Population Genetic Assessment of Anadromous and Resident Striped Bass (Morone saxatilis) in the Roanoke River System, Eastern United States
Abstract
1. Introduction
2. Results
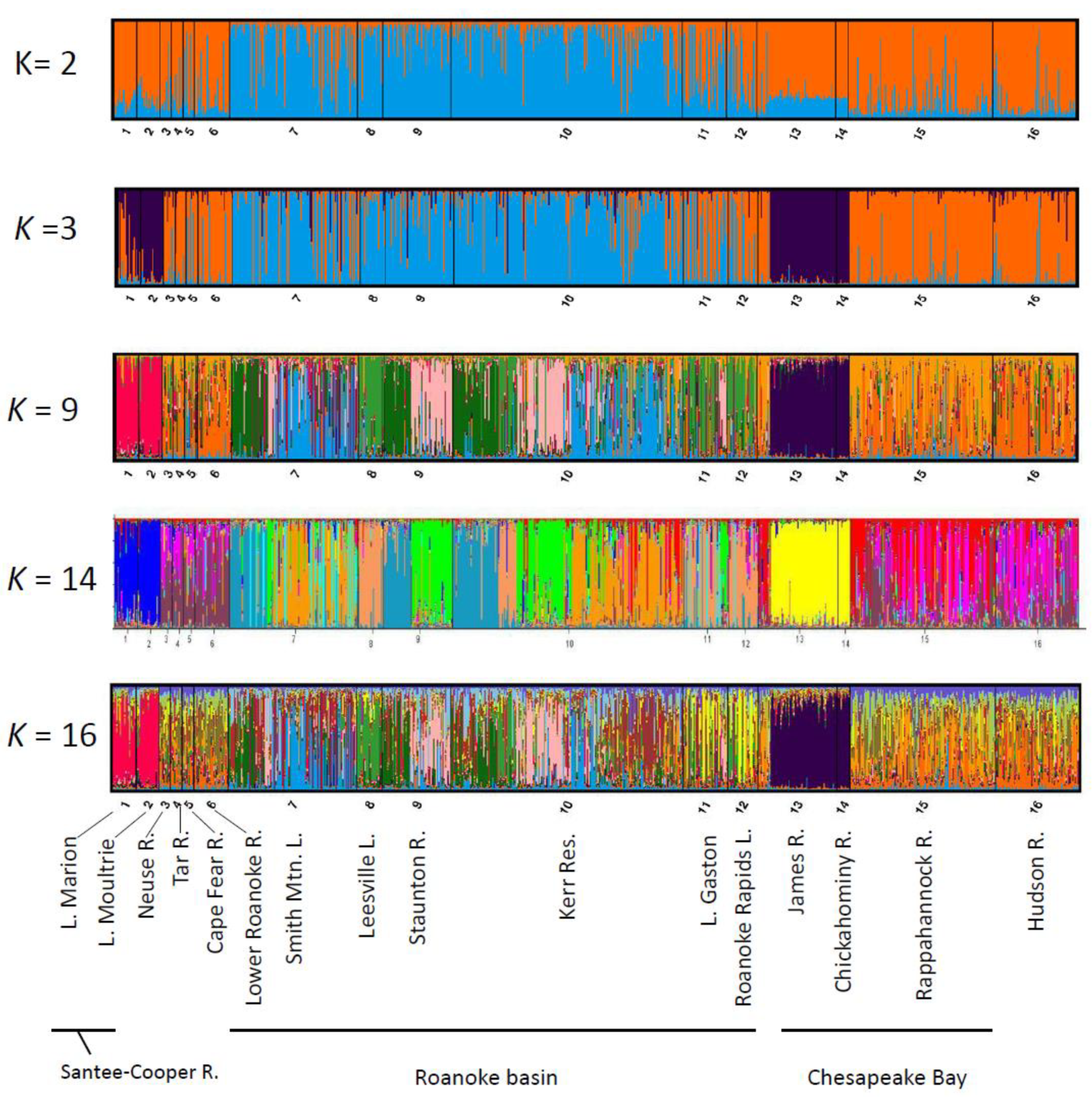
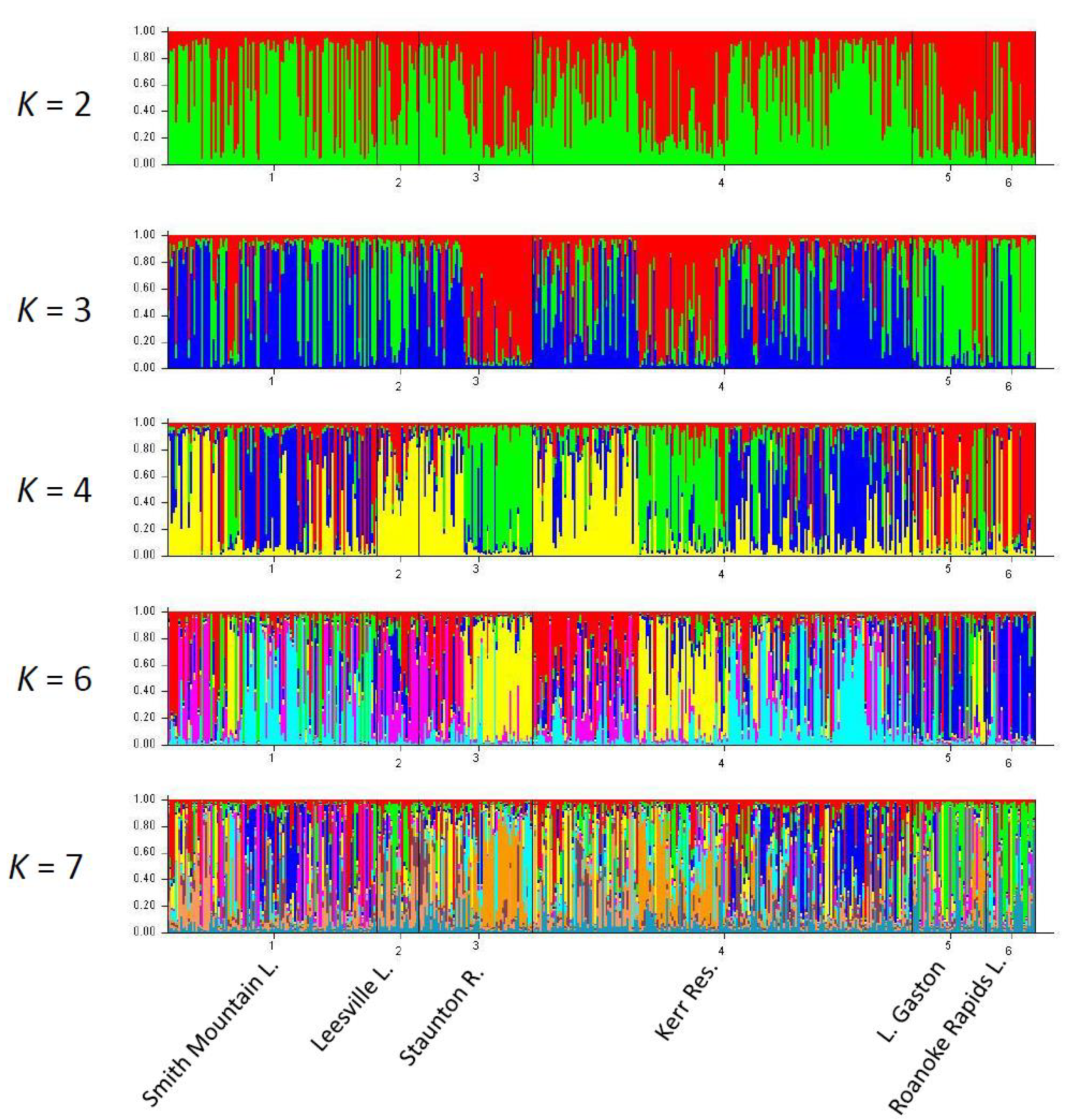
2.1. Assessment of U.S. Atlantic Coast Populations
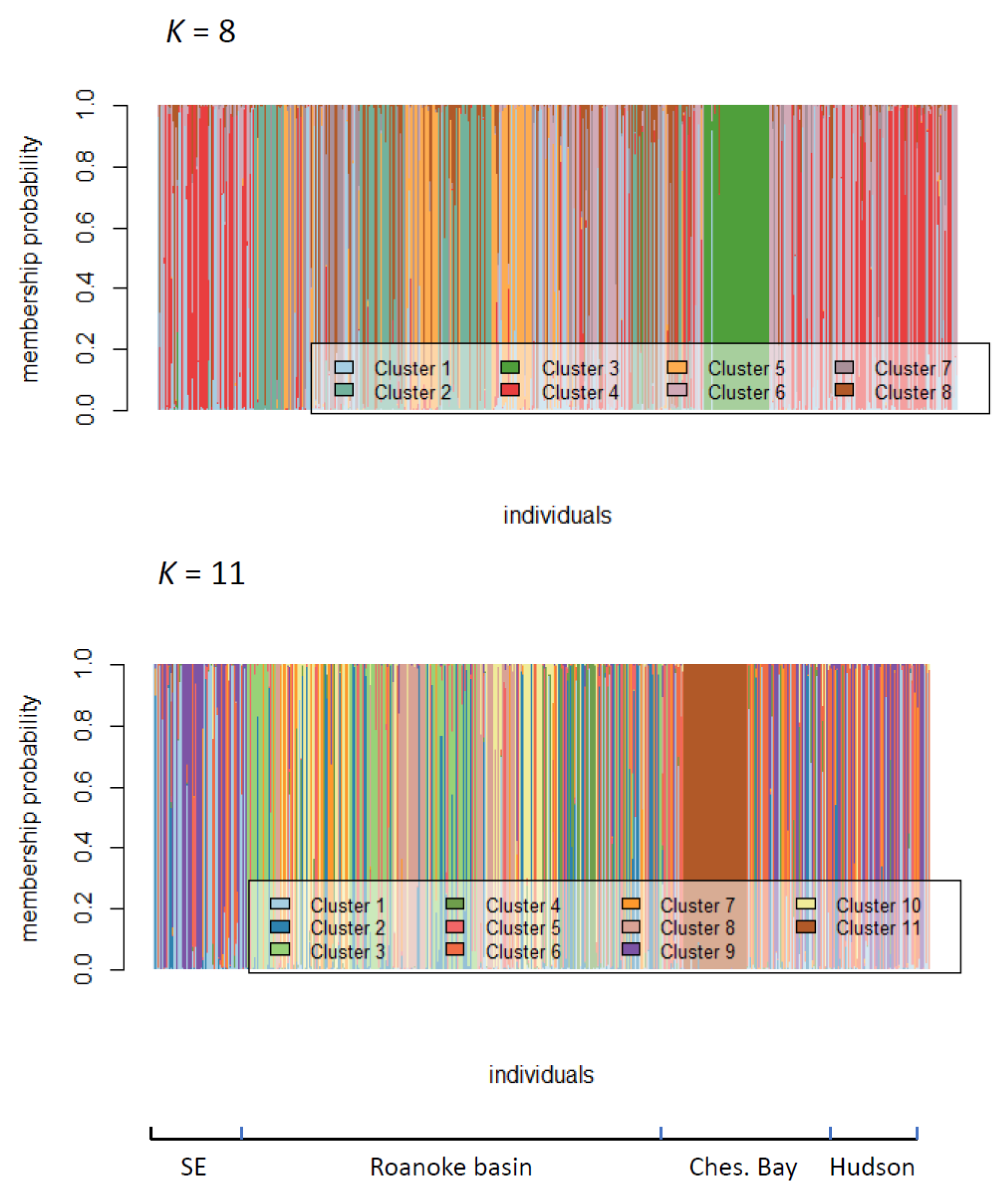
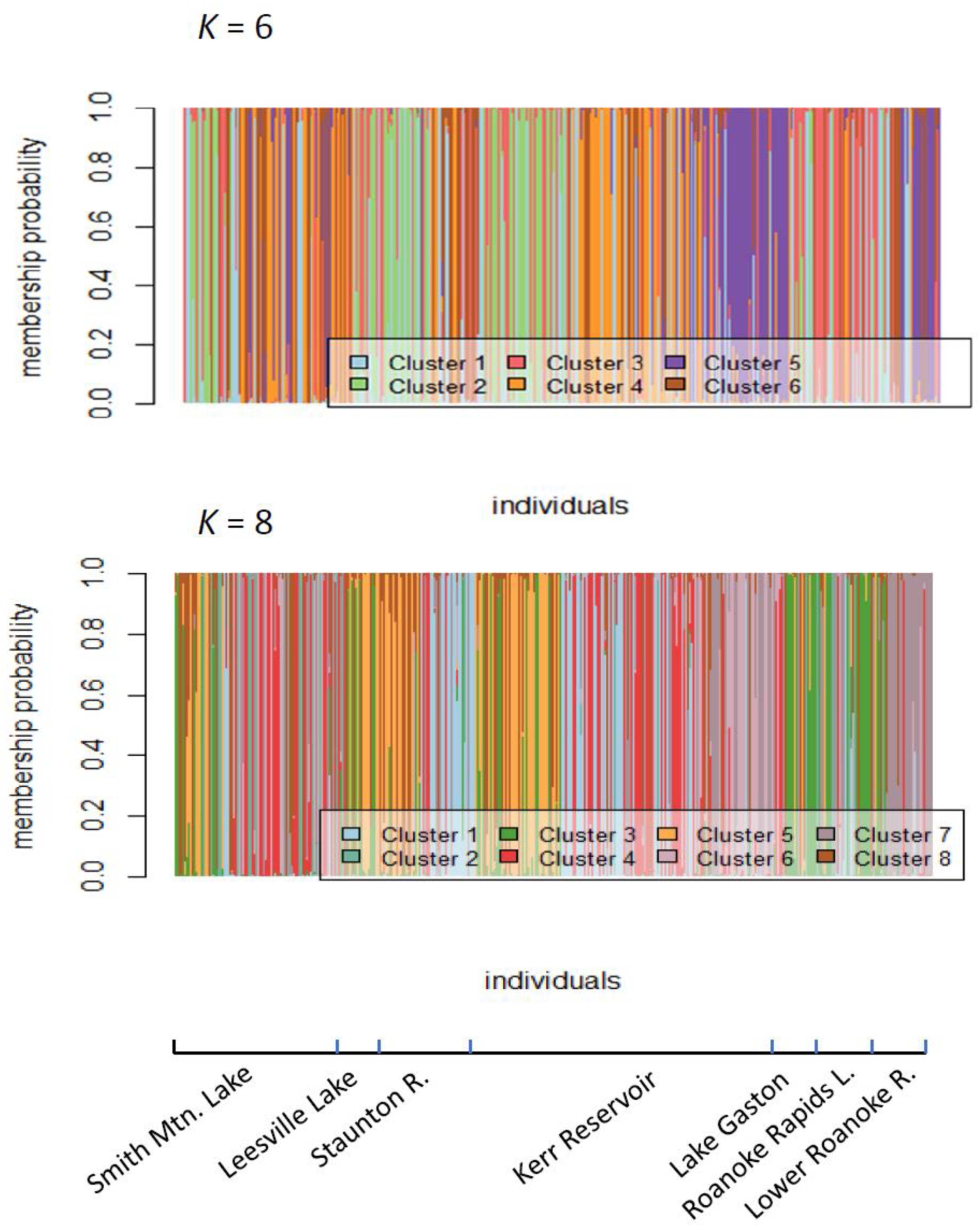
2.2. Assessment of Roanoke River Genetic Population Structure
3. Discussion
3.1. Range-Wide Genetic Differentiation of Striped Bass
3.2. Genetic Structure of Striped Bass Within the Roanoke River Basin
3.3. Recommendations for Genetically Cognizant Management of Roanoke River Striped Bass
4. Materials and Methods
4.1. Sampling and Study Area
4.2. Molecular Methods
4.3. Data Analysis
4.4. Permits and Ethical Aspects
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Whitehead, J.C. Economics of recreational fisheries for inland striped bass and hybrid striped bass. Am. Fish. Soc. Symp. 2013, 80, 551–558. [Google Scholar]
- Jenkins, R.E.; Burkhead, N.M. Freshwater Fishes of Virginia; American Fisheries Society: Bethesda, MD, USA, 1994. [Google Scholar]
- Raney, E.C.; Tresselt, E.F.; Hollins, E.H.; Vladykov, V.D.; Wallace, D.H. The striped bass (Roccus saxatilis). Bull. Bingham Oceanogr. Collect. Yale Univ. 1952, 14. [Google Scholar]
