Identification and Characterization of Differentially Expressed IgM Transcripts of Channel Catfish Vaccinated with Antigens of Virulent Aeromonas hydrophila
Abstract
:1. Introduction
2. Materials and Methods
2.1. RNA Isolation, cDNA Synthesis and Primers
2.2. Cloning of Full-Length IgM Transcripts
2.3. Quantitative Real-Time PCR
2.4. Cloning of the Variable Domain of IgM
2.5. Two-Dimensional Gel Electrophoresis of IgM Protein
2.6. Data Processing and Analysis
3. Results
3.1. Expression of IgM Transcripts
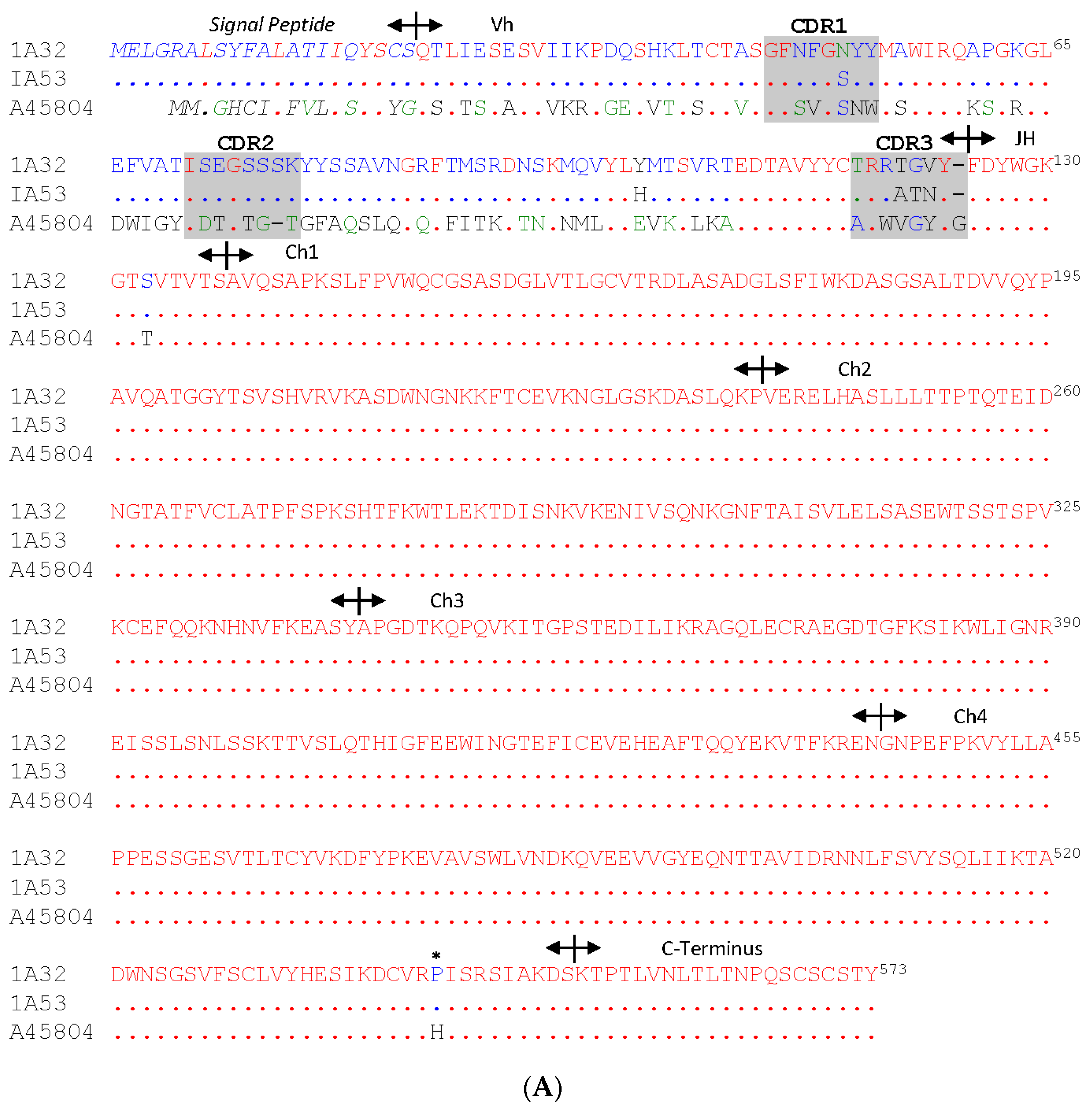
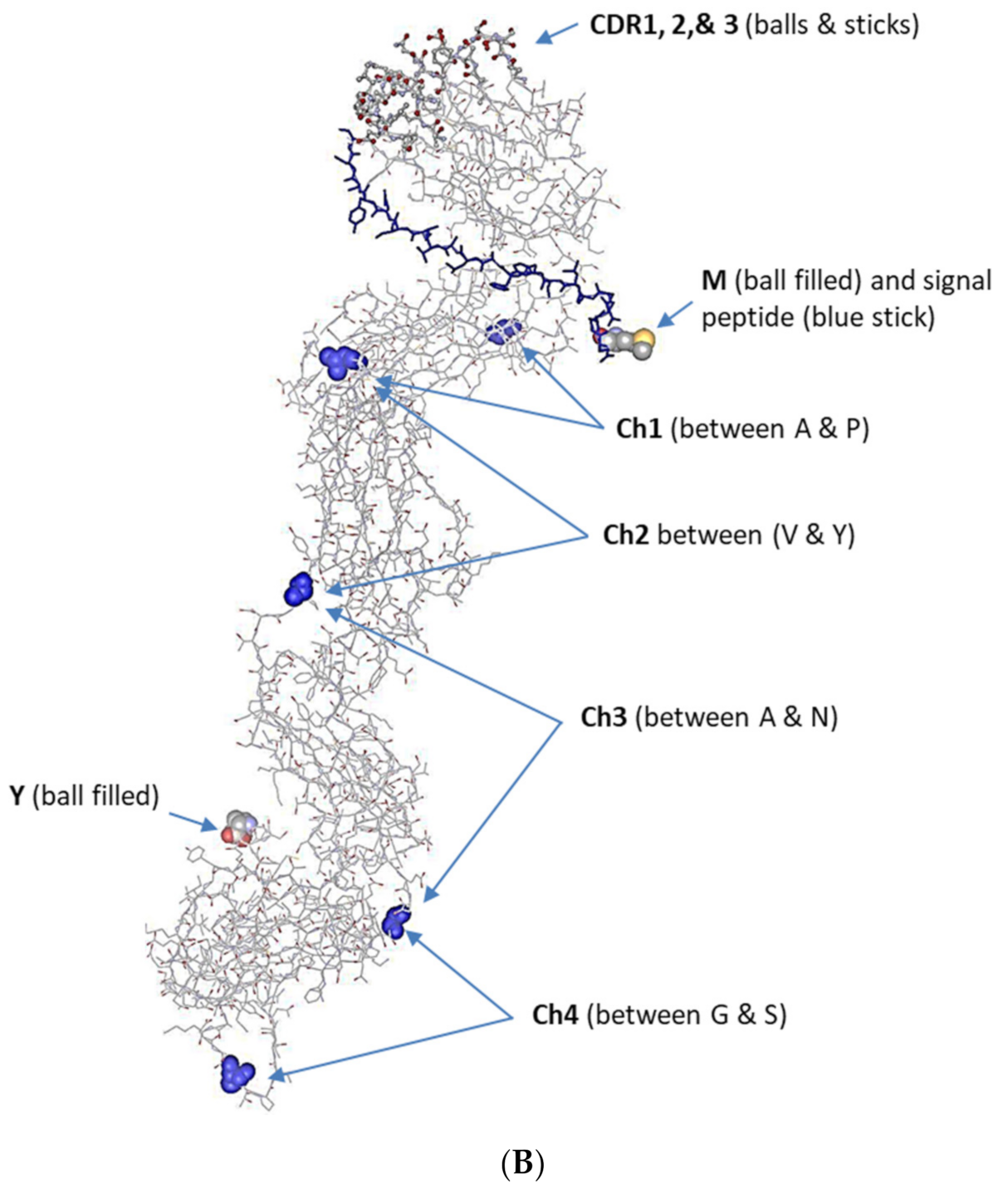
3.2. The Full-Length IgM cDNA
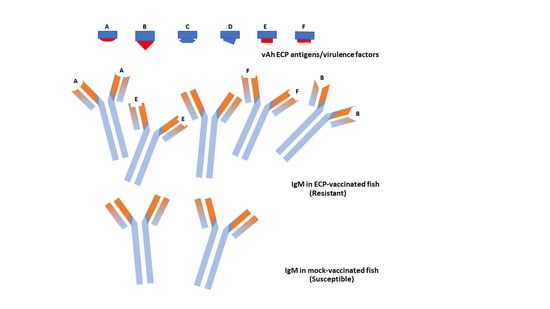
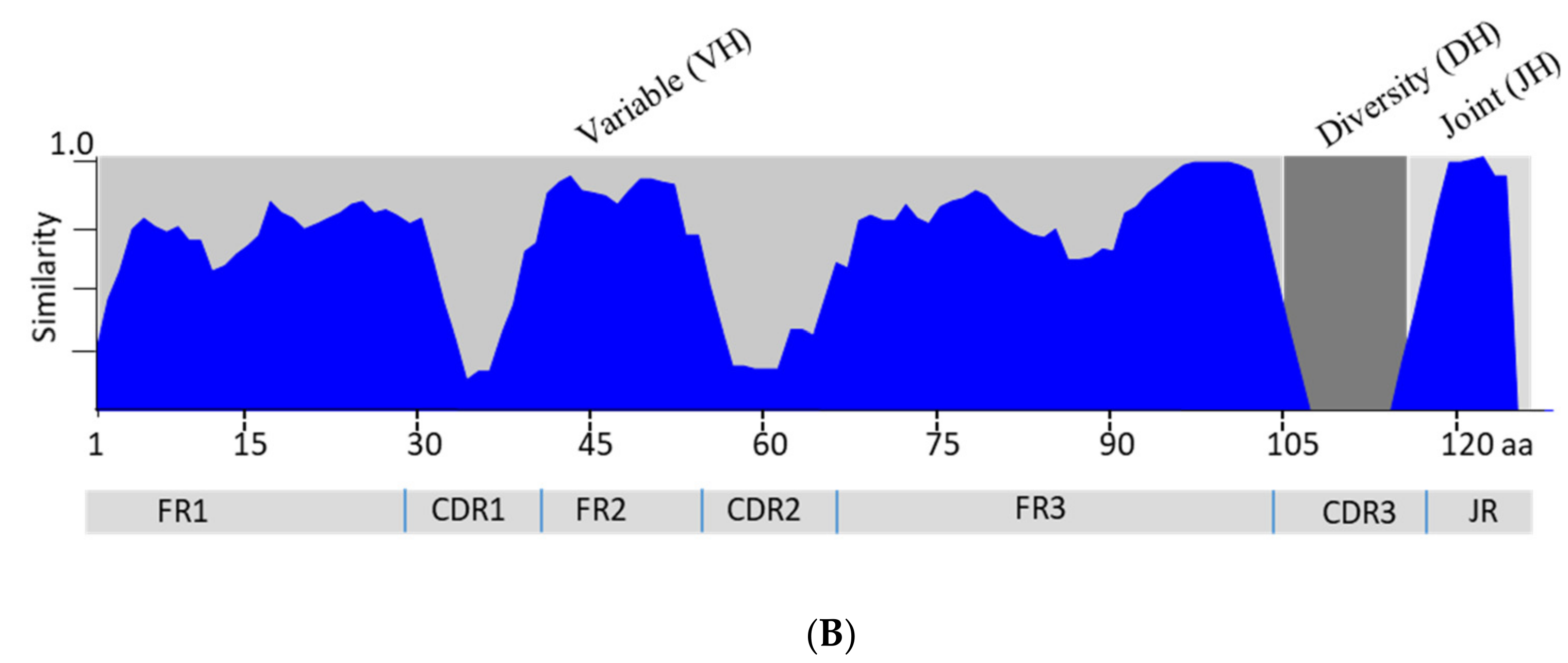
3.3. Analysis of the Variable Domain of IgM Heavy Chain
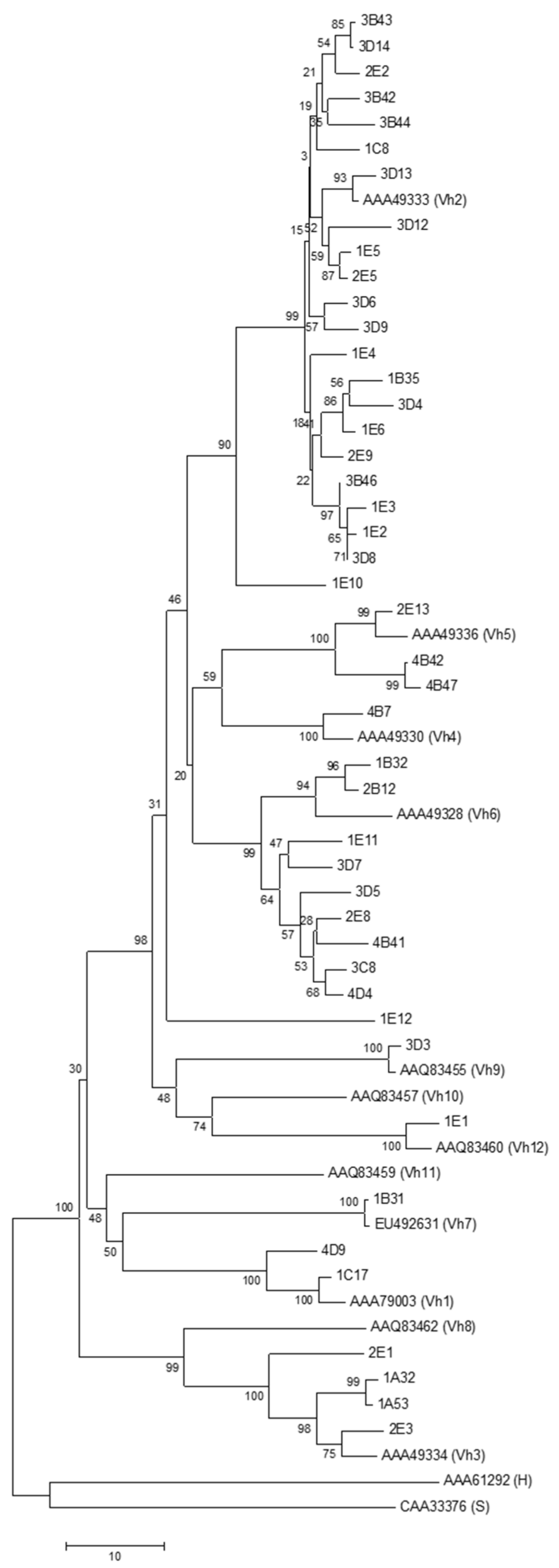
3.4. Phylogenetic Analysis
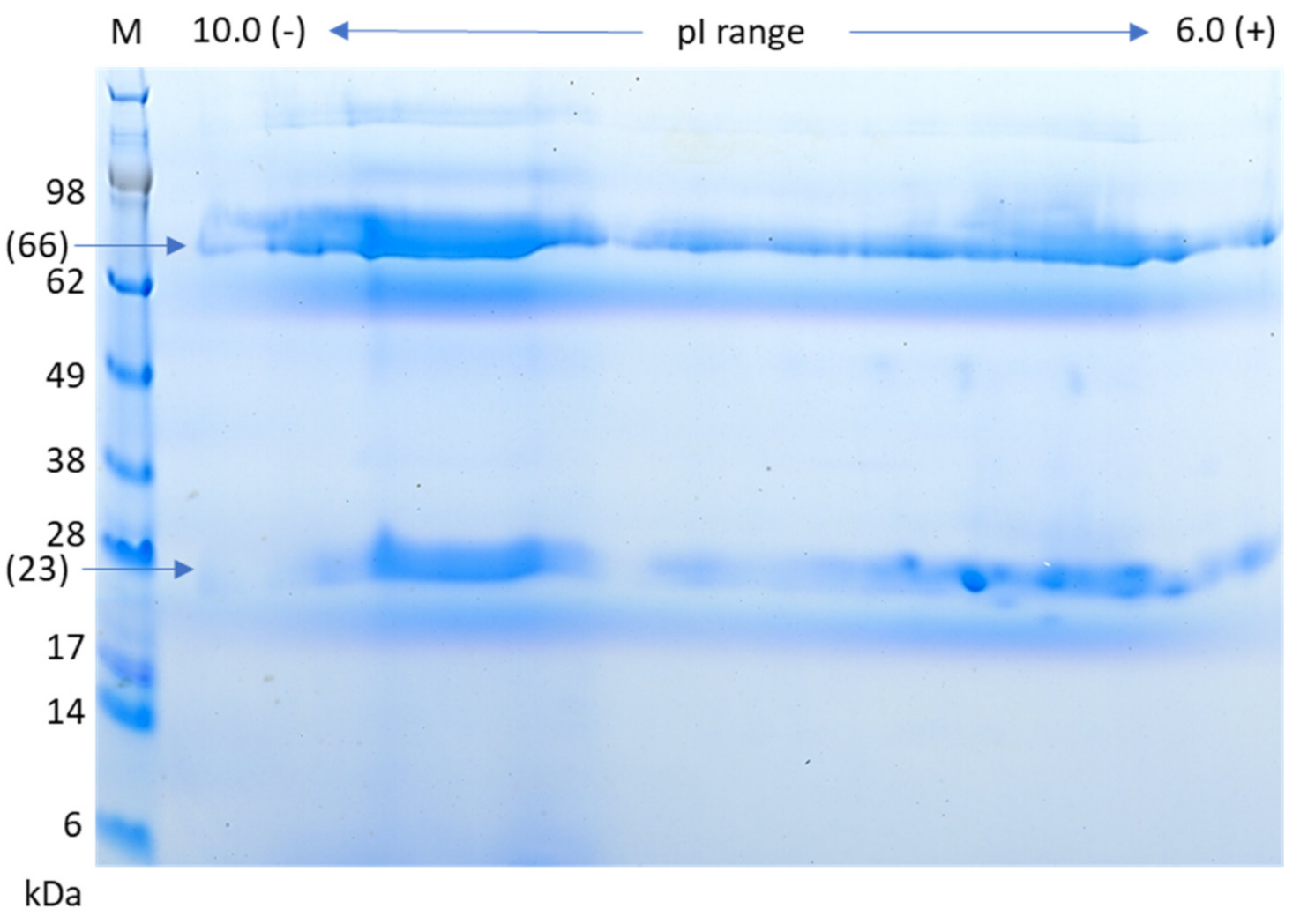
3.5. Two-Dimensional Gel Analysis of IgM Protein
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Salinas, I.; Zhang, Y.A.; Sunyer, J.O. Mucosal immunoglobulins and B cells of teleost fish. Dev. Comp. Immunol. 2011, 35, 1346–1365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hikima, J.; Jung, T.S.; Aoki, T. Immunoglobulin genes and their transcriptional control in teleosts. Dev. Comp. Immunol. 2011, 35, 924–936. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Wang, Q.; Huang, Z.; Ding, L.; Xu, Z. Immunoglobulins, mucosal immunity and vaccination in teleost fish. Front. Immunol. 2020, 11, 567941. [Google Scholar] [CrossRef]
