databases for functional classification and then performed GO enrichment analysis for all genes in the gene set using Goatools software [30]. For the KEGG pathway enrichment analysis, the goal was to discover new functional genes and pathways and to determine which genes were involved in determining gonad quality between the testis and ovary.
2.5. RT-qPCR Validation
We chose 12 DEGs between females and males for RT-qPCR validation, and the 18S rRNA was used as the reference gene [3]. The Primer 5.0 program [31] was used to design all primers, and samples were sent to Invitrogen (Shanghai) for synthesis (Table 1). Total RNA was extracted from three ovaries and three testes of S. intermedius using TRIzol reagent (Invitrogen, Shanghai, China). The cDNA was synthesized according to the instructions of a First Strand cDNA Synthesis Kit (Takara Biotech Co., Beijing, China). The resulting cDNA products were diluted five-fold as a template for the production of the RT-qPCR standard curve. Then, the best concentration of cDNA was selected for RT-qPCR amplification using TB Green™ Premix Ex Taq ™II (Tli-RNaseH Plus) (Takara, Beijing, China). All reactions consisted of three biological replicates and three technical replicates. After amplification, the Ct value of each reaction was obtained, and the relative expression level of each gene was calculated by the 2−ΔΔCt method [32]. RT-qPCR was performed using an ABI 7500 StepOneplus Real-time Detection System (Applied Biosystems, Waltham, MA, USA). The reaction was carried out in a total volume of 20 µL and contained 10 µL TB Green™ Premix Ex Taq ™II, 0.8 µL of each primer at a final concentration of 0.4 μM, 0.4 μLdye, 7 µL RNase-free ddH2O, and 1 µL of diluted cDNA template.

Table 1.
Primers for target genes for RT-qPCR.
3. Results
3.1. Illumina Sequencing and De Novo Assembly
The total numbers of raw reads from the Illumina HiSeq sequencing were 43,797,146 and 56,222,782 for S. intermedius males and females, respectively. To ensure the accuracy of the subsequent data assembly and bioinformatic analyses, the numbers of clean reads with Q30 values of 94.97% and 93.61% were 41,959,814 and 54,537,612, respectively. The specific information is shown in Table 2. Then, two cDNA libraries were assembled de novo by Trinityto [33] produce 128,979 transcripts and 85,754 unigenes. The largest unigenes were approximately 500 bp in length and accounted for 43% and 54% of transcripts and unigenes, respectively (Table 3). The RNA-Seq datasets have been uploaded to NCBI with accession numbers SRR8775067 and SRR8775077.

Table 2.
Statistical data of the RNA-Seq reads for two samples.

Table 3.
The Length distribution of the transcriptome and unigenes of S. intermedius.
3.2. Annotation and Functional Classification
A total of 24,143 (28.16% of the total) unigenes were annotated using six databases; 23,244 (27.11%) unigenes were annotated to the NR database; numbers annotated to the Swiss-Prot, Pfam, COG, KEGG, and GO databases were 14,800 (17.26%), 15,012 (17.51%), 2779 (3.24), 11,476 (13.38%), and 10,500 (12.25%), respectively. There was a large number of unigenes (71.84%) without annotation; this may be partly due to the overall short lengths of these unigenes. In addition, the coverage was relatively low for whole transcripts and may have been limited by the available sea urchin genome information [3,34]. As shown in Figure 1, the comparison with the NR protein database obtained sequence similarities between the gene sequences of this species and those of other species as well as functional information for this species. There were19,718 (85.01%) unigenes annotated in S. purpuratus, a number that may be due to the complete genome sequencing of the model sea urchin S. purpuratus [1]; S. intermedius is from the same class as S. purpuratus, and, thus, it is closely related to S. purpuratus. This was followed by 649 (2.80%) unigenes annotated to Acanthaster planci (Figure 1B). Results with smaller E-values have higher confidence, and 74.75% of the E-values were distributed between 0 and 1 × 10−10 (Figure 1C). A degree of similarity of 80% or more accounted for 73.66% of predicted protein, and the proportion with a similarity of 60%–80% was 21.02% (Figure 1A). This result showed the accuracy and reliability of the RNA-Seq sequences.
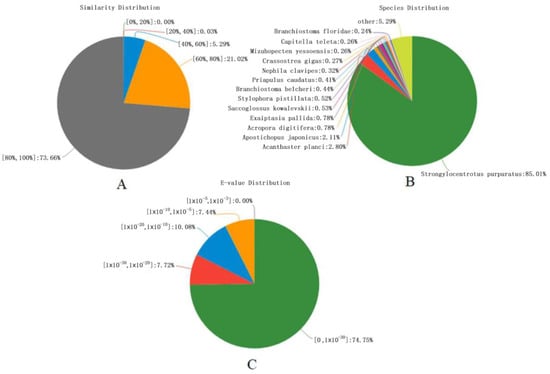
Figure 1.
Summary of NR protein database annotations. (A) Similarity Distribution, (B) Species Distribution, and (C) E-value Distribution.
The GO database provides classification and analysis of the functions of a large proportion of the transcriptome data from a macro perspective. That is classification from the GO database describes the cellular environment in which the gene product is located, possible molecular functions, and biological processes involved. A total of 12,348 unigenes were annotated into 16 molecular function classifications (Figure 2), and most unigenes were involved in binding (GO:0005488) followed by catalytic activity (GO:0003824), with 5408 and 4625 unigenes, respectively. The cellular component hada total of 20,778 unigenes annotated into 14 classifications, and the two most frequent categories of unigenes were cell (4049; GO:0005623) and cell part (4003; GO:0044464) functions; 18,122 unigenes were annotated to the classification of 24 biological processes. These results are similar to the annotated results of Chen et al. [3]; further emphasizing the differences between the male and female gonads of S. intermedius.