- Van Horn, S.L. A brief history of inland striped bass management. Am. Fish. Soc. Symp. 2013, 80, 1–13. [Google Scholar]
- Clark, J. Seasonal movements of Striped Bass contingents of Long Island Sound and the New York Bight. Trans. Am. Fish. Soc. 1968, 97, 320–343. [Google Scholar] [CrossRef]
- Secor, D. Specifying divergent migrations in the concept of stock: The contingent hypothesis. Fish. Res. 1999, 43, 13–34. [Google Scholar] [CrossRef]
- Secor, D.H.; Rooker, J.R.; Zlokovitz, E.; Zdanowicz, V.S. Identification of riverine, estuarine, and coastal contingents of Hudson River striped bass based upon otolith elemental fingerprints. Mar. Ecol. Prog. Ser. 2001, 211, 245–253. [Google Scholar] [CrossRef]
- Gahagan, B.; Fox, D.; Secor, D. Partial migration of striped bass: Revisiting the contingent hypothesis. Mar. Ecol. Prog. Ser. 2015, 525, 185–197. [Google Scholar] [CrossRef]
- Andrews, S.N.; Linnansaari, T.; Curry, R.A.; Dadswell, M.J. The misunderstood striped bass of the Saint John River, New Brunswick: Past, present, and future. N. Am. J. Fish. Manag. 2017, 37, 235–254. [Google Scholar] [CrossRef]
- Waldman, J.R.; Dunning, D.J.; Ross, Q.E.; Mattson, M.T. Range dynamics of Hudson River striped bass along the Atlantic Coast. Trans. Am. Fish. Soc. 1990, 119, 910–919. [Google Scholar] [CrossRef]
