Otolith Microchemistry and Demographic History Provide New Insight into the Migratory Behavior and Heterogeneous Genetic Divergence of Coilia grayii in the Pearl River
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Otolith Preparation and Microprobe Analysis
2.3. DNA Extraction, Library Construction, and Sequencing
2.4. Data Analysis
3. Results
3.1. Microchemistry Analysis
3.2. Phylogenetic Analyses
3.3. Population Genetics and Structure
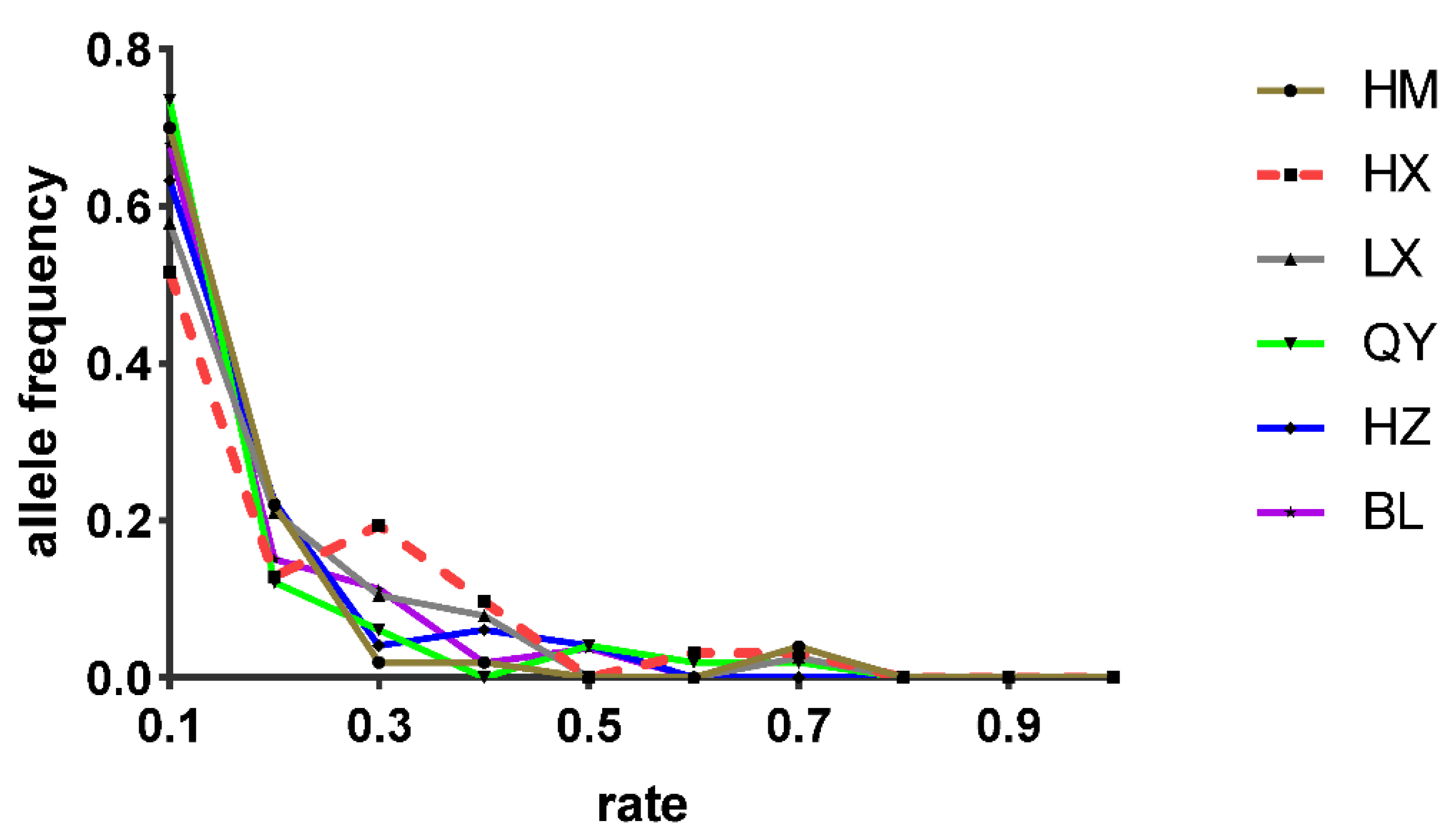
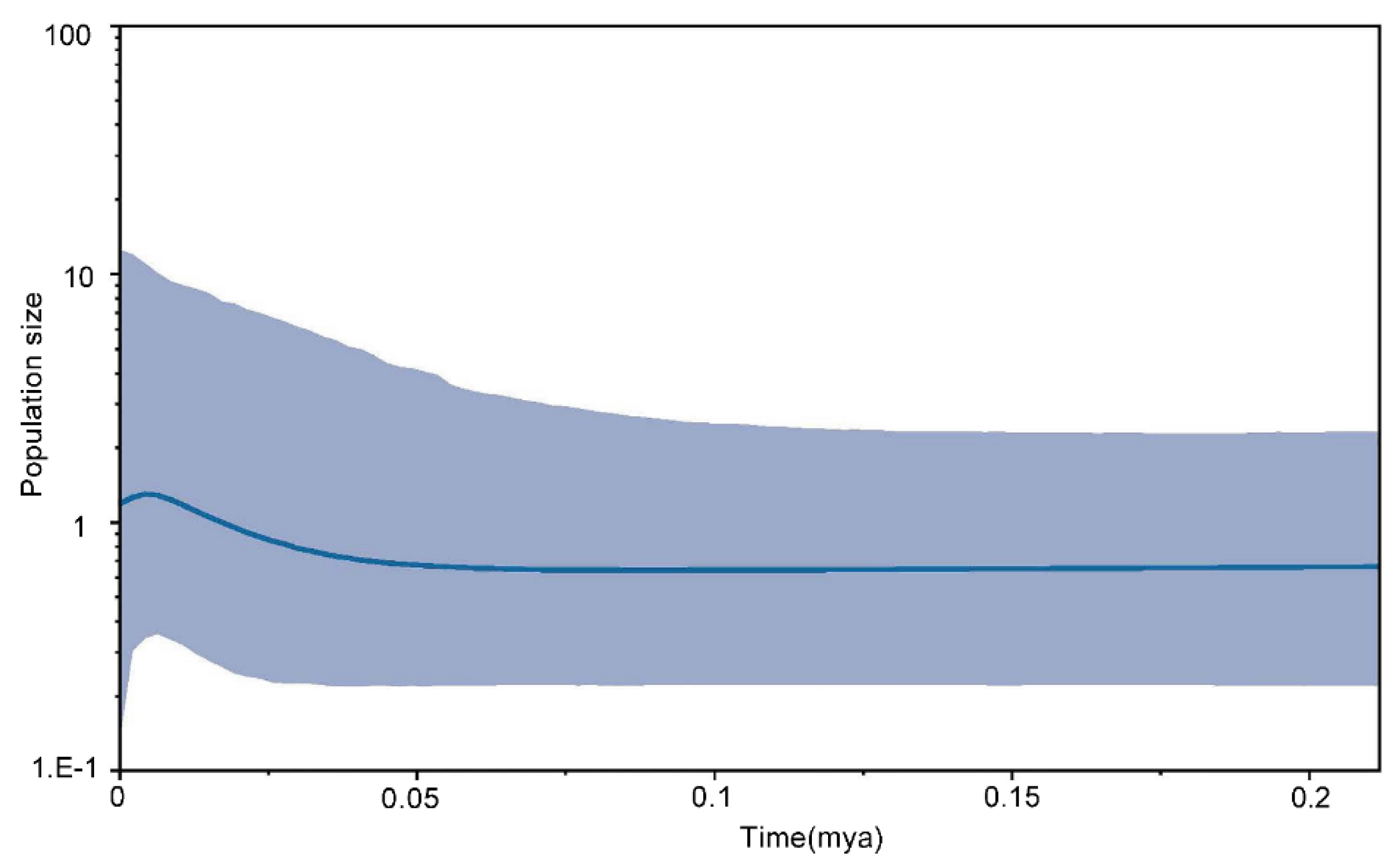
3.4. Demography History
4. Discussion
4.1. Migratory Behavioral Variations
4.2. Genetic Diversity
4.3. Population Structure and Habitat Fragmentation
4.4. Fluctuations in Demography and Migration
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yamane, K.; Shirai, K.; Nagakura, Y.; Otake, T. Assessing the usefulness of otolith elemental compositions for evaluating the population structure of the Pacific herring Clupea pallasii in northern Japan. Fish. Sci. 2012, 78, 295–307. [Google Scholar] [CrossRef]
- Bae, S.E.; Kim, J.-K. Otolith microchemistry reveals the migration patterns of the flathead grey mulletMugil cephalus(Pisces: Mugilidae) in Korean waters. J. Ecol. Environ. 2020, 44, 21. [Google Scholar] [CrossRef]
- Barradas, J.R.S.; Silva, L.G.; Harvey, B.C.; Fontoura, N.F. Estimating migratory fish distribution from altitude and basin area: A case study in a large Neotropical river. Freshw. Biol. 2012, 57, 2297–2305. [Google Scholar] [CrossRef]
- Roffe, A.J. Drift as constitutive: Conclusions from a formal reconstruction of population genetics. Hist. Philos. Life Sci. 2019, 41, 55. [Google Scholar] [CrossRef] [PubMed]
- Palstra, F.P.; Ruzzante, D.E. A temporal perspective on population structure and gene flow in Atlantic salmon (Salmo salar) in Newfoundland, Canada. Can. J. Fish. Aquat. Sci. 2010, 67, 225–242. [Google Scholar] [CrossRef]
- Crookes, S.; Shaw, P.W. Isolation by distance and non-identical patterns of gene flow within two river populations of the freshwater fish Rutilus rutilus (L. 1758). Conserv. Genet. 2016, 17, 861–874. [Google Scholar] [CrossRef]
- Beacham, T.D.; Withler, R.E. Population structure of sea-type and lake-type sockeye salmon and kokanee in the Fraser River and Columbia River drainages. PLoS ONE 2017, 12, e0183713. [Google Scholar] [CrossRef]
- Waldman, J.R.; King, T.; Savoy, T.; Lorraine, M. Stock origins of subadult and adult atlantic sturgeon, acipenser oxyrinchus, in a non-natal estuary, long island sound. Estuaries Coasts 2013, 36, 257–267. [Google Scholar] [CrossRef]
- Florentina, O.D.; Daniela, H.; Radu, S. Management applications of genetic structure of anadromous sturgeon populations in the lower danube river (ldr), romania. scientific annals of the danube delta institute. Sci. Ann. Danub. Delta Inst. 2013, 19, 129–138. [Google Scholar]
- Chiesa, S.; Piccinini, A.; Lucentini, L.; Filonzi, L.; Marzano, F.N. Genetic data on endangered twaite shad (clupeidae) assessed in landlocked and anadromous populations: One or more species? Rev. Fish Biol. Fish. 2014, 24, 659–670. [Google Scholar] [CrossRef]