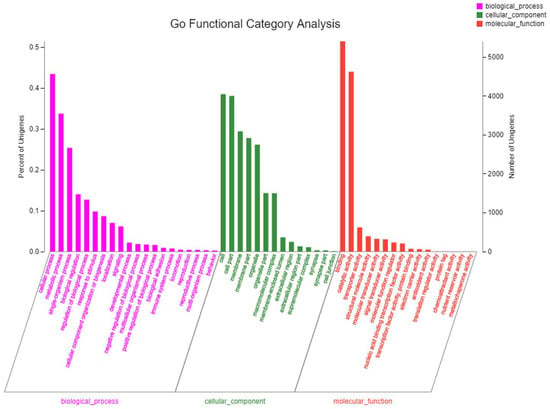
Figure 2.
Gene ontology (level 2) function for expressed genes listed by molecular function, cellular component, and biological process.
3.3. DEGs Identification and Enrichment Analysis
In total, we obtained 7597 female-specific transcripts and 4127 male-specific transcripts (Figure 3).There were 6736 DEGs between the female and male sea urchins, including 2950 up-regulated genes and 3786 down-regulated genes in males (Figure 4). At the same time, we annotated all DEGs using three large databases, COG, GO, and KEGG, to further describe and classify their common functions. There were 332 DEGs annotated in the COG database. Among these annotated functional classes, the largest subcategories were translation, ribosomal structure, and biogenesis function (90 DEGs); this was followed by post-translational modification, protein turnover, and chaperones (31 DEGs) (Figure 5).
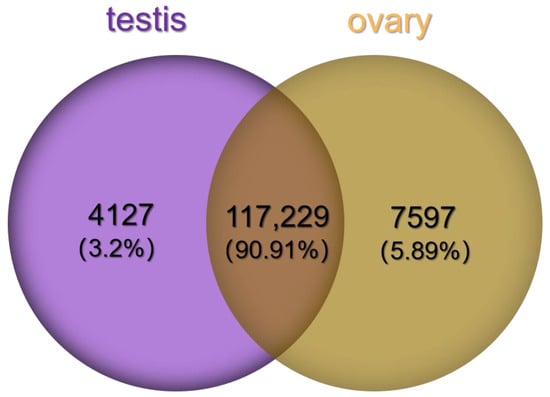
Figure 3.
Venn diagram of male-specific and female-specific transcripts.
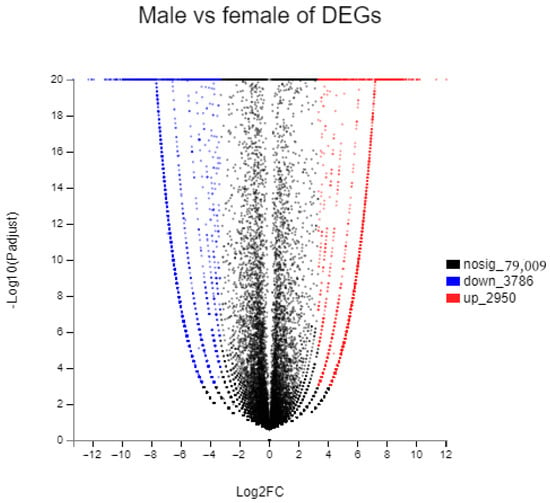
Figure 4.
Expression levels of all male and female unigenes (individual points represent individual genes as follows: black dots indicate genes with no significant differences between sexes; red dots indicate up-regulated genes with significant differences; blue dots indicate down-regulated genes with significant differences).
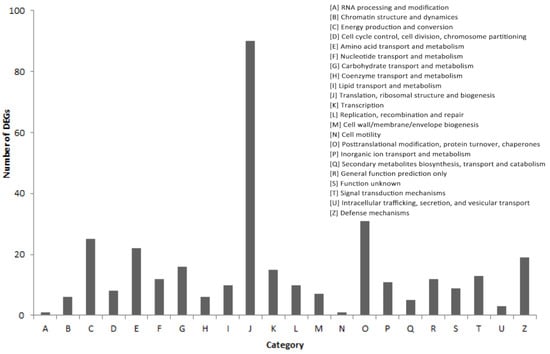
Figure 5.
COG classification of all DEGs.
To identify the GO functions and biological pathways of these DEGs, we also classified them according to the GO and KEGG databases. A total of 12,189 DEGs were annotated to 51 GO terms; similar to the previous results, the largest functional categories were binding (G0:0005488) and catalytic activity (GO:0003824). However, the total number of annotated DEG functions was greater than the number of DEGs, probably because one gene can have multiple functions. The results of the pathway analysis are shown in Figure 6. In the analysis, 4668 DEGs were annotated to 43 categories, of which 447 DEGs were annotated to signal transduction and 333 were annotated to cancers: an overview. To better understand the functional and biochemical pathways related to gonadal development and quality, all DEGs were used to conduct enrichment analyses using the GO and KEGG databases. In these results, 2669 DEGsand 2734DEGs were annotated into 80 GO ontologies and 310 pathways, respectively. Among GO terms, there were three terms with frequent annotations: molecular function (GO:0003674), cellular component (GO:0005575), and catalytic activity (GO:0003824). A total of 2734 DEGs were annotated into 310 pathways, among which the most annotated pathways involved cancer (ko05200) and microRNAs in cancer (ko05206) with 121 and 109 DEGs, respectively. After the enrichment analysis, the differential gene functions were found to be enriched in the GO term. In addition, we selected five genes related to delicious amino acids, fatty acids, and gonadal development from all annotated DEGs to reflect the differences in gonad quality between S. intermedius males and females.
