Transcriptomic Changes during Previtellogenic and Vitellogenic Stages of Ovarian Development in Wreckfish (Hāpuku), Polyprion oxygeneios (Perciformes)
Abstract
1. Introduction
2. Results
2.1. Stages of Ovarian Development
2.2. Illumina Sequencing and De Novo Assembly
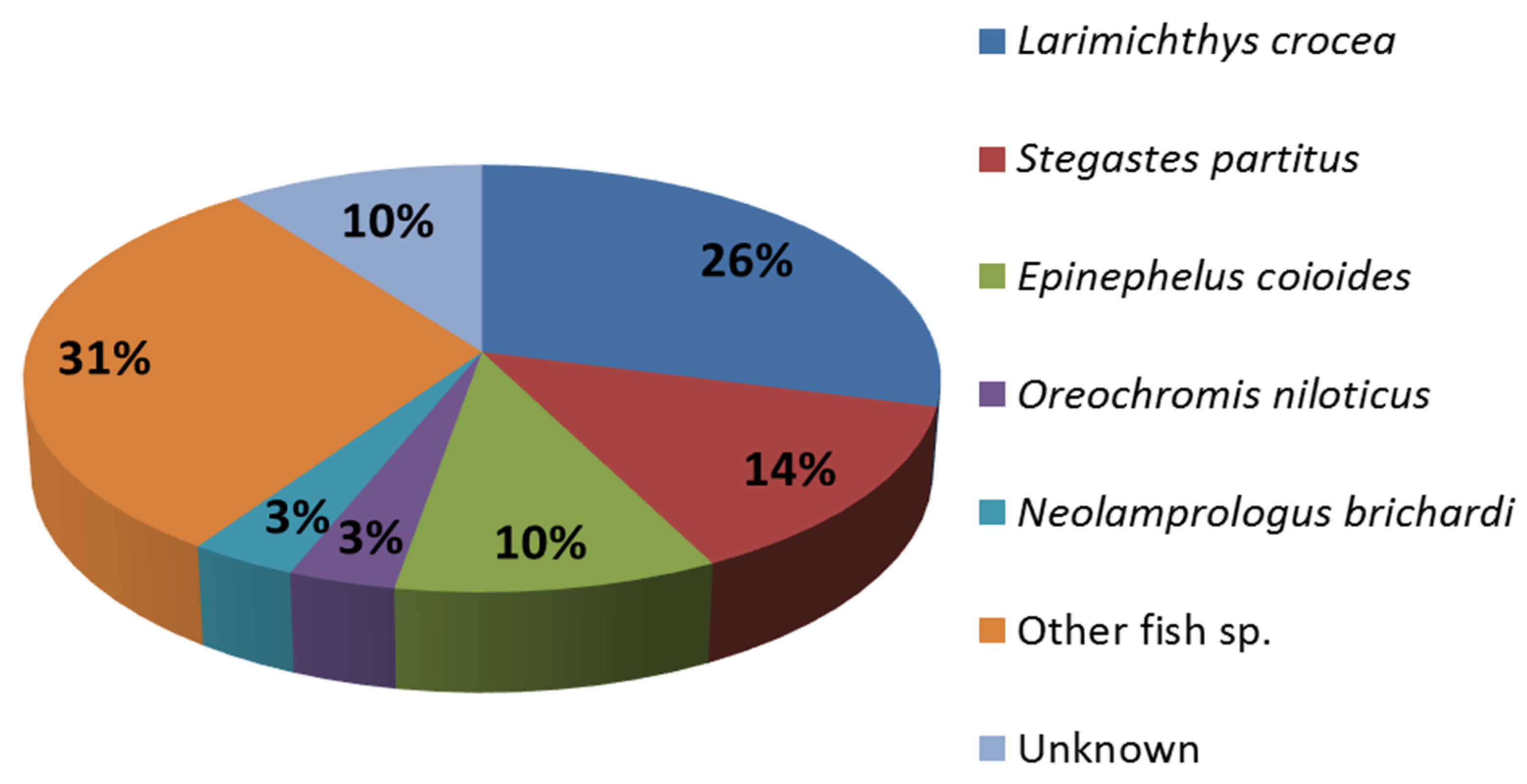
2.3. Identification of Differentially Expressed Genes and Transcriptome Annotation
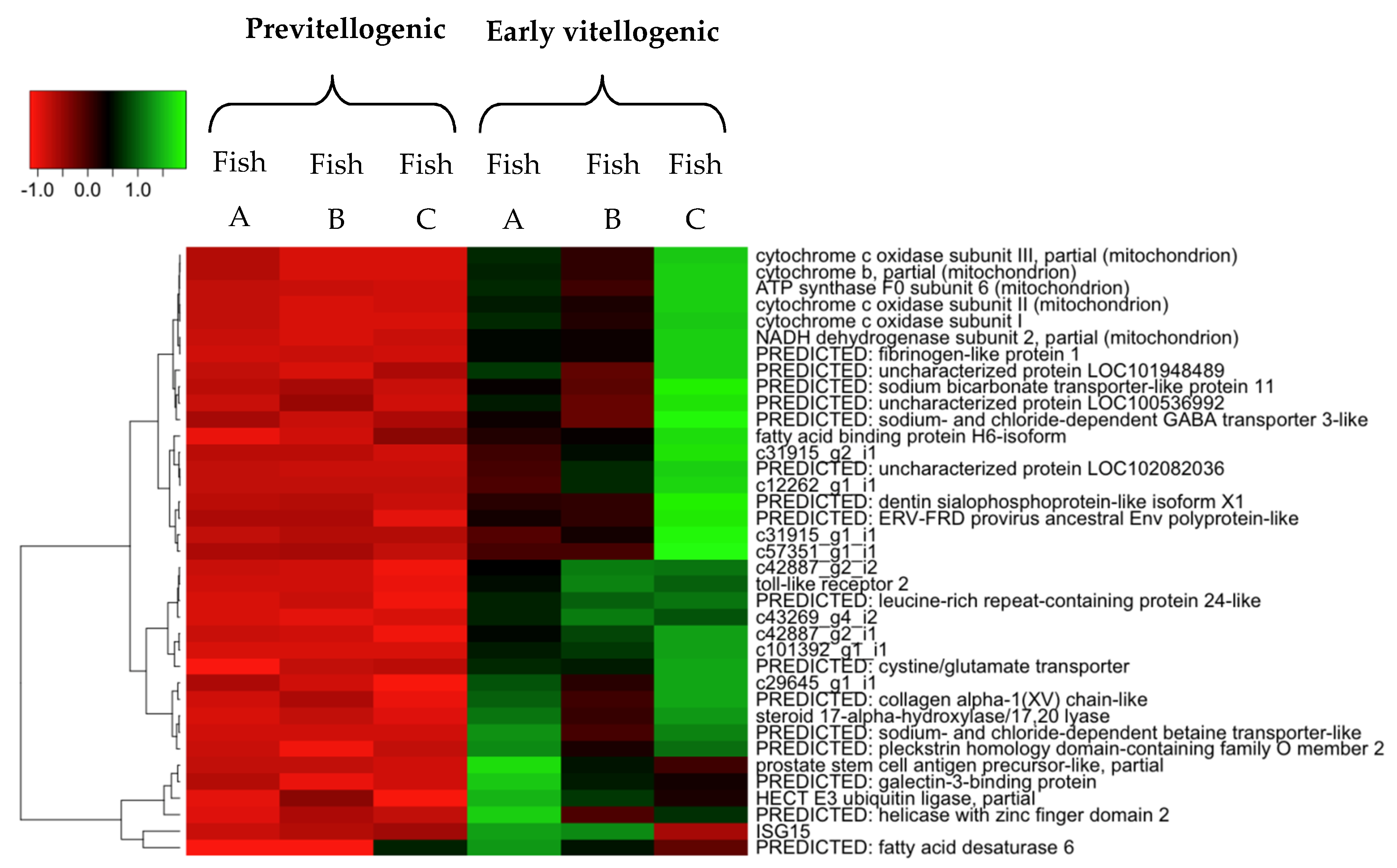
2.4. Hierarchical Cluster and Gene Ontology Enrichment/Pathway Analyses
3. Discussion
4. Materials and Methods
4.1. Broodstock and Experimental Design
4.2. Sample Collection
4.2.1. Anaesthesia
4.2.2. Blood Sampling and Measurement of E2 Levels by Radioimmunoassay
4.2.3. Ovarian Biopsy Collection
4.3. RNA Extraction and Preparation for RNA-Seq
4.4. Illumina Sequencing
Library Construction
4.5. De Novo Assembly and Annotation
4.5.1. Normalisation and Differential Expression Analysis
4.6. Hierarchical Cluster and Gene Ontology (GO) Enrichment/Pathway Analyses
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Migaud, H.; Bell, G.; Cabrita, E.; McAndrew, B.; Davie, A.; Bobe, J.; Herraez, M.P.; Carrillo, M. Gamete quality and broodstock management in temperate fish. Rev. Aquac. 2013, 5, S194–S223. [Google Scholar] [CrossRef]
- Lubzens, E.; Bobe, J.; Young, G.; Sullivan, C.V. Maternal investment in fish oocytes and eggs: The molecular cargo and its contributions to fertility and early development. Aquaculture 2017, 472, 107–143. [Google Scholar] [CrossRef]
- Patiño, R.; Sullivan, C.V. Ovarian follicle growth, maturation, and ovulation in teleost fish. Fish Physiol. Biochem. 2002, 26, 57–70. [Google Scholar] [CrossRef]
- Lubzens, E.; Young, G.; Bobe, J.; Cerdà, J. Oogenesis in teleosts: How fish eggs are formed. Gen. Comp. Endocrinol. 2010, 165, 367–389. [Google Scholar] [CrossRef] [PubMed]
- Pelegri, F. Maternal factors in zebrafish development. Dev. Dyn. 2003, 228, 535–554. [Google Scholar] [CrossRef] [PubMed]