- Callihan, J.L.; Godwin, C.H.; Buckel, J.A. Effect of demography on spatial distribution: Movement patterns of the Albemarle Sound-Roanoke River stock of Striped Bass (Morone saxatilis) in relation to their recovery. Fish. Bull. 2014, 112, 131–143. [Google Scholar] [CrossRef]
- Conroy, C.; Piccoli, P.; Secor, D. Carryover effects of early growth and river flow on partial migration in striped bass Morone saxatilis. Mar. Ecol. Prog. Ser. 2015, 541, 179–194. [Google Scholar] [CrossRef]
- Secor, D.H.; Piccoli, P.M. Oceanic migration rates of upper Chesapeake Bay striped bass (Morone saxatilis), determined by otolith microchemical analysis. Fish. Bull. 2007, 105, 62–73. [Google Scholar]
- Richards, R.A.; Rago, P.J. A case history of effective fishery management: Chesapeake Bay Striped Bass. N. Am. J. Fish. Manag. 1999, 19, 356–375. [Google Scholar] [CrossRef]
- Waldman, J.R.; Grossfield, J.; Wirgin, I.I. Review of stock discrimination techniques for striped bass. N. Am. J. Fish. Manag. 1988, 8, 410–425. [Google Scholar] [CrossRef]
- Coutant, C.C. Striped bass, temperature, and dissolved oxygen: A speculative hypothesis for environmental risk. Trans. Am. Fish. Soc. 1985, 114, 31–61. [Google Scholar] [CrossRef]
- VDGIF (Virginia Department of Game and Inland Fisheries). Fishing. 2017. Available online: https://www.dgif.virginia.gov/fishing/ (accessed on 17 June 2017).