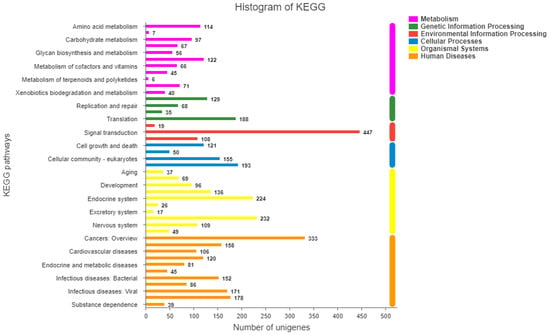
Figure 6.
KEGG pathways of DEGs.
3.4. RT-qPCR Validation of DEGs
The results for the 12 genes are shown in Figure 7. There were five DEGs related to gonad quality and sevenother genes. Although two DEGs were unable to be confirmed, one possible explanation is that there are differences between the analysis of transcriptome sequencing data and the actual experimental analysis, even if the experimental individuals of the two methods are the same [34]. Therefore, we believe that the transcriptome data of the sea urchin used in this study are accurate and reliable.
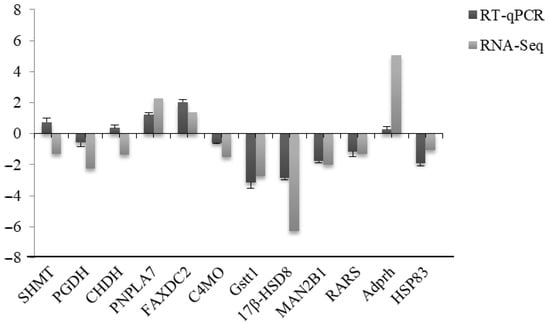
Figure 7.
RT-qPCR validation of 12 DEGs between males and females.
4. Discussion
The gonad of the sea urchin is the only edible part of the animal. Its appearance and quality are the main factors affecting the price of sea urchins, particularly the distinctive aroma and taste. Therefore, studying sea urchin gonads has always been important to researchers [35]. Previous studies have shown that the types of free amino acids in the gonads can determine their sweetness [6]. Murata et al. [36] found bitter amino acid in the mature ovaries of the sea urchin H. pulcherrimus. The concentration was in good agreement with the intensity of bitterness of the sea urchin ovaries, but this amino acid is not found in the testes. Some studies have also shown that there are differences in the types of free amino acids between testes and ovaries in thesea urchin species S. nudus, Hemicentrotus pulcherrimus, and Anthocidaris crassispina [14,37,38]. Anotherstudy on the amino acid content in the gonads of the male and female S. nudus indicated that amino acid contents were higher in males than in females, and, thus, it is widely believed that male sea urchin gonads have a better taste than female gonads. However, the molecular mechanisms involved have been rarely studied.
Therefore, this study analyzed the gonads of male and female S. intermedius by RNA-Seq. We showed that 6736 DEGs (2950 were up-regulated and 3786 were down-regulated in males), and a total of 164 pathways were enriched. We selected five DEGs related to gonad quality for discussion and analysis through KEGG pathway enrichment analysis: PGDH, C4MO, FAXDC2, 17β-HSD8, and PNPLA7 (Table 4). Among these genes, PGDH (D-3-phosphoglycerate dehydrogenase) was expressed at lower levels in testis, whereas PNPLA7 (patatin-like phospholipase domain-containing protein 7) was expressed at higher levels in testis. Previously, it was reported that PGDH is involved in the metabolism of serine, glycine, and threonine, all of which are sweet amino acids [15]. PGDH initiates the synthesis of serine by the glycolysis intermediate D-3-phosphoglycerate (3-PG), and Gly reversibly produces Ser via the gluconeogenesis pathway under the catalysis of the serine hydroxymethyl-transferase-like (SHMT) enzyme. L-Serine is necessary for the synthesis of various essential compounds, including glycine, cysteine, serine phospholipids, sphingomyelins, and cerebrosides, and is also abasic component of protein synthesis [39,40,41,42,43]. PGDH likely plays an important role in serine synthesis, and the binding portion of Ser has different regulatory effects; in addition, the enzyme is feedback inhibited by L-serine. Research on E. coli has demonstrated that PGDH is a tetramer enzyme, and the binding site between serine and the enzyme can lead to positive ornegative regulation [44,45,46,47,48]. As far as the urchin itself is concerned, size, feed, and seasonal variation of sea urchin gonads can all affect the color and taste of the gonads [49]. Some researchers have found that the gonad quality is higher when the sea urchin diet consists of higher levels of protein than when the sea urchin diet consists of seaweed and kelp; the overall taste is better and is less bitter [50,51]. Inomata et al. [15] found that the glycine content in the gonads of Mesocentrotus nudus that were fed red algae was significantly lower; in the ovaries, the threonine content of sea urchins that were fed red algae was significantly higher than sea urchins that were fed kelp. The free Gly content changes with the maturation of sea urchin gonads, but the bitter-tasting amino acids are basically unchanged, and this change is more pronounced in the ovary than in the testis. Thus, mature gonads taste the best, and late or overripe gonads are paste-like, have white pulp, and the taste and quality are poor [14].

Table 4.
DEG annotation between S. intermedius males and females.