- Danilova, N.; Bussmann, J.; Jekosch, K.; Steiner, L.A. The immunoglobulin heavy-chain locus in zebrafish: Identification and expression of a previously unknown isotype, immunoglobulin Z. Nat. Immunol. 2005, 6, 295–302. [Google Scholar] [CrossRef]
- Hansen, J.D.; Landis, E.D.; Phillips, R.B. Discovery of a unique Ig heavy-chain isotype (IgT) in rainbow trout: Implications for a distinctive B cell developmental pathway in teleost fish. Proc. Natl. Acad. Sci. USA 2005, 102, 6919–6924. [Google Scholar] [CrossRef] [Green Version]
- Bengtén, E.; Quiniou, S.; Hikima, J.; Waldbieser, G.; Warr, G.W.; Miller, N.W.; Wilson, M. Structure of the catfish IGH locus: Analysis of the region including the single functional IGHM gene. Immunogenetics 2006, 58, 831–844. [Google Scholar] [CrossRef]
- Zhang, Y.A.; Salinas, I.; Li, J.; Parra, D.; Bjork, S.; Xu, Z.; LaPatra, S.E.; Bartholomew, J.; Sunyer, J.O. IgT, a primitive immunoglobulin class specialized in mucosal immunity. Nat. Immunol. 2010, 11, 827–835. [Google Scholar] [CrossRef]