- Lanes, C.F.C.; Bizuayehu, T.T.; de Oliveira Fernandes, J.M.; Kiron, V.; Babiak, I. Transcriptome of Atlantic cod (Gadus morhua L.) early embryos from farmed and wild broodstocks. Mar. Biotechnol. 2013, 15, 677–694. [Google Scholar] [CrossRef] [PubMed]
- Bobe, J. Egg quality in fish: Present and future challenges. Anim. Front. 2015, 5, 66–72. [Google Scholar] [CrossRef]
- Johansen, S.D.; Karlsen, B.O.; Furmanek, T.; Andreassen, M.; Jørgensen, T.E.; Bizuayehu, T.T.; Breines, R.; Emblem, Å.; Kettunen, P.; Luukko, K.; et al. RNA deep sequencing of the Atlantic cod transcriptome. Comp. Biochem. Physiol. D 2011, 6, 18–22. [Google Scholar] [CrossRef] [PubMed]
- Wheat, C.W. Rapidly developing functional genomics in ecological model systems via 454 transcriptome sequencing. Genetica 2010, 138, 433–451. [Google Scholar] [CrossRef] [PubMed]
- Xuan, J.; Yu, Y.; Qing, T.; Guo, L.; Shi, L. Next-generation sequencing in the clinic: Promises and challenges. Cancer Lett. 2013, 340, 284–295. [Google Scholar] [CrossRef] [PubMed]
- Lokman, P.M.; Symonds, J.E. Molecular and biochemical tricks of the research trade:-omics approaches in finfish aquaculture. N. Z. J. Mar. Freshw. Res. 2014, 48, 492–505. [Google Scholar] [CrossRef]
- Taranger, G.L.; Carrillo, M.; Schulz, R.W.; Fontaine, P.; Zanuy, S.; Felip, A.; Weltzien, F.A.; Dufour, S.; Karlsen, Ø.; Norberg, B.; et al. Control of puberty in farmed fish. Gen. Comp. Endocrinol. 2010, 165, 483–515. [Google Scholar] [CrossRef] [PubMed]
- Mylonas, C.C.; Zohar, Y. Use of GnRHa-delivery systems for the control of reproduction in fish. Rev. Fish Biol. Fish. 2000, 10, 463–491. [Google Scholar] [CrossRef]
- Zohar, Y.; Mylonas, C.C. Endocrine manipulations of spawning in cultured fish: From hormones to genes. In Reproductive Biotechnology in Finfish Aquaculture; Donaldson, E.M., Lee, C.S., Eds.; Elsevier: Amsterdam, The Netherlands, 2001; pp. 99–136. [Google Scholar]
- Symonds, J.E.; Walker, S.P.; Pether, S.; Gublin, Y.; McQueen, D.; King, A.; Irvine, G.W.; Setiawan, A.N.; Forsythe, J.A.; Bruce, M. Developing yellowtail kingfish (Seriola lalandi) and hāpuku (Polyprion oxygeneios) for New Zealand aquaculture. N. Z. J. Mar. Freshw. Res. 2014, 48, 371–384. [Google Scholar] [CrossRef]