Studies have reported that other important substances in sea urchins are lipids, and that the lipid content of the sea urchin gonads is more than 20% of the dry weight. Phospholipids (PLs) are the principal lipids in sea urchin gonads [11]. We found that PNPLA7 is related to lipid metabolism from the transcriptome data of the gonad in S. intermedius. PNPLA7 is a member of the patatin-like phospholipase family (PNPLAs) that is involved in regulating adipocyte differentiation, and PNPLA7 (NTE-like) is relatively highly expressed in several human tissues [52]. However, there have been few results concerning the function of PNPLA7 in the past decades, althoughother gene functions of the family have been studied. PNPLA4 has previously demonstrated both lipase and transacylation activities, and PNPLA3, also known as adiponutrin, is regulated by the same hormonal pathways that regulate fat deposition in the liver; these pathways have been recognized as genetic determinants of the liver fat content [53,54]. Thus, we hypothesize that PNPLA7 is involved in phospholipid (PL) metabolism. A study by Zhou et al. [11] indicated that dried gonads of S. intermedius have 22.70% lipids; PLs (39.45–50.30% of total lipids) are some of the most important lipids, andt he content of PLs was 11.42% in the dried gonads of S. intermedius. It has also been suggested that the lipid content in female gonads is higher than that in male gonads, but the C20:4 (n-6) (AA) content was significantly higher in males than in female sea urchins. After ejaculation/ovulation, the content of most fatty acids is significantly reduced in females and males; this may be because female sea urchins accumulate more yolk protein to provide important nutrients and energy for developmental processes [7,55]. However, as described in other studies, almost all C18 to C20 unsaturated fatty acids in male urchin gonads were significantly higher than the corresponding unsaturated fatty acids in female gonads; in addition, the maturity stages ofthegonad and sex also have a certain impact on the accumulation and utilization of lipids in sea urchins [17,56]. These findings are partly in agreement with the present study, where the expression of PNPLA7 in the testis was higher than that in the ovary. Generally, the content of specific fatty acids in biological lipids can indirectly reflect the catalytic activity of related enzymes in the biosynthesis of fatty acids. Male sea urchins may have faster fatty acid conversion and synthesis abilities than female sea urchins, and we predict that PNPLA7 is involved in the synthesis of lipids.
In addition, two other factors affect the quality of sea urchins: gonad development and steroid content. Previous studies have shown that sex hormones are associated with gonadal development in mollusks; in addition, echinoderms also have estrogen in their ovaries [57,58]. There are two types of estrogen in the gonads of sea urchins, E1 and E2, and their contents vary with the season; E2 is the more active estrogen and induces the synthesis of main yolk protein (MYP) to promote ovarian growth, and E1 promotes testicular growth and sperm formation [59]. Studies have shown that 17β-hydroxysteroid dehydrogenases (17β-HSDs) are involved in the synthesis and metabolism of estrogen and that they catalyze the inactivation between estrone (E1) and estradiol (E2). Estradiol 17-beta-dehydrogenase 8 (17β-HSD type 8) catalyzes the oxidoreduction of androgens, converting E2 to E1 (E1 to E2 conversion is rare) [60,61,62,63]. Previous studies have suggested that the expression level of 17β-HSD8 was higher in the ovary and lower in the testis during the proliferation phase and that this changed with the different developmental stages in the ovary and testis in Chlamys farreri [64]. Further, the expression level in ovaries in the proliferative phase is significantly higher than that in the testis, and the expression in the testis is higher in the developing and mature stages; therefore, it is believed that 17β-HSD8 can regulate oocyte development by regulating E2 levels. Our results show that the expression of 17β-HSD8 was higher in the female sea urchin gonads, and, thus, it can be surmised that the gonads in these sea urchins were in the proliferative phase [64]. This also indicates that there are many estrogens in the ovary, and estrogen controls the synthesis of MYP, a protein that plays a role in ovarian development. However, in ovaries, high expression of 17β-HSD type 8 leads to increased estrogen synthesis, thereby causing breast cancer [65,66]. This can be further explained by the fact that the estrogen level of female sea urchins is higher than that of males in the early developmental stages. Research also shows that 17β-HSD8 is associated with mitochondrial fatty acid synthesis (FAS); however, while mechanism has been studied in humans, it has not been reported in lower animals such as sea urchin [67].
We found that two genes from our transcriptome analysis were enzymes related to steroid biosynthesis. FAXDC2 and C4MO are members of the fatty acid hydroxylase superfamily. FAXDC2 (fatty acid hydroxylase domain-containing protein 2) promotes megakaryocytic maturationis involved in the differentiation of megakaryocytes and is up-regulated during megakaryocyte maturation. There are also studies reporting that lipid biosynthesis plays a critical part in megakaryocyte differentiation and that FAXDC2 is involved in lipid oxidation [68,69,70]. The expression level of FAXDC2 in the sea urchin testis was higher than in the ovaries, a result that may be related to lipid oxidation; however, the lipid concentration of Arbacia dufresnii gonads was affected by the gonadal stage and location in the gonad [70]. However, the main function of FAXDC2 on lipids has not been reported in the literature, and thuswe cannot know its main role.
Another gene, C4MO (C-4 methyl sterol oxidase-like), catalyzes the conversion oftestis-meiosis activating sterol (T-MAS) to zymosterol, and then cholesterol is eventually produced by other enzymes. MAS is an important lipid molecule, and this intermediate of the cholesterol biosynthesis pathway accumulates in the gonads andisalso involved in sterol biosynthesis [28,71,72]. Previous studies have suggested that C4MO contributes to the synthesis and metabolism of gonadotropin MAS in rabbit gonads and is expressed at a higher level in sexually mature ovaries than in testes [73]. Our results supported this finding inthat the expression of C4MO was higher in the ovary; therefore, the synthesis of MAS was increased so that the steroid content was increased in the ovary. This has important effects on the growth and development of the urchin and may reduce the nutritional value of female gonads. In summary, free amino acids and fatty acids differ between the sexes and are related to the maturity of the gonads in S. intermedius; these results are the same as those from previous studies.
5. Conclusions
In this study, we investigated why the male sea urchin gonads taste better than those of female sea urchins, and found that the content of sweet amino acids and of fatty acids that decrease when the gonads mature may be more pronounced in males. The increase in cholesterol in females resulted in decreasing taste. In addition, we identified 85,745 unigenes and 6736 DEGs between the male and female gonads of the sea urchin S. intermedius. This transcriptome database could provide reference information for studying the molecular mechanisms underlying differences in gonad quality between the testes and ovaries of S. intermedius.