- Castro, R.; Jouneau, L.; Pham, H.P.; Bouchez, O.; Giudicelli, V.R.; Lefranc, M.P.; Quillet, E.; Benmansour, A.; Cazals, F.; Six, A.; et al. Teleost fish mount complex clonal IgM and IgT responses in spleen upon systemic viral infection. PLoS Pathog. 2013, 9, e1003098. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.R.; Warr, G.W. Fish immunoglobulins and the genes that encode them. Annu. Rev. Fish Dis. 1992, 2, 201–221. [Google Scholar] [CrossRef]
- Bengtén, E.; Clem, L.M.; Miller, N.W.; Warr, G.W.; Wilson, M. Channel catfish immunoglobulins: Repertoire and expression. Dev. Comp. Immunol. 2006, 30, 77–92. [Google Scholar] [CrossRef]
- Wilson, M.R.; Marcuz, A.; Van Ginkel, F.; Miller, N.W.; Clem, L.W.; Middleton, D.; Warr, G.W. The immunoglobulin M heavy chain constant region gene of the channel catfish, Ictalurus punctatus: An unusual mRNA splice pattern produces the membrane form of the molecule. Nucleic Acids Res. 1990, 18, 5227–5233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mashoof, S.; Pohlenz, C.; Chen, P.; Deiss, T.C.; Gatlin, D.; Buentelo, A.; Criscitiello, M. Expressed IgH µ and τ transcripts share diversity segment in ranched Thunnus orientalis. Dev. Comp. Immunol. 2014, 43, 76–86. [Google Scholar] [CrossRef] [PubMed]
- Ralph, D.; Frederick, A.; Matsen IV, F.A. Consistency of VDJ rearrangement and substitution parameters enable accurate B cells receptor sequence annotation. PLoS Comput. Biol. 2015, 12, e1004409. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.; Pridgeon, J.W.; Klesius, P.H. Vaccination of channel catfish with extracellular products of Aeromonas hydrophila provides protection against infection by the pathogen. Fish Shellfish Immunol. 2014, 36, 270–275. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Xu, D.-H.; Beck, B. Analysis of agglutinants elicited by antiserum of channel catfish immunized with extracellular proteins of virulent Aeromonas hydrophila. Fish Shellfish Immunol. 2019, 86, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Ghaffari, S.H.; Lobb, C.J. Nucleotide sequence of channel catfish heavy chain cDNA and genomic blot analysis: Implications for the phylogeny of Ig heavy chains. J. Immunol. 1989, 143, 2730–2739. [Google Scholar] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Solem, S.T.; Stenvik, J. Antibody repertoire development in teleosts—A review with emphasis on salmonids and Gadus morhua L. Dev. Comp. Immunol. 2006, 30, 57–76. [Google Scholar] [CrossRef]
- Perdiguero, P.; Martín-Martín, A.; Benedicenti, O.; Díaz-Rosales, P.; Morel, E.; Muñoz-Atienza, E.; García-Flores, M.; Simón, R.; Soleto, I.; Cerutti, A.; et al. Teleost IgD+ IgM- B Cells Mount Clonally Expanded and Mildly Mutated Intestinal IgD Responses in the Absence of Lymphoid Follicles. Cell Rep. 2019, 29, 4223–4235. [Google Scholar] [CrossRef] [Green Version]