- Wylie, M.J.; Setiawan, A.N.; Irvine, G.W.; Symonds, J.E.; Elizur, A.; Lokman, P.M. Effects of neuropeptides and sex steroids on the pituitary-gonadal axis of pre-pubertal F1 wreckfish (hāpuku) Polyprion oxygeneios in vivo: Evidence of inhibitory effects of androgens. Gen. Comp. Endocrinol. 2018, 257, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Wylie, M.J.; Setiawan, A.N.; Irvine, G.W.; Symonds, J.E.; Elizur, A.; Dos Santos, M.; Lokman, P.M. Ovarian development of captive F1 wreckfish (hāpuku) Polyprion oxygeneios under constant and varying temperature regimes—Implications for broodstock management. Gen. Comp. Endocrinol. 2018, 257, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Diaz de Cerio, O.; Rojo-Bartolomeé, I.; Bizarro, C.; Ortiz-Zarragoitia, M.; Cancio, I. 5S rRNA and accompanying proteins in gonads: Powerful markers to identify sex and reproductive endocrine disruption in fish. Environ. Sci. Technol. 2012, 46, 7763–7771. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Reading, B.J.; Chapman, R.W.; Schaff, J.E.; Scholl, E.H.; Opperman, C.H.; Sullivan, C.V. An ovary transcriptome for all maturational stages of the striped bass (Morone saxatilis), a highly advanced perciform fish. BMC Res. Notes 2012, 5, 111. [Google Scholar] [CrossRef] [PubMed]
- Chapman, R.W.; Reading, B.J.; Sullivan, C.V. Ovary transcriptome profiling via artificial intelligence reveals a transcriptomic fingerprint predicting egg quality in striped bass, Morone saxatilis. PLoS ONE 2014, 9, e96818. [Google Scholar] [CrossRef] [PubMed]
- Tyler, C.R.; Sumpter, J.P. Oocyte growth and development in teleosts. Rev. Fish Biol. Fish. 1996, 6, 287–318. [Google Scholar] [CrossRef]
- Lokman, P.; Kazeto, Y.; Ijiri, S.; Young, G.; Miura, T.; Adachi, S.; Yamauchi, K. Ovarian mitochondrial cytochrome b mRNA levels increase with sexual maturity in freshwater eels (Anguilla spp.). J. Comp. Physiol. B 2003, 173, 11–19. [Google Scholar] [PubMed]