- Lamprecht, S.D.; Mauck, M.D.; DiCenzo, V.J. Natural reproduction: How it has affected striped bass management of John H. Kerr and Santee-Cooper reservoirs and Lake Texoma. Am. Fish. Soc. Symp. 2013, 80, 413–430. [Google Scholar]
- Doyle, R.W. An approach to the quantitative analysis of domestication selection in aquaculture. Aquaculture 1983, 33, 167–185. [Google Scholar] [CrossRef]
- Ryman, N.; Laikre, L. Effects of supportive breeding on the genetically effective population size. Cons. Biol. 1991, 5, 325–329. [Google Scholar] [CrossRef]
- Wirgin, I.; Maceda, L.; Tozer, M.; Stabile, J.; Waldman, J. Atlantic coastwide population structure of striped bass Morone saxatilis using microsatellite DNA analysis. Fish. Res. 2020, 226, 105506. [Google Scholar] [CrossRef]
- Callihan, J.L.; Harris, J.E.; Hightower, J.E. Coastal migration and homing of Roanoke River striped bass. Mar. Coast. Fish. 2015, 7, 301–315. [Google Scholar] [CrossRef]
- Bjorgo, K.A.; Isely, J.J.; Thomason, C.S. Seasonal movement and habitat use by striped bass in the Combahee River, South Carolina. Trans. Am. Fish. Soc. 2000, 129, 1281–1287. [Google Scholar] [CrossRef]
- Wirgin, I.; Grunwald, C.; Garte, S.J.; Mesing, C. The use of DNA fingerprinting in the identification and management of striped bass populations in the southeastern United States. Trans. Am. Fish. Soc. 1991, 120, 273–282. [Google Scholar] [CrossRef]
- Wirgin, I.I.; Maceda, L.; Waldman, J.R.; Crittenden, R.N. Use of mitochondrial DNA polymorphisms to estimate the relative contributions of the Hudson River and Chesapeake Bay striped bass stocks to the mixed fishery on the Atlantic coast. Trans. Am. Fish. Soc. 1993, 122, 669–684. [Google Scholar] [CrossRef]
- Wirgin, I.; Ong, T.-L.; Maceda, L.; Waldman, J.R.; Moore, D.; Courtenay, S. Mitochondrial DNA variation in striped bass (Morone saxatilis) from Canadian rivers. Can. J. Fish. Aquat. Sci. 1993, 50, 80–87. [Google Scholar] [CrossRef]
- Gauthier, D.T.; Audemard, C.A.; Carlsson, J.E.L.; Darden, T.L.; Denson, M.R.; Reece, K.S.; Carlsson, J. Genetic population structure of U.S. Atlantic coastal striped bass (Morone saxatilis). J. Hered. 2013, 104, 510–520. [Google Scholar] [CrossRef]
- Anderson, A.P.; Denson, M.R.; Darden, T.L. Genetic structure of striped bass in the southeastern United States and effects from stock enhancement. N. Am. J. Fish. Manag. 2014, 34, 653–667. [Google Scholar] [CrossRef]
- LeBlanc, N.M.; Gahagan, B.I.; Andrews, S.N.; Avery, T.S.; Puncher, G.N.; Reading, B.J.; Buhariwalla, C.F.; Curry, R.A.; Whiteley, A.R.; Pavey, S.A. Genomic population structure of striped bass (Morone saxatilis) from the Gulf of St. Lawrence to Cape Fear River. Evol. Appl. 2020, in press. [Google Scholar] [CrossRef]
- Rogier, C.G.; Ney, J.J.; Turner, B.J. Electrophoretic analysis of genetic variability in a landlocked striped bass population. Trans. Am. Fish. Soc. 1985, 114, 244–249. [Google Scholar] [CrossRef]
- Blankenship, H.; Leber, K. A responsible approach to marine stock enhancement. Uses and effects of cultured fishes in aquatic ecosystems. Am. Fish. Soc. Symp. 1995, 15, 167–175. [Google Scholar]
- Miller, L.M.; Kapuscinski, A.R. Genetic guidelines for hatchery supplementation programs. In Population Genetics: Principles and Applications for Fisheries Scientists; Hallerman, E.M., Ed.; American Fisheries Society: Bethesda, MD, USA, 2003; pp. 329–355. [Google Scholar]
- Liu, Z.J.; Cordes, J.F. DNA marker technologies and their applications in aquaculture genetics. Aquaculture 2004, 238, 1–37. [Google Scholar] [CrossRef]
- Garza, J.C.; Williamson, E.G. Detection of reduction in population size using data from microsatellite loci. Mol. Ecol. 2001, 10, 305–318. [Google Scholar] [CrossRef] [PubMed]
- Jombart, T.; Devillard, S.; Balloux, F. Discriminant analysis of principal components: A new method for the analysis of genetically structured populations. BMC Genet. 2010, 11, 94. [Google Scholar] [CrossRef] [PubMed]
- Couch, C.R.; Garber, A.F.; Rexroad, C.E., III; Abrams, J.M.; Stannard, J.A.; Westerman, M.E.; Sullivan, C.V. Isolation and characterization of 149 novel microsatellite DNA markers for striped bass, Morone saxatilis, and cross-species amplification in white bass, Morone chrysops, and their hybrid. Mol. Ecol. Notes 2006, 6, 667–669. [Google Scholar] [CrossRef]
- Rexroad, C., III; Vallejo, R.; Coulibaly, I.; Couch, C.; Garber, A.; Westerman, M.; Sullivan, C. Identification and characterization of microsatellites for striped bass from repeat-enriched libraries. Cons. Genet. 2006, 7, 971–982. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar]
- Gilbert, K.J.; Andrew, R.L.; Bock, D.G.; Franklin, M.T.; Kane, N.C.; Moore, J.S.; Moyers, B.T.; Renaut, S.; Rennison, D.J.; Veen, T.; et al. Recommendations for utilizing and reporting population genetic analyses: The reproducibility of genetic clustering using the program structure. Mol. Ecol. 2012, 21, 4925–4930. [Google Scholar] [CrossRef]
- Harris, S.C. Genetic Marker-Assisted Management of Virginia Sport Fishes. Master’s Thesis, Virginia Polytechnic Institute and State University, Blacksburg, VA, USA, 2019. [Google Scholar]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Holm, S. A simple sequential rejective multiple test procedure. Scand. J. Stat. 1979, 6, 65–70. [Google Scholar]
- Do, C.; Waples, R.S.; Peel, D.; Macbeth, G.M.; Tillett, B.J.; Ovenden, J.R. NeEstimator v2: Re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol. Ecol. Resour. 2014, 14, 209–214. [Google Scholar] [CrossRef]
- Wang, J. A new method for estimating effective population size for multilocus genotypes. Mol. Ecol. 2009, 18, 2148–2164. [Google Scholar] [CrossRef]
- Waples, R.S.; Do, C. Linkage disequilibrium estimates of contemporary Ne using highly variable genetic markers: A largely untapped resource for applied conservation and evolution. Evol. Appl. 2010, 3, 244–262. [Google Scholar] [CrossRef] [PubMed]
- Chybicki, I.J.; Burczyk, J. Simultaneous estimation of null alleles and inbreeding coefficients. J. Hered. 2009, 100, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Jones, O.R.; Wang, J. Colony: A program for parentage and sibship inference from multilocus genotype data. Mol. Ecol. Resour. 2010, 10, 551–555. [Google Scholar] [CrossRef] [PubMed]
- Kalinowski, S.T.; Taper, M.L. ML-Relate: A computer program for maximum likelihood estimation of relatedness and relationship. Mol. Ecol. Notes 2006, 6, 576–579. Available online: http://www.montana.edu/kalinowski/Software/MLRelate.htm (accessed on 15 March 2019). [CrossRef]
- Pielou, E.C. After the Ice Age: The Return of Life to Glaciated North America; University of Chicago Press: Chicago, IL, USA, 1991. [Google Scholar]
- Hewitt, G. The genetic legacy of the Quaternary ice ages. Nature 2000, 405, 907–913. [Google Scholar] [CrossRef]
- Soltis, D.D.; Morris, A.B.; McLachlan, J.S.; Manos, P.S.; Soltis, P.S. Comparative phylogeography of unglaciated North America. Mol. Ecol. 2006, 15, 4261–4293. [Google Scholar] [CrossRef]
- Curry, R.A. Late glacial impacts on dispersal and colonization of Atlantic Canada and Maine by freshwater fishes. Quat. Res. 2007, 67, 225–233. [Google Scholar] [CrossRef]
- Benestan, L.M.; Ferchaud, A.; Hohenlohe, P.A.; Garner, B.A.; Naylor, G.J.P.; Baums, I.B.; Schwartz, M.K.; Kelley, J.L.; Luikart, G. Conservation genomics of natural and managed populations: Building a conceptual and practical framework. Mol. Ecol. 2016, 25, 2967–2977. [Google Scholar] [CrossRef]
- Jombart, T.; Collins, C. A Tutorial for Discriminant Analysis or Principal Components (DAPC) Using Adegenet 2.0.0. 2015. Available online: https://adegenet.r-forge.r-project.org/files/tutorial-dapc.pdf (accessed on 26 June 2019).