Author Contributions
X.Q. planned and designed the experiments; P.G. conducted all the experiments, analyzed the data, and wrote the manuscript; Z.W. analyzed the data; J.Y. and X.W. collected samples; X.Q. reviewed and edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the Science and Technology Innovation Fund Project of Dalian city, China (2018J12SN070) and the Innovation Team Project of Dalian Ocean University, China (B202102).
Institutional Review Board Statement
The individuals (Strongylocentrotus intermedius) were collected from Dalian, China. All experimental procedures were approved by the Animal Care and Use Committee (ACUC) of Dalian Ocean University, China. The Ethics Approval Code is DLOU-2019-036.
Data Availability Statement
The RNA-Seq datasets have been submitted to NCBI. The accession numbers are SRR8775067 and SRR8775077.
Conflicts of Interest
The authors declare that they have no conflict of interest.
References
- Sodergren, E.; Weinstock, G.M.; Davidson, E.H.; Cameron, R.A.; Gibbs, R.A.; Angerer, R.C.; Angerer, L.M.; Arnone, M.I.; Burgess, D.R.; Burke, R.D.; et al. The genome of the sea urchin Strongylocentrotus purpuratus. Science 2006, 314, 941–952. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.W.; Qin, L.; Zhou, D.Y.; Wu, H.T.; Jing, W.; Yang, J.F.; Li, D.M.; Dong, X.P.; Yoshiyuki, M. Extraction of lipid from sea urchin (Strongylocentrotus nudus) gonad by enzyme-assisted aqueous and supercritical carbon dioxide methods. Eur. Food Res. Techno. 2010, 230, 737–743. [Google Scholar] [CrossRef]
- Chen, Y.-D.; Chang, Y.-Q.; Wang, X.-L.; Qiu, X.-M.; Liu, Y. De novo assembly and analysis of tissue-specific transcriptomes revealed the tissue-specific genes and profile of immunity from Strongylocentrotus intermedius. Fish Shellfish Immunol. 2015, 46, 723–736. [Google Scholar] [CrossRef] [PubMed]
- Jia, Z.-Y.; Wang, Q.; Wu, K.-K.; Wei, Z.-L.; Zhou, Z.C.; Liu, X.-L. De novo transcriptome sequencing and comparative analysis to discover genes involved in ovarian maturity in Strongylocentrotus nudus. Comp. Biochem. Physiol. Part D Genom. Proteom. 2017, 23, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.-Y.; Pan, N.; Wu, J.-N.; Liu, S.; Liu, Z.; Chen, L. Analysis and evaluation of nutritional composition of sea urchin (Anthocidaris crassispina) gonad. Fish. Mod. 2017, 44, 50–55. (In Chinese) [Google Scholar]
- Zhao, C.; Sun, P.; Zhou, H.-S.; Tian, X.-F.; Feng, W.-P.; Chang, Y.-Q. Heritability and phenotypic correlations of gonad sweetness in the sea urchin Strongylocentrotus intermedius. Aquacult. Int. 2014, 22, 1737–1742. [Google Scholar] [CrossRef]
- Feng, W.-P.; Chang, Y.-Q.; Zhao, C.; Sun, P.; Wei, J.; Hou, S. Effects of inbreeding on growth, gametogenesis, gonad production, quality and MYP expression in the sea urchin Strongylocentrotus intermedius. Aquacult. Int. 2015, 23, 903–912. [Google Scholar] [CrossRef]
- Chen, G.-Q.; Xiang, W.-Z.; Lau, C.; Peng, J.; Qiu, J.W.; Chen, F.; Jiang, J. A comparative analysis of lipid and carotenoid composition of the gonads of Anthocidaris crassispina, Diadema setosum and Salmacis sphaeroides. Food Chem. 2010, 120, 973–977. [Google Scholar] [CrossRef]
- Murata, Y.; Yokoyama, M.; Unuma, T.; Sata, N.; Kuwahara, R.; Kaneniwa, M. Seasonal changes of bitterness and pulcherrimine content in gonads of green sea urchin Hemicentrotus pulcherrimus at Iwaki in Fukushima Prefecture. Fish. Sci. 2002, 68, 184–189. [Google Scholar] [CrossRef][Green Version]
- Ghisaura, S.; Loi, B.; Biosa, G.; Baroli, M.; Pagnozzi, D.; Roggio, T.; Uzzau, S.; Anedda, R.; Addis, M. Proteomic changes occurring along gonad maturation in the edible sea urchin Paracentrotus lividus. J. Proteomics 2016, 144, 63–72. [Google Scholar] [CrossRef]
- Zhou, X.; Zhou, D.-Y.; Lu, T.; Liu, Z.-Y.; Zhao, Q.; Liu, Y.-X.; Hu, X.-P.; Zhang, J.-H.; Shasidi, F. Characterization of lipids in three species of sea urchin. Food Chem. 2018, 241, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Dincer, T.; Cakli, S. Chemical composition and biometrical measurements of the Turkish sea urchin (Paracentrotus lividus, lamarck, 1816). Crit. Rev. Food Sci. Nutr. 2007, 47, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Komata, Y. Study of the extractives of ‘‘Uni” IV. Taste of each component in the extractives. Bull. Jpn. Soc. Fish. Sci. 1964, 30, 749–756. [Google Scholar] [CrossRef]
- Osako, K.; Kiriyama, T.; Ruttanapornvareesakul, Y. Free amino acid compositions of the gonad of the wild and cultured sea urchins Anthocidaris crassispina. Aquacult. Sci. 2006, 54, 301–304. [Google Scholar]