- Wahab, M.A.; Rahman, M.M.; Mariom; Wong, L. Morpho-genetic divergence and adaptation of anadromous hilsa shad (Tenualosa ilisha) along their heterogenic migratory habitats. Front. Mar. Sci. 2020, 7, 554. [Google Scholar]
- Waples, R.S. Separating the wheat from the Chaff: Patterns of genetic differentiation in high gene flow species. J. Hered. 1998, 89, 438–450. [Google Scholar] [CrossRef]
- Hauser, L.; Carvalho, G.R. Paradigm shifts in marine fisheries genetics: Ugly hypotheses slain by beautiful facts. Fish Fish. 2008, 9, 333–362. [Google Scholar] [CrossRef]
- Schlotfeldt, B.E.; Kleindorfer, S. Adaptive divergence in the Superb Fairy-wren (Malurus cyaneus): A mainland versus island comparison of morphology and foraging behaviour. Emu-Austral Ornithol. 2006, 106, 309–319. [Google Scholar] [CrossRef][Green Version]
- Hsieh, Y.C.; Chung, J.D.; Wang, C.N.; Chang, C.T.; Chen, C.Y.; Hwang, S.Y. Historical connectivity, contemporary isolation and local adaptation in a widespread but discontinuously distributed species endemic to Taiwan, Rhododendron oldhamii (Ericaceae). Heredity 2013, 111, 147–156. [Google Scholar] [CrossRef][Green Version]
- Wang, I.J.; Summers, K. Genetic structure is correlated with phenotypic divergence rather than geographic isolation in the highly polymorphic strawberry poison-dart frog. Mol. Ecol. 2010, 19, 447–458. [Google Scholar] [CrossRef]
- Beheregaray, L.B.; Sunnucks, P. Fine-scale genetic structure, estuarine colonization and incipient speciation in the marine silverside fish Odontesthes argentinensis. Mol. Ecol. 2001, 10, 2849–2866. [Google Scholar] [CrossRef]
- Jiang, T.; Zheng, C.; Huang, H.; Xuan, Z.; Qiu, C.; Liu, H.; Yang, J. Landmark-based morphometric comparison of otolith for Coilia mystus and C. grayii from Pearl River estuary and Jiulong River estuary. South China Fish. Sci. 2018, 14, 10–16. [Google Scholar]
- Einfeldt, A.L.; Jesson, L.K.; Addison, J.A. Historical human activities reshape evolutionary trajectories across both native and introduced ranges. Ecol. Evol. 2020, 10, 6579–6592. [Google Scholar] [CrossRef]
- Zhang, H.X.; Wang, Q.; Jia, S.W. Genomic Phylogeography of Gymnocarpos przewalskii (Caryophyllaceae): Insights into Habitat Fragmentation in Arid Northwestern China. Divers.-Basel 2020, 12, 335. [Google Scholar] [CrossRef]
- Yang, Q.L.; Han, Z.Q.; Sun, D.R.; Xie, S.G.; Lin, L.S.; Gao, T.X. Genetics and phylogeny of genus Coilia in China based on AFLP markers. Chin. J. Oceanol. Limnol. 2010, 28, 795–801. [Google Scholar] [CrossRef]
- Jiang, T.; Liu, H.; Huang, H.; Yang, J. Otolith microchemistry of coilia grayii from the pearl river estuary, china. Acta Hydrobiol. Sin. 2015, 39, 816–821. [Google Scholar]
- Lucas, M.C.; Frear, P.A. Effects of a flow-gauging weir on the migratory behaviour of adult barbel, a riverine cyprinid. J. Fish Biol. 1997, 50, 382–396. [Google Scholar] [CrossRef]