- Feng, J.; Guan, R.; Lin, P.; Guo, S. Molecular cloning and characterization analysis of immunoglobulin M heavy chain gene in European ell (Anguilla anguilla). Vet. Immunol. Immunopathol. 2009, 127, 144–147. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Ventura-Holman, T.; Waldbieser, G.C.; Lobb, C.J. Structure, genomic organization, and phylogenetic implications of six new VH families in channel catfish. Molec. Immunol. 2003, 40, 247–260. [Google Scholar] [CrossRef]
- Yang, F.; Waldbieser, G.C.; Lobb, C.J. The nucleotide targets of somatic mutation and the role of selection in immunoglobulin heavy chains of a teleost fish. J. Immunol. 2006, 176, 1655–1667. [Google Scholar] [CrossRef]
- Mashoof, S.; Criscitiello, M.F. Fish immunoglobulins. Biology 2016, 5, 45. [Google Scholar] [CrossRef] [Green Version]
- Ye, J.; Ma, N.; Madden, T.L.; Ostell, J.M. IgBLAST: An immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 2013, 41, w34–w40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghaffari, S.H.; Lobb, C.J. Structure and Genomic Organization of a Second Cluster of Immunoglobulin Heavy Chain Gene Segments in the Channel Catfish. J. Immunol. 1999, 162, 1519–1529. [Google Scholar]
- Magnadóttir, B.; Crispin, M.; Royle, L.; Colominas, C.; Harvey, D.J.; Dwek, R.A.; Rudd, P.M. The carbohydrate moiety of serum IgM from Atlantic cod (Gadus morhua L.). Fish Shellfish Immunol. 2002, 12, 209–227. [Google Scholar] [CrossRef]