- Waldman, J.R.; Fabrizio, M.C. Problems of stock definition in estimating relative contributions of Atlantic striped bass to the coastal fishery. Trans. Am. Fish. Soc. 1994, 123, 766–778. [Google Scholar] [CrossRef]
- Waldman, J.R.; Maceda, L.; Wirgin, I. Mixed-stock analysis of wintertime aggregations of Striped Bass along the Mid-Atlantic coast. J. Appl. Ichthyol. 2012, 28, 1–6. [Google Scholar] [CrossRef]
- Fountain, J.; Darden, T.; Jenkins, W.; Denson, M. Three multiplexed microsatellite panels for striped bass. Southeast. Nat. 2009, 8, 671–676. [Google Scholar] [CrossRef]
- Neal, W.E. Landlocked striped bass survey. In Dingell-Johnson Project Report F-19-R; Virginia Commission of Game and Inland Fisheries: Richmond, VA, USA, 1976; p. 30. [Google Scholar]
- Whitehurst, D.K. Kerr reservoir striped bass population study. In Final Report, Virginia Dingell-Johnson Project F-34; Virginia Department of Fish and Game: Richmond, VA, USA, 1981; p. 243. [Google Scholar]
- Frankham, R. Effective population size/adult population size ratios in wildlife: A review. Genet. Res. 1995, 66, 95–107. [Google Scholar] [CrossRef]
- Allendorf, F.W.; Luikart, G. Conservation and the Genetics of Populations; Blackwell Publishing: Malden, MA, USA, 2007. [Google Scholar]
- Moritz, C. Defining ‘evolutionary significant units’ for conservation. Trends Ecol. Evol. 1994, 9, 373–375. [Google Scholar] [CrossRef]
- Busack, C.A.; Currens, K.P. Genetic risks and hazards in hatchery operations: Fundamental concepts and issues. In Uses and Effects of Cultured Fishes in Aquatic Ecosystems; Schramm, H.L., Piper, R.G., Eds.; American Fisheries Society: Bethesda, MD, USA, 1993; pp. 337–380. [Google Scholar]
- Franklin, I.R. Evolutionary change in small populations. In Conservation Biology: The Science of Scarcity and Diversity; Soule, M.E., Wilcox, B.A., Eds.; Sinauer Associates: Sunderland, MA, USA, 1980; pp. 135–140. [Google Scholar]
- Harmon, L.; Braude, S. Conservation of small populations: Effective population sizes, inbreeding, and the 50/500 rule. In An Introduction to Methods and Models in Ecology, Evolution, and Conservation Biology; Princeton University Press: Princeton, NJ, USA, 2010; pp. 125–138. [Google Scholar]
- Bartley, D.; Bagley, M.; Gall, G.; Bentley, B. Use of linkage disequilibrium data to estimate effective size of hatchery and natural fish populations. Cons. Biol. 1992, 6, 365–375. [Google Scholar] [CrossRef]
- Hara, M.; Sekino, M. Efficient detection of parentage in a cultured Japanese flounder Paralichthys olivaceus using microsatellite DNA marker. Aquaculture 2003, 217, 107–114. [Google Scholar] [CrossRef]
- Sekino, M.; Saitoh, K.; Yamada, T.; Kumagai, A.; Hara, M.; Yamashita, Y. Microsatellite-based pedigree tracing in a Japanese flounder Paralichthys olivaceus hatchery strain: Implications for hatchery management related to stock enhancement program. Aquaculture 2003, 221, 255–263. [Google Scholar] [CrossRef]
- Brown, R.C.; Woolliams, J.A.; McAndrew, B.J. Factors influencing effective population size in commercial populations of gilthead seabream Sparus aurata. Aquaculture 2005, 247, 219–225. [Google Scholar] [CrossRef]
- Frost, L.A.; Evans, B.S.; Jerry, D.R. Loss of genetic diversity due to hatchery culture practices in barramundi (Lates calcarifer). Aquaculture 2006, 261, 1056–1064. [Google Scholar] [CrossRef]
- Loughnan, S.R.; Domingos, J.A.; Smith-Keune, C.; Forrester, J.P.; Jerry, D.R.; Beheregaray, L.B.; Robinson, N.A. Broodstock contribution after mass spawning and size grading in barramundi (Lates calcarifer, Bloch). Aquaculture 2013, 404, 139–149. [Google Scholar] [CrossRef]
- Herlin, M.; Delghandi, M.; Wesmajervi, M.; Taggart, J.B.; McAndrew, B.J.; Penman, D.J. Analysis of the parental contribution to a group of fry from a single day of spawning from a commercial Atlantic cod (Gadus morhua) breeding tank. Aquaculture 2008, 274, 218–224. [Google Scholar] [CrossRef]
- Darden, T.L.; Sessions, F.; Denson, M.R. Use of genetic microsatellite markers to identify factors affecting stocking success in striped bass. Am. Fish. Symp. 2013, 80, 395–411. [Google Scholar]
- Frankham, R.; Ballou, J.D.; Eldridge, M.D.; Lacy, R.C.; Ralls, K.; Dudash, M.R.; Fenster, C.B. Predicting the probability of outbreeding depression. Cons. Biol. 2011, 25, 465–475. [Google Scholar] [CrossRef]
- Ricks, B.R.; Buckley, C.A. Neuse River striped bass monitoring, 2016–2017. In Federal Aid in Sport Fish Restoration Project F-108 Report; North Carolina Wildlife Resources Commission: Raleigh, NC, USA, 2018. [Google Scholar]
- Cushman, B.; O’Donnell, T.; Farrae, D. 2017 Striped Bass Genotyping and Parentage Analysis; Final Report to the North Carolina Wildlife Resources Commission; North Carolina Wildlife Resources Commission: Raleigh, NC, USA, 2018. [Google Scholar]
- Steele, C.A.; Hess, M.; Narum, S.; Campbell, M. Parentage-based tagging: Reviewing the implementation of a new tool for an old problem. Fisheries 2019, 44, 412–422. [Google Scholar] [CrossRef]
- Letcher, B.H.; King, T.L. Targeted stock identification using multilocus genotype ‘familyprinting’. Fish. Res. 1999, 43, 99–111. [Google Scholar] [CrossRef]
- Van Oosterhout, C.; Hutchinson, W.F.; Wills, D.P.M.; Shipley, P. Microchecker: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Excoffier, L.; Laval, G.; Schneider, S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evol. Bioinformat. Online 2005, 1, 47–50. [Google Scholar] [CrossRef]
- Jombart, T.; Kamvar, Z.N.; Collins, C.; Lustrik, R.; Beugin, M.-P.; Knaus, B.J.; Solymos, P.; Mikryukov, V.; Schliep, K.; Maié, T.; et al. Package ‘Adegenet’. 2020. Available online: https://github.com/thibautjombart/adegenet (accessed on 26 June 2019).