- Geeraerts, C.; Ovidio, M.; Verbiest, H.; Buysse, D.; Coeck, J.; Belpaire, C.; Philippart, J.C. Mobility of individual roach Rutilus rutilus (L.) in three weir-fragmented Belgian rivers. Hydrobiologia 2007, 582, 143–153. [Google Scholar] [CrossRef]
- Esguicero, A.L.H.; Arcifa, M. Fragmentation of a Neotropical migratory fish population by a century-old dam. Hydrobiologia 2010, 638, 41–53. [Google Scholar] [CrossRef]
- Yamamoto, S.; Morita, K.; Koizumi, I.; Maekawa, K. Genetic differentiation of white-spotted charr (Salvelinus leucomaenis) populations after habitat fragmentation: Spatial-temporal changes in gene frequencies. Conserv. Genet. 2004, 5, 529–538. [Google Scholar] [CrossRef]
- Hollatz, C.; Vilaca, S.T.; Redondo, R.A.F.; Marmontel, M.; Baker, C.S.; Santos, F.R. The Amazon River system as an ecological barrier driving genetic differentiation of the pink dolphin (Inia geoffrensis). Biol. J. Linn. Soc. 2011, 102, 812–827. [Google Scholar] [CrossRef]
- Catarino, L.; Martins, E.S.; Diniz, M.A. Vegetation structure and ecology of the Cufada Lagoon (Guinea-Bissau). Afr. J. Ecol. 2002, 40, 252–259. [Google Scholar] [CrossRef]
- Irwin, D.E.; Irwin, J.H.; Smith, T.B. Genetic variation and seasonal migratory connectivity in Wilson’s warblers (Wilsonia pusilla): Species-level differences in nuclear DNA between western and eastern populations. Mol. Ecol. 2011, 20, 3102–3115. [Google Scholar] [CrossRef]
- Jacobson, B.; Peres-Neto, P.R. Quantifying and disentangling dispersal in metacommunities: How close have we come? How far is there to go? Landsc. Ecol. 2010, 25, 495–507. [Google Scholar] [CrossRef]
- Christie, M.R.; Meirmans, P.G.; Gaggiotti, O.E.; Toonen, R.J.; White, C. Disentangling the relative merits and disadvantages of parentage analysis and assignment tests for inferring population connectivity. Ices J. Mar. Sci. 2017, 74, 1749–1762. [Google Scholar] [CrossRef]
- Hedrick, P.W. Perspective: Highly variable loci and their interpretation in evolution and conservation. Evolution 1999, 53, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Estoup, A.; Jarne, P.; Cornuet, J.M. Homoplasy and mutation model at microsatellite loci and their consequences for population genetics analysis. Mol. Ecol. 2002, 11, 1591–1604. [Google Scholar] [CrossRef] [PubMed]
- Bohonak, A.J. Dispersal, gene flow, and population structure. Q. Rev. Biol. 1999, 74, 21–45. [Google Scholar] [CrossRef] [PubMed]
- Ramos, R.; Song, G.; Navarro, J.; Zhang, R.Y.; Symes, C.T.; Forero, M.G.; Lei, F.M. Population genetic structure and long-distance dispersal of a recently expanding migratory bird. Mol. Phylogenet. Evol. 2016, 99, 194–203. [Google Scholar] [CrossRef]
- Yang, J.; Arai, T.; Liu, H.; Miyazaki, N.; Tsukamoto, K. Reconstructing habitat use of Coilia mystus and Coilia ectenes of the Yangtze River Estuary, and of Coilia ectenes of Taihu Lake, based on otolith strontium and calcium. J. Fish. Biol. 2006, 69, 1120–1135. [Google Scholar] [CrossRef]