| Primer | Gene/Function | Sequence (5′ to 3′) |
|---|---|---|
| Ch1f-IF | IgM/3′-RACE | gattacgccaagcttCGTCGTGCAATACCCGGCGGTGCAAGC |
| Ch1r-IF | IgM/5′-RACE | gattacgccaagcttGCAGACGAAGGTAGCTGTTCCATTGTC |
| Ch2f | IgM/qPCR | ATCCCTGTTTCCCGTGTGGCA |
| Ch2r | IgM/qPCR | TCCCGCTCGCATCCTTCCATA |
| Ch2r-IF | IgM/V-domain | gattacgccaagcttCTCCCGCTCGCATCCTTCCATATG |
| Ch3r-IF | IgM/full | gattacgccaagcttTCAAGCATTAATAGCAGACAACACA |
| 18Sf | 18S rRNA/qPCR | TTGATAACCTCGGGCCGATCG |
| 18Sr | 18S rRNA/qPCR | CGTTACCCGTGGTCACCATG |
| Tissues | Immunization | Average CT ± SD | Relative to Calibrator (ΔCT) | Fold Change (2−ΔΔCT) |
|---|---|---|---|---|
| Head Kidney | ECP | 18.78 ± 0.06 | 9.13 | 1.60 * |
| Mock | 18.52 ± 0.03 | 9.81 | - | |
| Liver | ECP | 20.12 ± 0.13 | 10.11 | 1.47 * |
| Mock | 20.67 ± 0.09 | 10.67 | - |
| IGVH Family (Reference) 1 | Head Kidney | Liver | Variants | |||
|---|---|---|---|---|---|---|
| Immunized | Mock | Immunized | Mock | Unique | Total | |
| 1 (AAA79003) | 1 | 1 (3) 2 | 2 | 4 | ||
| 2 (AAA49333) | 9 (14) | 3 (6) | 11 (18) | 23 | 38 | |
| 3 (AAA49334) | 2 (3) | 2 | 4 | 5 | ||
| 4 (AAA49330) | 1 (2) | 1 | 2 | |||
| 5 (AAA49336) | 1 (3) | 2 (5) | 3 | 8 | ||
| 6 (AAA49328) | 2 | 2 (5) | 3 (4) | 2 (6) | 9 | 17 |
| 7 (EU492631) | 1 (2) | 1 | 2 | |||