- Dent, E.A.; von Holdt, B.M. Structure harvester: A website and program for visualizing structure output and implementing the Evanno method. Cons. Genet. Resour. 2012, 4, 359–361. [Google Scholar]
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.A.; Mayrose, I. Clumpak: A program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 2015, 5, 1179–1191. [Google Scholar] [CrossRef]
- Chybicki, I.J.; Oleksa, A.; Burczyk, J. Increased inbreeding and strong kinship structure in Taxus baccata estimated for both AFLP and SSR data. Heredity 2011, 107, 589–600. [Google Scholar] [CrossRef]
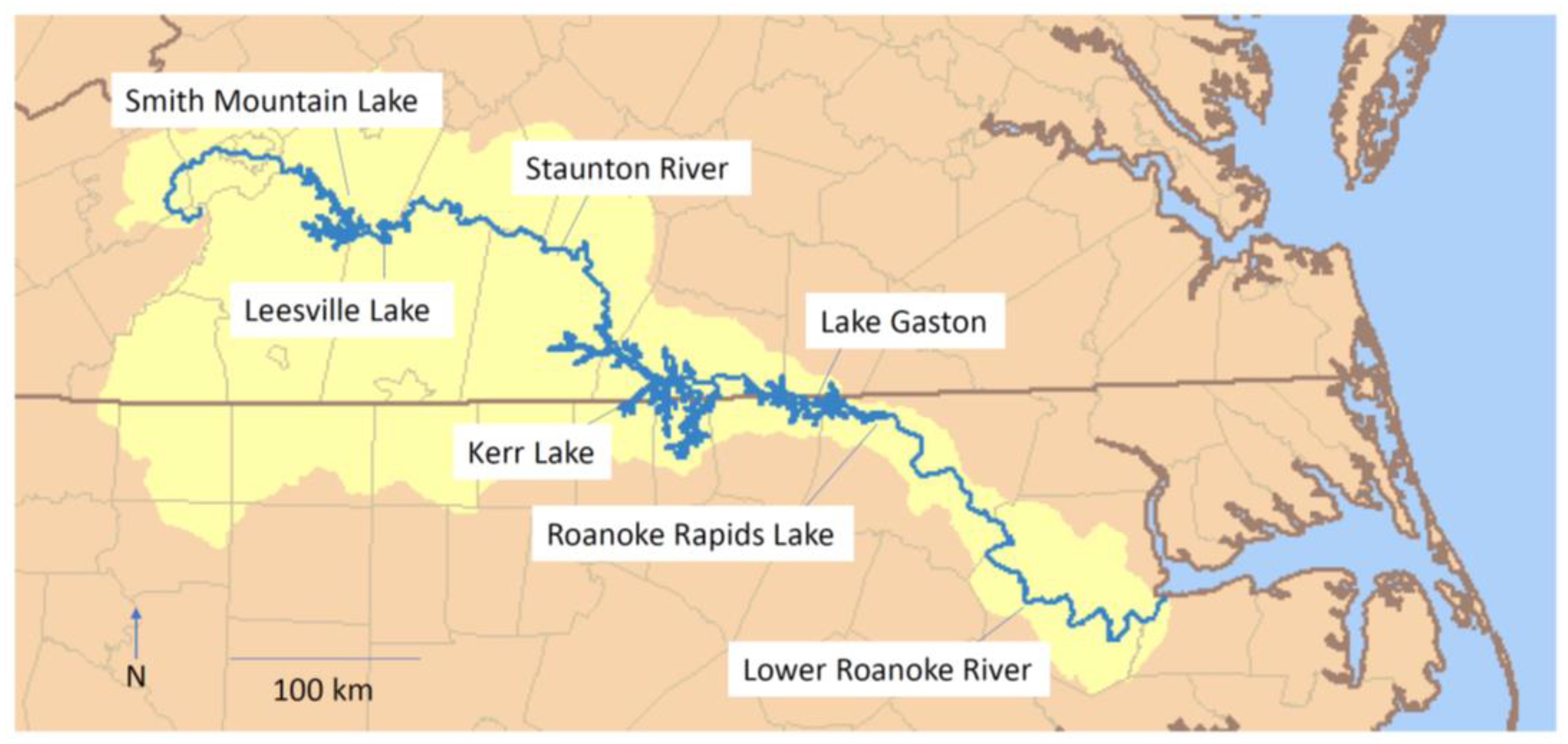
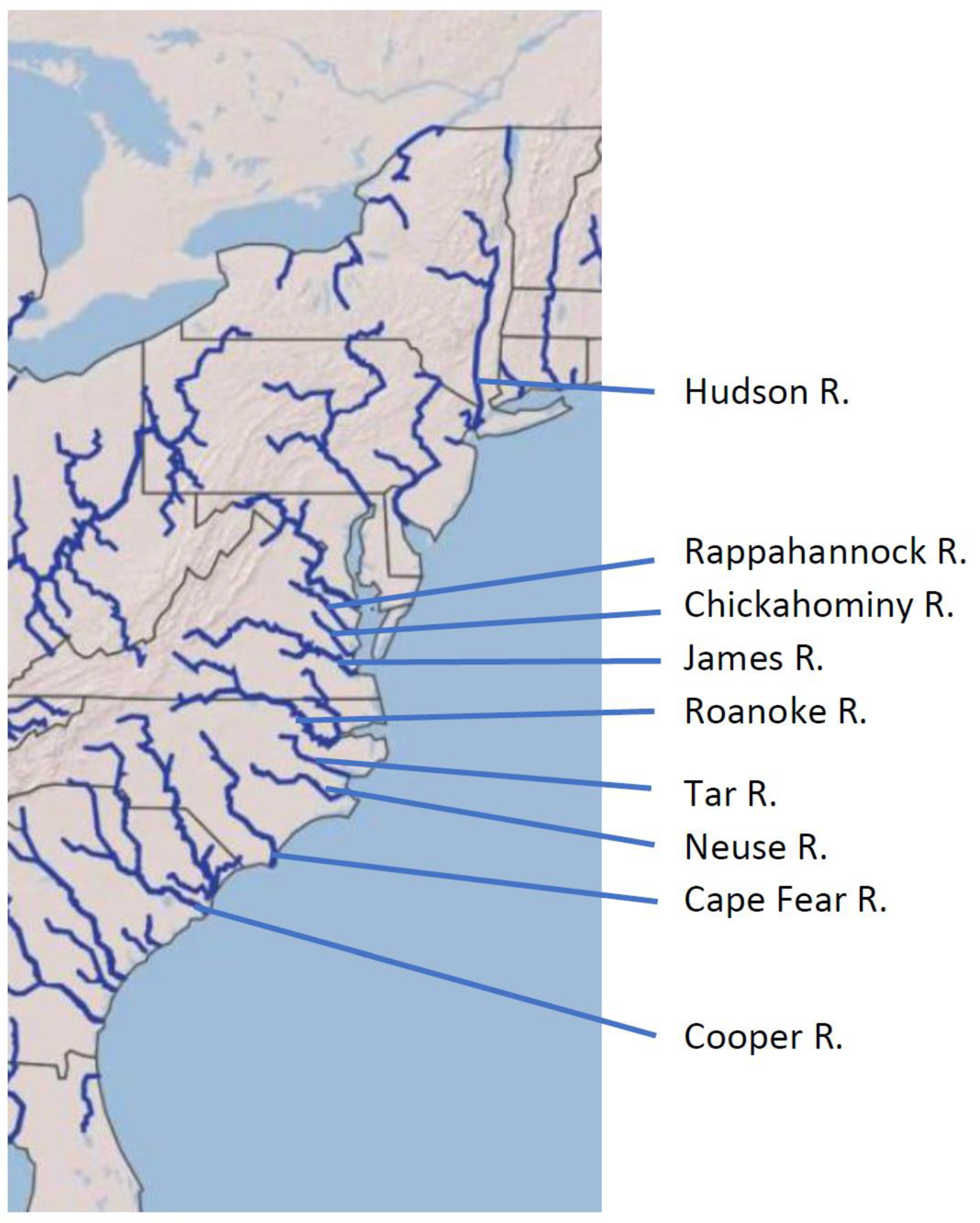
| Major Drainage | River | Location | Number (Year) | Provided by |
|---|---|---|---|---|
| Roanoke 1 | Upper Roanoke R. | Smith Mountain Lake Leesville Lake Staunton River Kerr Res.-Island Creek Park Kerr Res.-Island Creek Park Kerr Res.-Ivy Hill Park Kerr Res.-Ivy Hill Park Lake Gaston Roanoke Rapids Lake | 22 (2015) 60 (2016) 47 (2015) 45 (2017) 57 (2015) 52 (2017) 39 (2016) 26 (2016) | Virginia Department of Game and Inland Fisheries |
| Chesapeake Bay 2 | James River Rappahannock R. Chickahominy R. | Upper tidal-Richmond Upper tidal-Richmond Tidal James River at I-95 Fredericksburg Fredericksburg Chickahominy River | 7 (2017) 110 (2018) 17 (2018) 37 (2017) 25 (2018) 11 (2018) | Virginia Department of Game and Inland Fisheries |
| Hudson River 3 | Hudson River | Various | 72 (various) | Isaac Wirgin |
| Southeast 4 | Neuse River Tar River Cape Fear R. Lower Roanoke R. Santee-Cooper R. Santee-Cooper R. | Neuse River Tar River Cape Fear River North Carolina Lake Marion 5 Lake Moultrie 5 | 10 10 10 30 20 20 | South Carolina Department of Natural Resources |
| Locus | Repeat | Size Range (bp) | Number of Alleles | Source |
|---|---|---|---|---|
| MSM 1144 | (CA)25 | 169–235 | 25 | [36] |
| MSM 1095 | (TG)28 | 156–198 | 20 | [36] |
| MSM 1096 | (CA)25 | 156–204 | 20 | [36] |
| MSM 1243 | (CA)18 | 227–273 | 13 | [36] |
| MSM 1094 | (CA)25 | 110–154 | 25 | [36] |
| MSM 1526 | (GT)26 | 124–238 | 31 | [37] |
| MSM 1208 | (TA)31 | 153–221 | 25 | [36] |
| MSM 1067 | (CA)14(GT)11 | 177–213 | 16 | [36] |
| MSM 1168 | (CA)27 | 131–159 | 15 | [36] |
| MSM 1139 | (AC)34 | 137–219 | 27 | [36] |
| MSM 1592 | (TAGA)33 | 144–242 | 35 | [37] |
| MSM 1357 | (GATA)21 | 203–283 | 35 | [37] |
| Population | N | A | Ar | Ho | He | Allelic Range | M |
|---|---|---|---|---|---|---|---|
| Southeast | |||||||
| Lake Marion | 40 | 8.3 | 1.78 | 0.75 | 0.79 | 96.3 | 0.23 |
| Lake Moultrie | 40 | 8.5 | 1.75 | 0.61 | 0.76 | 146.4 | 0.14 |
| Neuse River | 20 | 7.1 | 1.79 | 0.82 | 0.8 | 79.4 | 0.2 |
| Tar River | 20 | 6.6 | 1.78 | 0.79 | 0.8 | 74.6 | 0.22 |
| Cape Fear River | 20 | 5.9 | 1.76 | 0.65 | 0.69 | 74.6 | 0.2 |
| Roanoke River, NC | 60 | 9 | 1.77 | 0.76 | 0.78 | 95.1 | 0.24 |
| Upper Roanoke R. | |||||||
| Smith Mountain L. | 222 | 12.2 | 1.75 | 0.67 | 0.77 | 203.6 | 0.06 |
| Leesville Lake | 44 | 6.2 | 1.71 | 0.67 | 0.71 | 27 | 0.28 |