- Yang, J.; Jiang, T.; Liu, H. Are there habitat salinity markers of the Sr:Ca ratio in the otolith of wild diadromous fishes? A literature survey. Ichthyol. Res. 2011, 58, 291–294. [Google Scholar] [CrossRef]
- Xiao, W.H.; Zhang, Y.P.; Liu, H.Z. Molecular systematics of Xenocyprinae (Teleostei: Cyprinidae): Taxonomy, biogeography, and coevolution of a special group restricted in east Asia. Mol. Phylogenet. Evol. 2001, 18, 163–173. [Google Scholar] [CrossRef]
- Liu, K.; Duan, J.-r.; Xu, D.-p.; Zhang, M.-y.; Fang, D.-a.; Shi, W.-g. Present situation of Coilia nasus population features and yield in Yangtze River estuary waters in fishing season. Shengtaixue Zazhi 2012, 31, 3138–3143. [Google Scholar]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef] [PubMed]
- Talavera, G.; Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef] [PubMed]
- Nylander, J.A.A.; Ronquist, F.; Huelsenbeck, J.P.; Nieves-Aldrey, J.L. Bayesian phylogenetic analysis of combined data. Syst. Biol. 2004, 53, 47–67. [Google Scholar] [CrossRef] [PubMed]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Bandelt, H.J.; Alves-Silva, J.; Guimaraes, P.E.M.; Santos, M.S.; Brehm, A.; Pereira, L.; Coppa, A.; Larruga, J.M.; Rengo, C.; Scozzari, R.; et al. Phylogeography of the human mitochondrial haplogroup L3e: A snapshot of African prehistory and Atlantic slave trade. Ann. Hum. Genet. 2001, 65, 549–563. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Hohna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Sokal, R.R. Genetic, geographic, and linguistic distances in Europe. Proc. Natl. Acad. Sci. USA 1988, 85, 1722–1726. [Google Scholar] [CrossRef] [PubMed]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Fu, Y.-X. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 1997, 147, 915–925. [Google Scholar] [CrossRef]
- Rogers, A.R.; Harpending, H. Population growth makes waves in the distribution of pairwise genetic differences. Mol. Biol. Evol. 1992, 9, 552–569. [Google Scholar]
- Bouckaert, R.; Heled, J.; Kuhnert, D.; Vaughan, T.; Wu, C.H.; Xie, D.; Suchard, M.A.; Rambaut, A.; Drummond, A.J. BEAST 2: A Software Platform for Bayesian Evolutionary Analysis. PLoS Comput. Biol. 2014, 10, e1003537. [Google Scholar] [CrossRef] [PubMed]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian Phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef]
- Heled, J.; Drummond, A.J. Bayesian inference of population size history from multiple loci. BMC Evol. Biol. 2008, 8, 289. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Rosenberg, N.A.; Donnelly, P. Association mapping in structured populations. Am. J. Hum. Genet. 2000, 67, 170–181. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.A.; Vonholdt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Jakobsson, M.; Rosenberg, N.A. CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 2007, 23, 1801–1806. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, N.A. DISTRUCT: A program for the graphical display of population structure. Mol. Ecol. Notes 2004, 4, 137–138. [Google Scholar] [CrossRef]