- Inomata, E.; Murata, Y.; Matsui, T.; Agatsuma, Y. Gonadal production and quality in the sea urchin Mesocentrotus nudus fed a high-protein concentrated red alga Pyropia yezoensis. Aquaculture 2015, 454, 184–191. [Google Scholar] [CrossRef]
- Ayyagari, A.; Babu, K. Nutrient composition and antioxidant activity of gonads of sea urchin Stomopneustes variolaris. Food Chem. 2016, 197, 597–602. [Google Scholar]
- Li, L.; Yang, D.; Qi, S.-B.; Zuo, R.-T.; Wang, H.; Ding, J.; Chang, Y.-Q. Changes in lipids and fatty acids in gonads of sea urchin Strongylocentrotus intermedius before and after reproduction. J. Dalian Ocean. Univ. 2018, 33, 423–429. (In Chinese) [Google Scholar]
- Ding, J.; Chang, Y.Q.; Hao, Z.L.; Zhang, B. Comparative studies on urchin gonad fatty acid composition and β-carotene content in north China sea section. J. Agric. Sci. Technol. 2011, 13, 122–128. (In Chinese) [Google Scholar]
- Zhao, C.; Zhang, W.-J.; Chang, Y.Q.; Liu, P. Test and gonad characteristics in different genders of cultivated sea urchins (Strongylocentrotus intermedius, agassiz): First insight into sexual identification. Afr. J. Biotechnol. 2010, 9, 7560–7563. [Google Scholar]
- Phillips, K.; Bremer, P.; Silcock, P.; Hamid, N.; Delahunty, C.; Barker, M.; Kissick, J. Effect of gender, diet and storage time on the physical properties and sensory quality of sea urchin (Evechinus chloroticus) gonads. Aquaculture 2009, 288, 205–215. [Google Scholar] [CrossRef]
- Kong, D.-R.; Cui, J.; Wang, Z.-C.; Sun, H.; Hu, Z.-W.; Li, X.; Qiu, X.-M.; Jiang, C.; Liu, H.-Y.; Zhang, T.; et al. The regulatory networks conferred by IFN-γ in the kidney of Takifugu rubripes. Int. J. Agric. Biol. 2018, 20, 2189–2195. [Google Scholar]
- Cui, J.; Liu, S.-K.; Zhang, B.; Wang, H.-D.; Sun, H.-J.; Song, S.-H.; Qiu, X.-M.; Liu, Y.; Wang, X.-L.; Jiang, Z.-Q.; et al. Transciptome analysis of the gill and swimbladder of Takifugu rubripes by RNA-Seq. PLoS ONE 2014, 9, e85505. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.; Haas, B.; Yassour, M.; Levin, J.; Thompson, D.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.-D.; et al. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Eddy, S. Profile hidden Markov models. Bioinformatics 1998, 14, 755–763. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Mao, X.-Z.; Huang, J.-J.; Ding, Y.; Wu, J.-M.; Dong, S.; Kong, L.; Gao, G.; Li, C.-Y.; Wei, L.-P. Kobas 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef]
- Wang, Z.-C.; Qiu, X.-M.; Kong, D.-R.; Zhou, X.-X.; Guo, Z.-B.; Gao, C.-F.; Ma, S.; Hao, W.-W.; Jiang, Z.-Q.; Liu, S.-C.; et al. Comparative RNA-Seq analysis of differentially expressed genes in the testis and ovary of Takifugu rubripes. Comp. Biochem. Physiol. Part DGenom. Proteom. 2017, 22, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Dillies, M.; Rau, A.; Aubert, J.; Hennequet-Antier, C.; Jeanmougin, M.; Servant, N.; Keime, C.; Marot, G.; Castel, D.; Estelle, J.; et al. Comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Brief. Bioinform. 2013, 14, 671–683. [Google Scholar] [CrossRef]
- Wang, F.-C.; Yang, J.; Wang, H.-B.; Xia, G.-L. Gonadotropin-regulated expressions of lanosterol 14α-demethylase, sterol Δ14-reductase and C-4 sterol methyl oxidase contribute to the accumulation of meiosis-activating sterol in rabbit gonads. Prostag. Oth. Lipid Mediat. 2010, 92, 25–32. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, 106. [Google Scholar] [CrossRef]
- Klopfenstein, D.V.; Zhang, L.; Pedersen, B.S.; Ramírez, F.; Vesztrocy, A.W.; Naldi, A.; Mungall, C.J.; Yunes, J.M.; Botvinnik, O.; Weigel, M.; et al. GOATOOLS: A Python library for Gene Ontology analyses. Sci. Rep. 2018, 18, 10872. [Google Scholar] [CrossRef]
- Lalitha, S. Primer Premier 5. Biotech. Softw. Internet Rep. 2000, 1, 270–272. [Google Scholar] [CrossRef]
- Livak, K.; Schmittgen, T. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I. Trinity: Reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 2013, 29, 644. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.-W.; Cui, X.; Shen, X.-F.; Wang, L.-S.; Jiang, L.; Liu, H.-Y.; Liu, Y.; Liu, Q.; Jiang, C. De novo transcriptome analysis and differentially expressed genes in the ovary and testis of the Japanese mantis shrimp Oratosquilla oratoria by RNA-Seq. Comp. Biochem. Physiol. Part D Genom. Proteom. 2018, 26, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Mcbride, S.; Price, R.; Tom, P.; Lawrence, J.; Lawrence, A. Comparison of gonad quality factors: Color, hardness and resilience, of Strongylocentrotus franciscanus between sea urchins fed prepared feed or algal diets and sea urchins harvested from the northern California fishery. Aquaculture 2004, 233, 405–422. [Google Scholar] [CrossRef]