| Staunton River | 120 | 9.5 | 1.74 | 0.62 | 0.75 | 102.1 | 0.2 |
| Kerr Reservoir | 402 | 12.3 | 1.76 | 0.67 | 0.78 | 190.3 | 0.11 |
| Lake Gaston | 78 | 10.1 | 1.75 | 0.67 | 0.76 | 124 | 0.19 |
| Roanoke Rapids | 52 | 8 | 1.74 | 0.66 | 0.74 | 44.1 | 0.29 |
| Chesapeake Bay | |||||||
| James River | 138 | 17.4 | 1.84 | 0.75 | 0.85 | 159.1 | 0.18 |
| Chickahominy R. | 22 | 7.9 | 1.8 | 0.74 | 0.8 | 55.1 | 0.23 |
| Rappahannock R. | 252 | 13.5 | 1.8 | 0.82 | 0.8 | 91.4 | 0.26 |
| Hudson River | 144 | 12.4 | 1.81 | 0.7 | 0.82 | 207.2 | 0.06 |
| Source of Variation | d.f. | Sum of Squares | Variance Components 1 | Percentage of Variation |
|---|---|---|---|---|
| (A) 1 | ||||
| Among groups | 3 | 149.77 | 0.08 Va | 2.1 |
| Among populations within groups | 12 | 169.96 | 0.11 Vb | 2.8 |
| Among individuals within popns. | 821 | 3536.83 | 0.39 Vc | 9.5 |
| Within individuals | 837 | 2951 | 3.53 Vd | 85.7 |
| Total | 1673 | 6807.56 | 4.09 | 100 |
| (B) 2 | ||||
| Among groups | 4 | 111.59 | 0.07 Va | 2.3 |
| Among populations within groups | 11 | 106.14 | 0.07 Vb | 2.3 |
| Among individuals within popns. | 819 | 2573.2 | 0.20 Vc | 6.6 |
| Within individuals | 835 | 2286 | 2.74 Vd | 88.9 |
| Total | 1669 | 8076.9 | 3.08 | 100.6.60 |
| Population | FIS | FSC | FCT | FIT |
|---|---|---|---|---|
| Full range | 0.1 | 0.03 | 0.02 | 0.14 |
| Virginia populations | 0.11 | 0.03 | 0.02 | 0.16 |
| Roanoke River drainage | 0.13 | 0.02 | 0.03 | 0.18 |
| Chesapeake Bay and Hudson River | 0.08 | 0.03 | 0.01 | 0.11 |
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0.000 | |||||||||||||||
| 2 | 0.039 | 0.000 | ||||||||||||||
| 3 | 0.062 | 0.092 | 0.000 | |||||||||||||
| 4 | 0.049 | 0.083 | −0.008 | 0.000 | ||||||||||||
| 5 | 0.070 | 0.098 | 0.055 | 0.041 | 0.000 | |||||||||||
| 6 | 0.057 | 0.081 | 0.005 | 0.000 | 0.035 | 0.000 | ||||||||||
| 7 | 0.095 | 0.117 | 0.021 | 0.033 | 0.086 | 0.045 | 0.000 | |||||||||
| 8 | 0.093 | 0.121 | 0.044 | 0.031 | 0.092 | 0.056 | 0.043 | 0.000 | ||||||||
| 9 | 0.100 | 0.128 | 0.029 | 0.045 | 0.103 | 0.055 | 0.013 | 0.050 | 0.000 | |||||||
| 10 | 0.075 | 0.100 | 0.017 | 0.022 | 0.073 | 0.039 | 0.011 | 0.029 | 0.010 | 0.000 | ||||||
| 11 | 0.067 | 0.113 | 0.025 | 0.018 | 0.079 | 0.038 | 0.035 | 0.022 | 0.033 | 0.019 | 0.000 | |||||
| 12 | 0.089 | 0.116 | 0.031 | 0.026 | 0.082 | 0.046 | 0.038 | 0.026 | 0.035 | 0.020 | 0.009 | 0.000 | ||||
| 13 | 0.065 | 0.085 | 0.039 | 0.037 | 0.081 | 0.050 | 0.056 | 0.075 | 0.066 | 0.049 | 0.051 | 0.055 | 0.000 | |||
| 14 | 0.112 | 0.121 | 0.096 | 0.085 | 0.136 | 0.094 | 0.095 | 0.122 | 0.112 | 0.096 | 0.103 | 0.108 | 0.029 | 0.000 | ||
| 15 | 0.064 | 0.099 | 0.026 | 0.012 | 0.058 | 0.025 | 0.039 | 0.053 | 0.047 | 0.030 | 0.030 | 0.027 | 0.040 | 0.084 | 0.000 | |
| 16 | 0.053 | 0.075 | 0.026 | 0.018 | 0.039 | 0.022 | 0.055 | 0.069 | 0.061 | 0.047 | 0.048 | 0.051 | 0.045 | 0.081 | 0.027 | 0.000 |
| Source of Variation | d.f. | Sum of Squares | Variance Components 1 | Percentage of Variation |
|---|---|---|---|---|
| Among groups | 1 | 21.48 | 0.12 Va | 3.4 |