- Piry, S.; Luikart, G.; Cornuet, J.M. BOTTLENECK: A computer program for detecting recent reductions in the effective population size using allele frequency data. J. Hered. 1999, 90, 502–503. [Google Scholar] [CrossRef]
- Gahagan, B.I.; Vokoun, J.C.; Whitledge, G.W.; Schultz, E.T. Evaluation of Otolith Microchemistry for Identifying Natal Origin of Anadromous River Herring in Connecticut. Mar. Coast. Fish. 2012, 4, 358–372. [Google Scholar] [CrossRef]
- Thomas, O.R.B.; Thomas, K.V.; Jenkins, G.P.; Swearer, S.E. Spatio-temporal resolution of spawning and larval nursery habitats using otolith microchemistry is element dependent. Mar. Ecol. Prog. Ser. 2020, 636, 169–187. [Google Scholar] [CrossRef]
- Jiang, T.; Zhou, X.; Liu, H.; Liu, H.; Yang, J. Two microchemistry patterns in otoliths of Coilia nasus from Poyang Lake, China. J. Fish. China 2013, 37, 239–244. [Google Scholar] [CrossRef]
- Nachon, D.J.; Bareille, G.; Drouineau, H.; Tabouret, H.; Taverny, C.; Boisneau, C.; Berail, S.; Pecheyran, C.; Claverie, F.; Daverat, F. 1980s population-specific compositions of two related anadromous shad species during the oceanic phase determined by microchemistry of archived otoliths. Can. J. Fish. Aquat. Sci. 2020, 77, 164–176. [Google Scholar] [CrossRef]
- Chittaro, P.M.; Usseglio, P.; Sale, P. Variation in fish density, assemblage composition and relative rates of predation among mangrove, seagrass and coral reef habitats. Environ. Biol. Fishes 2005, 72, 175–187. [Google Scholar] [CrossRef]
- Ruttenberg, B.I.; Hamilton, S.L.; Warner, R.R. Spatial and temporal variation in the natal otolith chemistry of a Hawaiian reef fish: Prospects for measuring population connectivity. Can. J. Fish. Aquat. Sci. 2008, 65, 1181–1192. [Google Scholar] [CrossRef]
- Elsdon, T.S.; Gillanders, B.M. Reconstructing migratory patterns of fish based on environmental influences on otolith chemistry. Rev. Fish Biol. Fish. 2003, 13, 219–235. [Google Scholar] [CrossRef]
- Hsu, C.C.; Tzeng, W.N. Validation of Annular Deposition in Scales and Otoliths of Flathead Mullet Mugil cephalus. Zool. Stud. 2009, 48, 640–648. [Google Scholar]
- Ewens, W.J. Concepts of substitutional load in finite populations. Theor. Popul. Biol. 1972, 3, 153–161. [Google Scholar] [CrossRef]
- Bucklin, A.; Wiebe, P.H. Low mitochondrial diversity and small effective population sizes of the copepods Calanus finmarchicus and Nannocalanus minor: Possible impact of climatic variation during recent glaciation. J. Hered. 1998, 89, 383–392. [Google Scholar] [CrossRef]
- Bowen, B.W.; Grant, W.S. Phylogeography of the sardines (Sardinops spp.): Assessing biogeographic models and population histories in temperature upwelling zones. Evolution 1997, 51, 1601–1610. [Google Scholar] [CrossRef] [PubMed]
- Duan, L.J.; Li, S.Y.; Liu, Y.; Jiang, T.; Failler, P. A trophic model of the Pearl River Delta coastal ecosystem. Ocean Coast. Manag. 2009, 52, 359–367. [Google Scholar] [CrossRef]