- Murata, Y.; Sata, N.; Yokoyama, M.; Kuwahara, R.; Oohara, I. Determination of a novel bitter amino acid, pulcherrimine, in the gonad of the green sea urchin Hemicentrotus pulcherrimus. Fish. Sci. 2001, 67, 341–345. [Google Scholar] [CrossRef]
- Xu, W.-D.; Han, P.; Wang, L.-M.; Zhou, Z.-C.; Han, J.-B. The amino acid compositions of gonads in sea urchin Strongylocentrotus nudus. J. Dalian Ocean. Univ. 2009, 24, 583–586. (In Chinese) [Google Scholar]
- Murata, Y.; Yamamoto, T.; Kaneniwa, M.; Kuwahara, R.; Yokoyama, M. Occurrence of bitter gonad in Hemicentrotus pulcherrimus. Nippon. Suisan Gakkaishi 1998, 64, 477–478. [Google Scholar] [CrossRef][Green Version]
- Li, Y.; Chen, G.-K.; Tong, X.-W.; Zhang, H.-T.; Liu, X.-G.; Liu, Y.-H.; Lu, F.-P. Construction of Escherichia coli strains producing L-serine from glucose. Biotechnol. Lett. 2012, 34, 1525–1530. [Google Scholar] [CrossRef]
- Deng, Y.-H.; Ran, W.; Man, S.-Q.; Li, X.-P.; Gao, H.-J.; Tang, T.; Tachibana, H.; Cheng, X.-J. Artemether exhibits amoebicidal activity against acanthamoeba castellanii through inhibition of the serine biosynthesis pathway. Antimicrob. Agents Chemother. 2015, 59, 4680–4688. [Google Scholar] [CrossRef]
- Basurko, M.J.; Marche, M.; Darriet, M.; Cassaigne, A. Phosphoserine aminotra-nsferase, the second step-catalyzing enzyme for serine biosynthesis. IUBMB Life 1999, 48, 525–529. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, Y.; Sakuraba, H.; Doi, K.; Ohshima, T. Molecular and functional characterization of D-3-phosphoglycerate dehydrogenase in the serine biosynthetic pathway of the hyperthermophilic archaeon Sulfolobus tokodaii. Arch. Biochem. Biophys. 2008, 470, 120–128. [Google Scholar] [CrossRef] [PubMed]
- MacFarlane, A.; Liu, X.-W.; Perry, C.; Flodby, P.; Allen, R.; Stabler, S.; Stover, P. Cytoplasmic serine hydroxymethyltransferase regulates the metabolic partitioning of methylenetetrahydrofolate but is not essential in mice. J. Biol. Chem. 2008, 283, 25846–25853. [Google Scholar] [CrossRef]
- Grant, G.; Hu, Z.-Q.; Xu, X.-L. Amino acid residue mutations uncouple cooperative effects in Escherichia coli D-3-phosphoglycerate dehydrogenase. J. Bio. Chem. 2001, 276, 17844–17850. [Google Scholar] [CrossRef]
- Grant, G.; Xu, X.L.; Hu, Z. The relationship between effector binding and inhibition of activity in D-3-phosphoglycerate dehydrogenase. Protein Sci. 1999, 8, 2501–2505. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Grant, G. Transient kinetic analysis of L-serine Interaction with Escherichia coli D-3-phosphoglycerate dehydrogenase containing amino acid mutations in the hinge regions. Biochemistry 2011, 50, 2900–2906. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dey, S.; Grant, G.; Sacchettini, J. Crystal structure of mycobacterium tuberculosis D-3-phosphoglycerate dehydrogenase: Extreme asymmetry in a tetramer of identical subunits. J. Bio. Chem. 2005, 280, 14892–14899. [Google Scholar] [CrossRef]
- Zhang, W.; Zhang, M.; Gao, C.; Zhang, Y.-P.; Ge, Y.-S.; Guo, S.-T.; Guo, X.-T.; Zhou, Z.-K.; Liu, Q.-Y.; Zhang, Y.-X.; et al. Coupling between d-3-phosphoglycerate dehydrogenase and d-2-hydroxyglutarate dehydrogenase drives bacterial l-serine synthesis. Proc. Natl. Acad. Sci. USA 2017, 114, E7574–E7582. [Google Scholar] [CrossRef]
- Kaneko, K.; Matsumoto, H.; Shirai, T.; Kamei, M.; Okazaki, E.; Osako, K. Seasonal variations in free amino acid composition and taste aspects of black sea urchin, Diadema setosum, gonad. Food Sci. Technol. Res. 2012, 18, 835–842. [Google Scholar] [CrossRef][Green Version]
- Woods, C.; James, P.; Moss, G.; Wright, J.; Siikavuopio, S. A comparison of the effect of urchin size and diet on gonad yield and quality in the sea urchin. Aquac. Int. 2008, 16, 49–68. [Google Scholar] [CrossRef]
- Azad, A.; Pearce, C.; Mckinley, R. Effects of diet and temperature on ingestion, absorption, assimilation, gonad yield, and gonad quality of the purple sea urchin (Strongylocentrotus purpuratus). Aquaculture 2011, 317, 187–196. [Google Scholar] [CrossRef]
- 52 Wilson, P.; Gardner, S.; Lambie, N.; Commans, S.; Crowther, D. Characterization of the human patatin-like phospholipase family. J. Lipid R. 2006, 47, 1940–1949. [Google Scholar] [CrossRef] [PubMed]
- Rotman, Y.; Koh, C.; Zmuda, J.; Kleiner, D.; Liang, T.; Nash, C. The association of genetic variability in patatin-like phospholipase domain-containing protein 3 (PNPLA3) with histological severity of nonalcoholic fatty liver disease. Hepatology 2010, 52, 894–903. [Google Scholar] [CrossRef] [PubMed]
- Viganò, M.; Valenti, L.; Lampertico, P.; Facchetti, F.; Motta, B.; D’Ambrosio, R.; Romagnoli, S.; Dongiovanni, P.; Donati, B.; Fargion, S.; et al. Patatin-like phospholipase domain-containing 3 I148M affects liver steatosis in patients with chronic hepatitis B. Hepatology 2013, 58, 1245–1252. [Google Scholar] [CrossRef]