| 9 (AAQ83455) | 1 (2) | 1 | 2 | |||
| 12 (AAA83460) | 1 (2) | 1 | 2 | |||
| 13 (pseudo) | [1] | [2] | [2] | [5] | [5] | |
| (Total) | 16 (26) | 8 (18) | 15 (19) | 6 (22) | 45 | 85 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, D.; Lange, M.D.; Shoemaker, C.A.; Beck, B.H. Identification and Characterization of Differentially Expressed IgM Transcripts of Channel Catfish Vaccinated with Antigens of Virulent Aeromonas hydrophila. Fishes 2022, 7, 24. https://doi.org/10.3390/fishes7010024
Zhang D, Lange MD, Shoemaker CA, Beck BH. Identification and Characterization of Differentially Expressed IgM Transcripts of Channel Catfish Vaccinated with Antigens of Virulent Aeromonas hydrophila. Fishes. 2022; 7(1):24. https://doi.org/10.3390/fishes7010024
Chicago/Turabian StyleZhang, Dunhua, Miles D. Lange, Craig A. Shoemaker, and Benjamin H. Beck. 2022. "Identification and Characterization of Differentially Expressed IgM Transcripts of Channel Catfish Vaccinated with Antigens of Virulent Aeromonas hydrophila" Fishes 7, no. 1: 24. https://doi.org/10.3390/fishes7010024
APA StyleZhang, D., Lange, M. D., Shoemaker, C. A., & Beck, B. H. (2022). Identification and Characterization of Differentially Expressed IgM Transcripts of Channel Catfish Vaccinated with Antigens of Virulent Aeromonas hydrophila. Fishes, 7(1), 24. https://doi.org/10.3390/fishes7010024