| Among populations within groups | 5 | 59.12 | 0.06 Vb | 1.7 |
| Among individuals within popns. | 482 | 1812.36 | 0.44 Vc | 12.6 |
| Within individuals | 489 | 1407 | 2.88 Vd | 82.3 |
| Total | 977 | 3299.95 | 3.5 |
| Collection | N | Ne1 | 95% CI 1 | Ne2 | 95% CI 2 |
|---|---|---|---|---|---|
| Smith Mountain L. | 111 | 24.0 | 20.9–27.6 | 56 | 39–82 |
| Leesville Lake | 22 | 31.9 | 18.8–73.6 | 23 | 13–45 |
| Staunton River | 60 | 34.7 | 27.7–44.6 | 43 | 20–70 |
| Kerr Reservoir | 201 | 75.2 | 64.2–88.7 | 125 | 98–162 |
| Lake Gaston | 39 | 31.9 | 24.4–43.5 | 36 | 23–60 |
| Roanoke Rapids Lake | 26 | 40.4 | 25.5–80.5 | 40 | 23–74 |
| Lower Roanoke River | 30 | 282.2 | 79.1–undefined 3 | 51 | 31–89 |
| Sibship Assignment | Maximum Likelihood | |||||||
|---|---|---|---|---|---|---|---|---|
| Collection | N | Number of Families | Numbers of Members | Percent Full-Sibs | Percent Half-Sibs | Percent Full-Sibs | Percent Half-Sibs | Percent Parent-Offspring |
| Smith Mountain L. | 111 | 90 | 1–4 | 0.26 | 2.09 | 3.88 | 13.12 | 1.27 |
| Leesville Lake | 22 | 17 | 1–2 | 1.08 | 2.96 | 2.16 | 12.12 | 0.13 |
| Staunton River | 60 | 57 | 1–2 | 0.08 | 6.28 | 1.92 | 12.59 | 0.96 |
| Kerr Reservoir | 201 | 177 | 1–4 | 0.07 | 1.00 | 2.34 | 12.91 | 0.90 |
| Lake Gaston | 39 | 31 | 1–5 | 1.01 | 2.56 | 2.43 | 9.59 | 0.14 |
| Roanoke Rapids L. | 26 | 24 | 1–2 | 0.30 | 3.50 | 2.15 | 8.31 | 0.00 |
| Lower Roanoke R. | 30 | 30 | 1 | 0.00 | 3.79 | 0.23 | 8.51 | 0.23 |
| Collection | FIS | p-Value 1 | Mean F 2 | Avg. Fi 2 |
|---|---|---|---|---|
| Smith Mountain L. | 0.12 | 0 | 0.0025 | 0.0018 |
| Leesville Lake | 0.06 | 0.13 | 0.009 | 0.0084 |
| Staunton River | 0.18 | 0 | 0.0191 | 0.0284 |
| Kerr Reservoir | 0.14 | 0 | 0.0336 | 0.0223 |
| Lake Gaston | 0.12 | 0 | 0.0198 | 0.0173 |
| Roanoke Rapids | 0.11 | 0 | 0.1317 | 0.1092 |
| Lower Roanoke River | 0.03 | 0.18 | 0.0043 | 0 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Harris, S.C.; Cope, W.R.; Wirgin, I.; Hallerman, E.M. Population Genetic Assessment of Anadromous and Resident Striped Bass (Morone saxatilis) in the Roanoke River System, Eastern United States. Fishes 2020, 5, 24. https://doi.org/10.3390/fishes5030024
Harris SC, Cope WR, Wirgin I, Hallerman EM. Population Genetic Assessment of Anadromous and Resident Striped Bass (Morone saxatilis) in the Roanoke River System, Eastern United States. Fishes. 2020; 5(3):24. https://doi.org/10.3390/fishes5030024
Chicago/Turabian StyleHarris, Sheila C., W. Robert Cope, Isaac Wirgin, and Eric M. Hallerman. 2020. "Population Genetic Assessment of Anadromous and Resident Striped Bass (Morone saxatilis) in the Roanoke River System, Eastern United States" Fishes 5, no. 3: 24. https://doi.org/10.3390/fishes5030024
APA StyleHarris, S. C., Cope, W. R., Wirgin, I., & Hallerman, E. M. (2020). Population Genetic Assessment of Anadromous and Resident Striped Bass (Morone saxatilis) in the Roanoke River System, Eastern United States. Fishes, 5(3), 24. https://doi.org/10.3390/fishes5030024