- Chang, Y.-Q.; Zhang, W.-J.; Ding, J.; Shi, S.-B.; Zhao, C.; Zhang, B. Comparison of gonad traits among families and between genders of the sea urchin Strongylocentrotus intermedius. J. Fish. China 2010, 34, 1080–1088. [Google Scholar]
- Tong, S.-Y.; Chen, W.; You, X.-C.; Liu, H.-L. Study on lipid and fatty acids composition of three kinds of echinoidea’s gonad. J. Fish. China 1998, 22, 247–252. (In Chinese) [Google Scholar]
- Hathaway, R. Conversion of estradiol-17β by sperm preparations of sea urchins and oysters. Gen. Comp. Endocrinol. 1965, 5, 504–508. [Google Scholar] [CrossRef]
- Wang, C.; Croll, R. Effects of sex steroids on gonadal development and gender determination in the sea scallop, Placopectenmagellanicus. Aquaculture 2004, 238, 483–498. [Google Scholar] [CrossRef]
- Kiyomoto, M.; Kikuchi, A.; Morinaga, S.; Unuma, T.; Yokota, Y. Exogastrulation and interference with the expression of major yolk protein by estrogens administered to sea urchins. Cell Biol. Toxicol. 2008, 24, 611–620. [Google Scholar] [CrossRef]
- Leenders, F.; Adamski, J.; Husen, B.; Thole, H.; Jungblut, P. Molecular cloning and amino acid sequence of the porcine 17beta-estradiol dehydrogenase. Eur. J. Biochem. 1994, 222, 221–227. [Google Scholar] [CrossRef]
- Ohno, S.; Nishikawa, K.; Honda, Y.; Nakajin, S. Expression in E. coli and tissue distribution of the human homologue of the mouse Ke 6 gene, 17beta-hydroxysteroid dehydrogenase type 8. Mol. Cell Biochemi. 2008, 309, 209–215. [Google Scholar] [CrossRef]
- Rotinen, M.; Celay, J.; Alonso, M.; Arrazola, A.; Encio, I.; Villar, J. Estradiol induces type 8 17beta-hydroxysteroid dehydrogenase expression: Crosstalk between estrogen receptor alpha and C/EBPbeta. J. Endocrinol. 2009, 200, 85–92. [Google Scholar] [CrossRef] [PubMed]
- 63 Smuc, T.; Rizner, T. Expression of 17beta-hydroxysteroid dehydrogenases and other estrogen-metabolizing enzymes in different cancer cell lines. Chem. Biol. Interact. 2009, 178, 228–233. [Google Scholar] [CrossRef] [PubMed]
- 64 Liu, J.-G.; Zhang, Z.-F.; Ma, X.-S.; Liang, S.-S.; Yang, D.-D. Characteristics of 17β-hydroxysteroid dehydrogenase 8 and its potential role in gonad of Zhikong scallop Chlamys farreri. J. Steroid. Biochem. Mol. Biol. 2014, 141, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Motohara, K.; Tashiro, H.; Taura, Y.; Ohba, T.; Katabuchi, H. Immunohistochemical analysis of 17β-hydroxysteroid dehydrogenase isozymes in human ovarian surface epithelium and epithelial ovarian carcinoma. Med. Mol. Morphol. 2010, 43, 197–203. [Google Scholar] [CrossRef]
- Hilborn, E.; Stål, O.; Jansson, A. Estrogen and androgen-converting enzymes 17β-hydroxysteroid dehydrogenase and their involvement in cancer: With a special focus on 17β-hydroxysteroid dehydrogenase type 1, 2, and breast cancer. Oncotarget 2017, 8, 30552–30562. [Google Scholar] [CrossRef]
- Chen, Z.-J.; Kastaniotis, A.; Miinalainen, I.; Rajaram, V.; Wierenga, R.; Hiltunen, J. 17β-hydroxysteroid dehydrogenase type 8 and carbonyl reductase type 4 assemble as a ketoacyl reductase of human mitochondrial FAS. FASEB J. 2009, 23, 3682–3691. [Google Scholar] [CrossRef]
- Jin, Q.; Ren, Y.; Wang, M.; Suraneni, P.; Crispino, J.; Fan, J.; Huang, Z. Novel function of faxdc2 in megakaryopoiesis. Blood Cancer J. 2016, 6, 478. [Google Scholar] [CrossRef]
- Machlus, K.; Italiano, J. The incredible journey: From megakaryocyte development to platelet formation. J. Cell Biol. 2013, 201, 785–796. [Google Scholar] [CrossRef]
- Parra, M.; Rubilar, T.; Latorre, M.; Epherra, L.; Gil, D. Nutrient allocation in the gonads of the sea urchin Arbacia dufresnii in different stages of gonadal development. Invertebr. Reprod. Dev. 2014, 59, 26–36. [Google Scholar] [CrossRef]
- Bard, M.; Bruner, D.; Pierson, C.; Lees, N.; Biermann, B.; Frye, L.; Koegel, C.; Barbuch, R. Cloning and characterization of ERG25, the Saccharomyces cerevisiae gene encoding C-4 sterol methyl oxidase. Proc. Natl. Acad. Sci. USA 1966, 93, 186–190. [Google Scholar] [CrossRef]
- Yao, J.N.; Liao, N.Q.; Li, H.M. Characterization and bioinformatics analysis of c-4 sterol methyl oxidase from Monascus purpureus. Appl. Mech. Mater. 2014, 522–524, 247–250. [Google Scholar] [CrossRef]
- Pollier, J.; Vancaester, E.; Kuzhiumparambil, U.; Vickers, C.; Klaas, V.K.; Goossens, A.; Fabris, M.; Kuzhiumparambil, U. A widespread alternative squalene epoxidase participates in eukaryote steroid biosynthesis. Nat. Microbiol. 2019, 4, 226–233. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).