- Dudgeon, C.L.; Gust, N.; Blair, D. No apparent genetic basis to demographic differences in scarid fishes across continental shelf of the Great Barrier Reef. Mar. Biol. 2000, 137, 1059–1066. [Google Scholar] [CrossRef]
- Pedrosa-Gerasmio, I.R.; Agmata, A.B.; Santos, M.D. Genetic diversity, population genetic structure, and demographic history of Auxis thazard (Perciformes), Selar crumenophthalmus (Perciformes), Rastrelliger kanagurta (Perciformes) and Sardinella lemuru (Clupeiformes) in Sulu-Celebes Sea inferred by mitochondrial DNA sequences. Fish. Res. 2015, 162, 64–74. [Google Scholar] [CrossRef]
- Grant, W.S.; Clark, A.M.; Bowen, B.W. Why restriction fragment length polymorphism analysis of mitochondrial DNA failed to resolve sardine (Sardinops) biogeography: Insights from mitochondrial DNA ochrome b sequences. Can. J. Fish. Aquat. Sci. 1998, 55, 2539–2547. [Google Scholar] [CrossRef]
- Avise, J.C.; Zink, R.M. Molecular Genetic Divergence between Avian Sibling Species: King and Clapper Rails, Long-billed and Short-billed Dowitchers, Boat-tailed and Great-tailed Grackles, and Tufted and Black-crested Titmice. Auk 2008, 105, 516–528. [Google Scholar] [CrossRef]
- Nei, M. Molecular Evolutionary Genetics; Columbia University Press: New York, NY, USA, 1987. [Google Scholar]
- Sharma, R.; Kumar, B.; Arora, R.; Ahlawat, S.; Mishra, A.K.; Tantia, M.S. Genetic diversity estimates point to immediate efforts for conserving the endangered Tibetan sheep of India. Meta Gene 2016, 8, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Scribner, K.T.; Soiseth, C.; McGuire, J.; Sage, G.K.; Thorsteinson, L.; Nielsen, J.L.; Knudsen, E. Genetic assessment of the effects of streamscape succession on coho salmon Oncorhynchus kisutch colonization in recently deglaciated streams. J. Fish Biol. 2017, 91, 195–218. [Google Scholar] [CrossRef]
- Saastamoinen, M.; Bocedi, G.; Cote, J.; Legrand, D.; Guillaume, F.; Wheat, C.W.; Fronhofer, E.A.; Garcia, C.; Henry, R.; Husby, A.; et al. Genetics of dispersal. Biol. Rev. 2018, 93, 574–599. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.L.; Tang, S.Q.; Cai, F.J.; Lin, Y.F.; Wu, Z.J. Microsatellite evidence of dispersal mechanism of red swamp crayfish (Procambarus clarkii) in the Pearl River basin and implications for its management. Sci. Rep. 2017, 7, 8272. [Google Scholar] [CrossRef]
- Camak, D.T.; Piller, K.R. Going with the Flow: Testing the Role of Habitat Isolation among Three Ecologically Divergent Darter Species. Copeia 2018, 106, 375–387. [Google Scholar] [CrossRef]
- Neraas, L.P.; Spruell, P. Fragmentation of riverine systems: The genetic effects of dams on bull trout (Salvelinus confluentus) in the Clark Fork River system. Mol. Ecol. 2001, 10, 1153–1164. [Google Scholar] [CrossRef]
- Thorstad, E.B.; Okland, F.; Aarestrup, K.; Heggberget, T.G. Factors affecting the within-river spawning migration of Atlantic salmon, with emphasis on human impacts. Rev. Fish Biol. Fish. 2008, 18, 345–371. [Google Scholar] [CrossRef]
- Neville, H.M.; Dunham, J.B.; Peacock, M.M. Landscape attributes and life history variability shape genetic structure of trout populations in a stream network. Landsc. Ecol. 2006, 21, 901–916. [Google Scholar] [CrossRef]
- Torterotot, J.B.; Perrier, C.; Bergeron, N.E.; Bernatchez, L. Influence of Forest Road Culverts and Waterfalls on the Fine-Scale Distribution of Brook Trout Genetic Diversity in a Boreal Watershed. Trans. Am. Fish. Soc. 2014, 143, 1577–1591. [Google Scholar] [CrossRef]
- Nason, J.D.; Aldrich, P.R.; Hamrick, J.L. Dispersal and the dynamics of genetic structure in fragmented tropical tree populations. Trop. For. Remn. Ecol. Manag. Conserv. Fragm. Communities 1997, 304–320. [Google Scholar]
- Meirmans, P.G. The trouble with isolation by distance. Mol. Ecol. 2012, 21, 2839–2846. [Google Scholar] [CrossRef]
- Lowe, W.H. Factors affecting stage-specific distribution in the stream salamander Gyrinophilus porphyriticus. Herpetologica 2005, 61, 135–144. [Google Scholar] [CrossRef]
- Lewallen, E.A.; Bohonak, A.J.; Bonin, C.A.; van Wijnen, A.J.; Pitmans, R.L.; Lovejoy, N.R. Population Genetic Structure of the Tropical Two-Wing Flyingfish (Exocoetus volitans). PLoS ONE 2016, 11, e0163198. [Google Scholar] [CrossRef]
- Whitfield, J.A. Studies of the Diversity and Evolution of Soldier Aphids; University of Cambridge: Cambridge, UK, 1998. [Google Scholar]
- Lu, K.X. Fishes and Fishery Resources of Pearl River System; Guangdong Science of Technology Press: Guangzhou, China, 1990; (In Chinese with English Abstract). [Google Scholar]
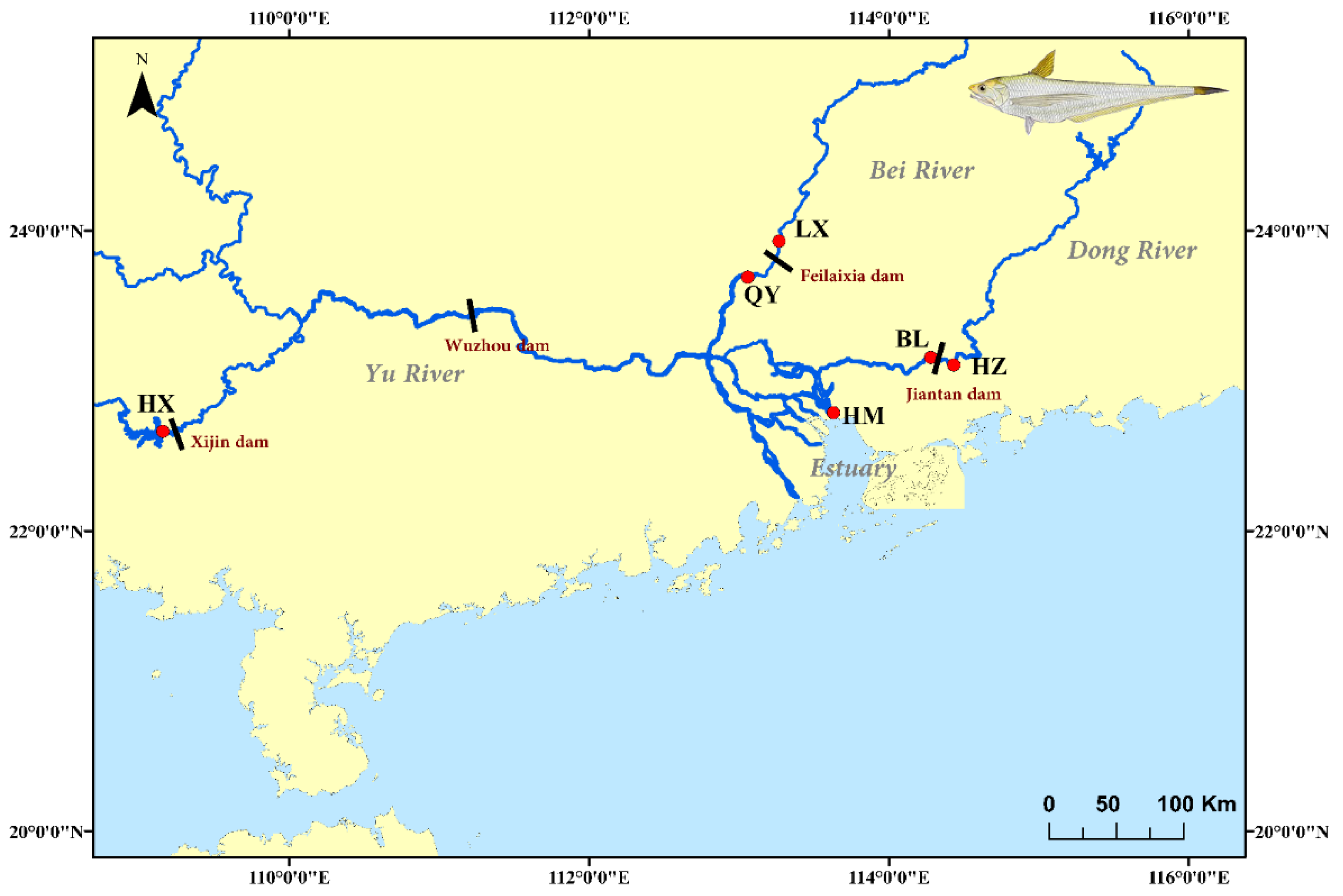

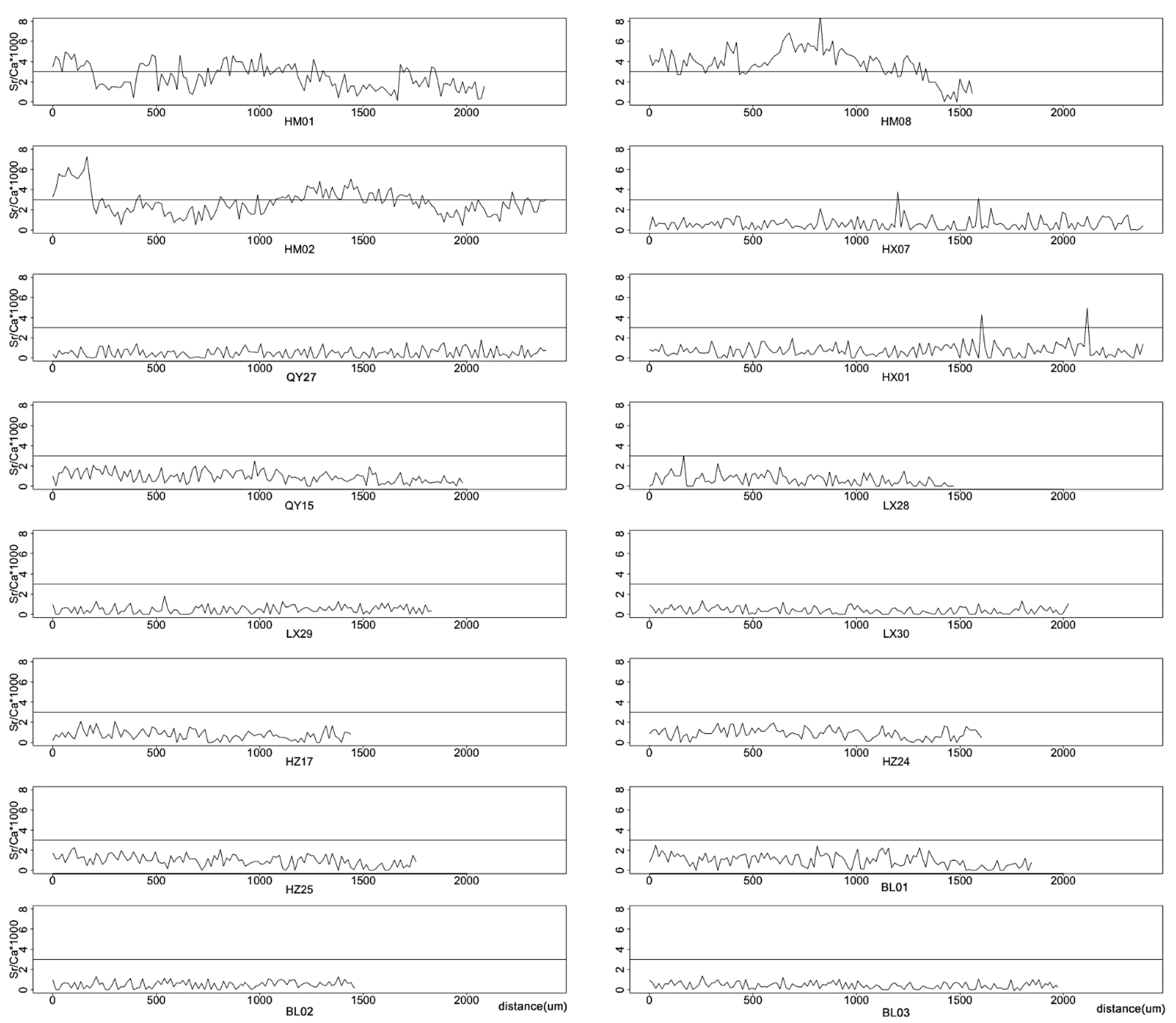
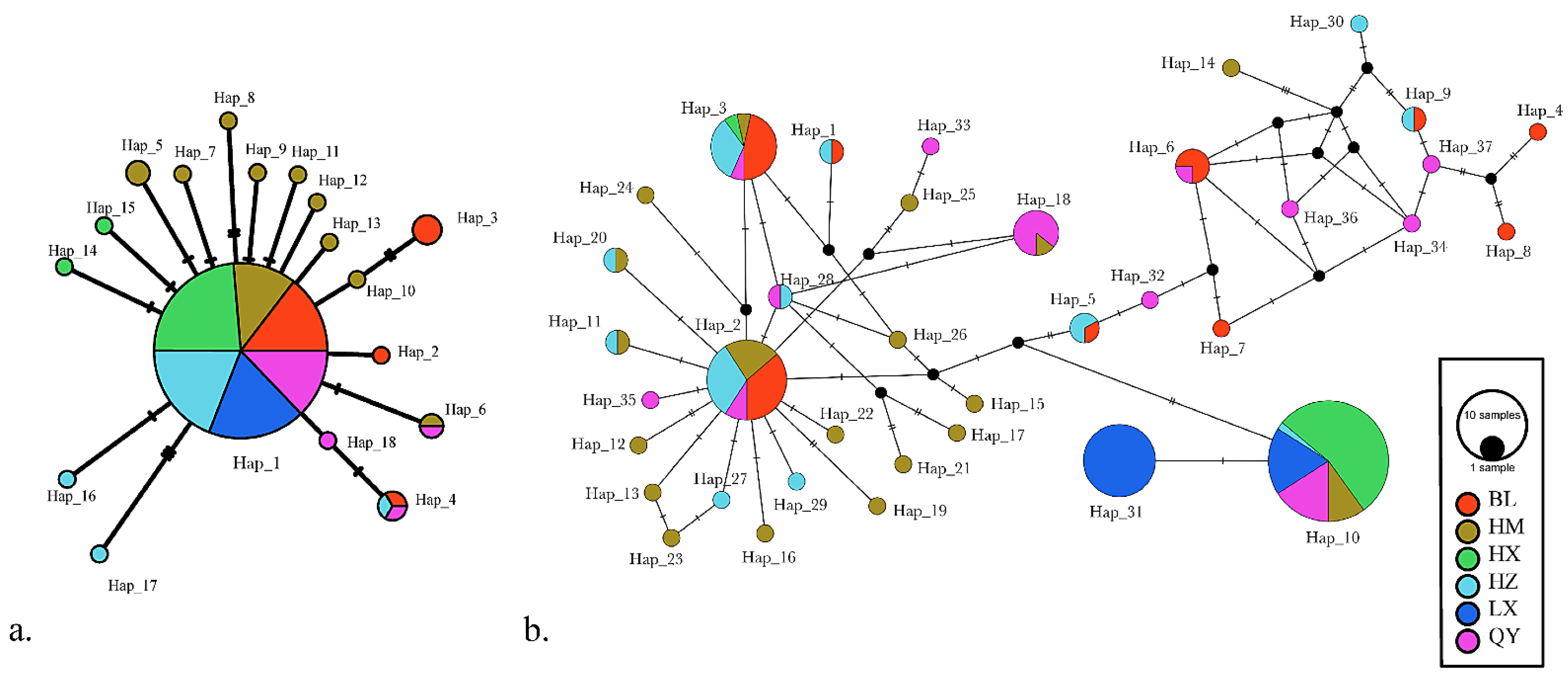
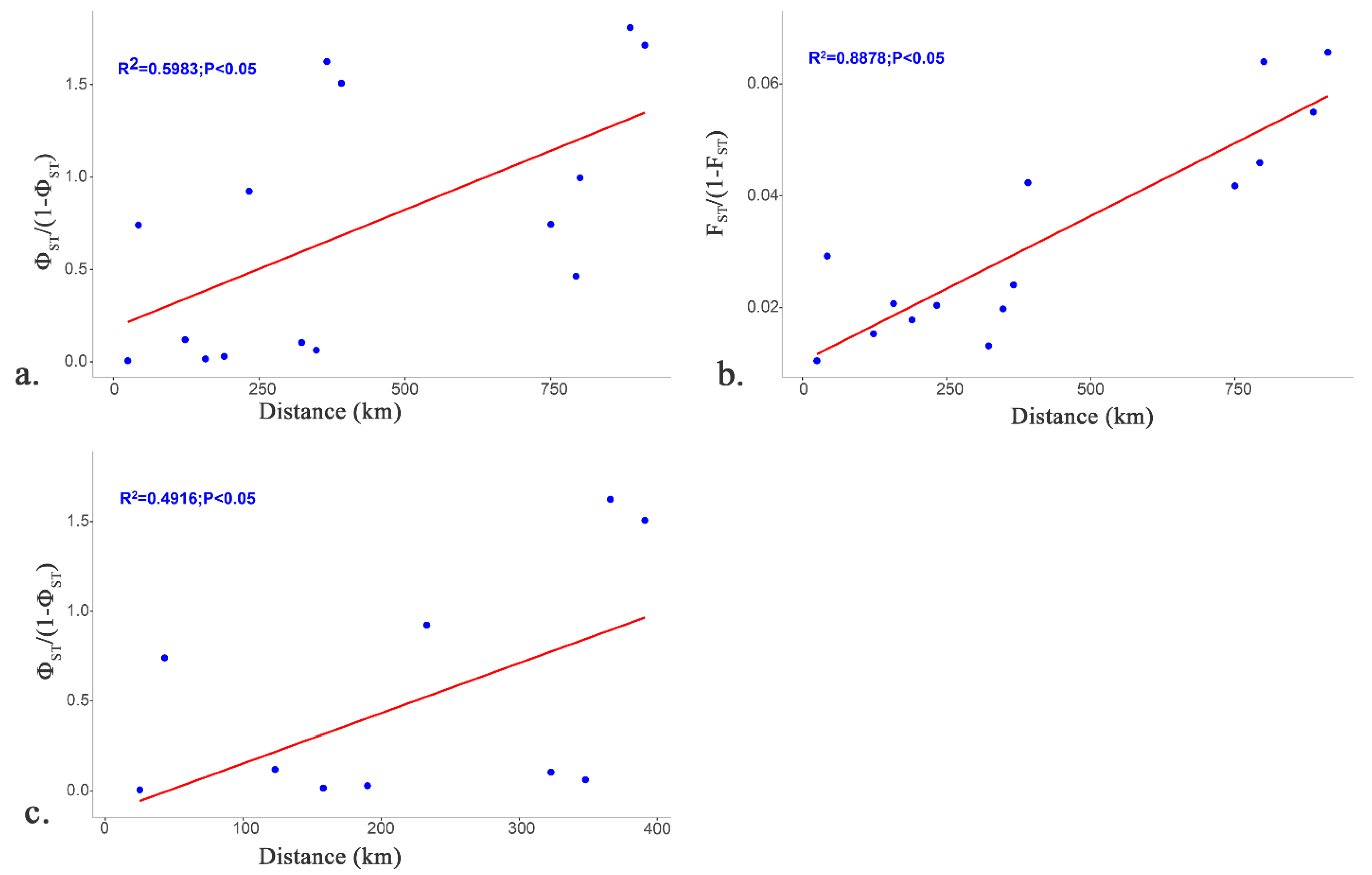

| Site | Sample | Number | Longitude | Latitude | Location | Region | Dam | Building Time |
|---|---|---|---|---|---|---|---|---|
| HM | November 2015 | 30 | 22.80° N | 113.67° E | estuary | Estuary | Xijin, Wuzhou | 1958 |
| HX | Semptember 2015 | 30 | 22.07° N | 109.25° E | upstream | Yu River | 1993 | |
| LX | May 2015 | 30 | 23.92° N | 113.27° E | upstream | Bei River | Feilaixia | 1994 |
| QY | March 2016 | 30 | 23.68° N | 113.05° E | dowmstream | Bei River | ||
| HZ | December 2015 | 30 | 23.10° N | 114.40° E | upstream | Dong River | Jiantan | 2004 |
| BL | December 2015 | 30 | 23.17° N | 114.28° E | downstream | Dong River |
| Population | N | Na | Ne | Ho | He | PIC |
|---|---|---|---|---|---|---|
| HM | 30 | 10 | 5.3453 | 0.755 | 0.8081 | 0.7546 |
| HX | 30 | 6.2 | 3.4408 | 0.6416 | 0.6955 | 0.6379 |
| LX | 28 | 7.6 | 4.3612 | 0.5742 | 0.7508 | 0.7059 |
| QY | 26 | 9.8 | 4.5307 | 0.752 | 0.7263 | 0.6785 |
| BL | 30 | 10.6 | 5.1144 | 0.738 | 0.8046 | 0.7674 |
| HZ | 30 | 9.8 | 5.2632 | 0.74 | 0.8126 | 0.7718 |
| Source of D-Loop | d.f. | Sum of Squares | Mean of Squares | Est. Var. | Variation (%) |
| Among Pops | 5 | 114.805 | 22.961 | 0.835 | 37% |
| Within Pops | 149 | 212.447 | 1.426 | 1.426 | 63% |
| Total | 154 | 327.252 | 2.261 | 100% | |
| Fixation Indices | ΦST = 0.369, p = 0.001 ** | ||||
| Source of cyt-b | d.f. | Sum of Squares | Mean of Squares | Est. Var. | Variation (%) |
| Among Pops | 5 | 2.255 | 0.451 | 0.008 | 3% |
| Within Pops | 126 | 35.033 | 0.278 | 0.278 | 97% |
| Total | 131 | 37.288 | 0.286 | 100% | |
| Fixation Indices | ΦST = 0.028, p = 0.036 * | ||||
| Source of Microsates | d.f. | Sum of Squares | Mean of Squares | Est. Var. | Variation (%) |
| Among tributaries | 3 | 12.869 | 1.46 | 0.02078 | 1.46% |
| Among populations | 2 | 5.02 | 1.39 | 0.01977 | 1.34% |
| Within populations | 342 | 472.85 | 97.15 | 1.3826 | 97.20% |
| Total | 347 | 490.739 | 1.42315 | ||
| Fixation Indices | FST = 0.02849, p = 0.016 * | ||||
| Sequence | Population | HM | HX | QY | LX | BL | HZ |
| D-loop | HM | 0 | |||||
| HX | 0.4988 ** | 0 | |||||
| QY | 0.0281 | 0.4264 ** | 0 | ||||
| LX | 0.4799 ** | 0.3164 ** | 0.4253 ** | 0 | |||
| BL | 0.1065 | 0.6439 ** | 0.0944 | 0.6189 ** | 0 | ||
| HZ | 0.0154 | 0.6314 ** | 0.0585 | 0.6011 ** | 0.0057 | 0 | |
| Sequence | Population | HM | HX | QY | LX | BL | HZ |
| cyt b | HM | 0 | |||||
| HX | 0.014 | 0 | |||||
| QY | 0.002 | 0 | 0 | ||||
| LX | 0.021 | 0.070 * | 0.073 * | 0 | |||
| BL | 0.032 | 0.079 * | 0.066 * | 0.027 | 0 | ||
| HZ | 0.006 | 0.04 | 0.000 | 0.000 | 0.037 | 0 |
| Population | HM | HX | LX | QY | HZ | BL |
|---|---|---|---|---|---|---|
| Sign test | ||||||
| IAM | 0.67321 | 0.31878 | 0.32888 | 0.67744 | 0.33788 | 0.68019 |
| TPM | 0.0899 | 0.33133 | 0.66868 | 0.33835 | 0.64685 | 0.09749 |
| SMM | 0.32886 | 0.09396 | 0.65422 | 0.09764 | 0.32851 | 0.10088 |
| Wilcoxon signed-rank test | ||||||
| IAM | 0.5000 | 0.03125 * | 0.04688 * | 0.6875 | 0.04688 * | 0.3125 |
| TPM | 0.95313 | 0.6875 | 0.5 | 0.89063 | 0.40625 | 0.95313 |
| SMM | 0.96875 | 0.95313 | 0.6875 | 0.96875 | 0.95313 | 0.98438 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, G.; Tang, Q.; Chen, Z.; Guo, D.; Zhou, L.; Lai, H.; Li, G. Otolith Microchemistry and Demographic History Provide New Insight into the Migratory Behavior and Heterogeneous Genetic Divergence of Coilia grayii in the Pearl River. Fishes 2022, 7, 23. https://doi.org/10.3390/fishes7010023
Wang G, Tang Q, Chen Z, Guo D, Zhou L, Lai H, Li G. Otolith Microchemistry and Demographic History Provide New Insight into the Migratory Behavior and Heterogeneous Genetic Divergence of Coilia grayii in the Pearl River. Fishes. 2022; 7(1):23. https://doi.org/10.3390/fishes7010023
Chicago/Turabian StyleWang, Gongpei, Qindong Tang, Zhi Chen, Dingli Guo, Lei Zhou, Han Lai, and Guifeng Li. 2022. "Otolith Microchemistry and Demographic History Provide New Insight into the Migratory Behavior and Heterogeneous Genetic Divergence of Coilia grayii in the Pearl River" Fishes 7, no. 1: 23. https://doi.org/10.3390/fishes7010023
APA StyleWang, G., Tang, Q., Chen, Z., Guo, D., Zhou, L., Lai, H., & Li, G. (2022). Otolith Microchemistry and Demographic History Provide New Insight into the Migratory Behavior and Heterogeneous Genetic Divergence of Coilia grayii in the Pearl River. Fishes, 7(1), 23. https://doi.org/10.3390/fishes7010023