- Martyniuk, C.J.; Prucha, M.S.; Doperalski, N.J.; Antczak, P.; Kroll, K.J.; Falciani, F.; Barber, D.S.; Denslow, N.D. Gene expression networks underlying ovarian development in wild largemouth bass (Micropterus salmoides). PLoS ONE 2013, 8, e59093. [Google Scholar] [CrossRef] [PubMed]
- Kagawa, H.; Young, G.; Adachi, S.; Nagahama, Y. Estradiol-17β production in amago salmon (Oncorhynchus rhodurus) ovarian follicles: Role of the thecal and granulosa cells. Gen. Comp. Endocrinol. 1982, 47, 440–448. [Google Scholar] [CrossRef]
- Uno, T.; Ishizuka, M.; Itakura, T. Cytochrome P450 (CYP) in fish. Environ. Toxicol. Pharmacol. 2012, 34, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Gohin, M.; Bobe, J.; Chesnel, F. Comparative transcriptomic analysis of follicle-enclosed oocyte maturational and developmental competence acquisition in two non-mammalian vertebrates. BMC Genom. 2010, 11, 18. [Google Scholar] [CrossRef] [PubMed]
- Su, T.; Ijiri, S.; Kanbara, H.; Hagihara, S.; Wang, D.S.; Adachi, S. Characterization and expression of cDNAs encoding P450c17-II (cyp17a2) in Japanese eel during induced ovarian development. Gen. Comp. Endocrinol. 2015, 221, 134–143. [Google Scholar] [CrossRef] [PubMed]
- Kagawa, H. Oogenesis in teleost fish. Aqua BioSci. Monogr. 2013, 6, 99–127. [Google Scholar] [CrossRef]
- Sampath Kumar, R.; Ijiri, S.; Trant, J.M. Changes in the expression of genes encoding steroidogenic enzymes in the channel catfish (Ictalurus punctatus) ovary throughout a reproductive cycle. Biol. Reprod. 2000, 63, 1676–1682. [Google Scholar] [CrossRef] [PubMed]
- Nunez, B.S.; Applebaum, S.L. Tissue-and sex-specific regulation of CYP19A1 expression in the Atlantic croaker (Micropogonias undulatus). Gen. Comp. Endocrinol. 2006, 149, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, L.R.; Chittenden, M.E., Jr.; Lowerre-Barbieri, S.K. Maturity, spawning, and ovarian cycle of Atlantic croaker, Micropogonias undulatus, in the Chesapeake Bay and adjacent coastal waters. Fish. Bull. 1994, 92, 671–685. [Google Scholar]
- Matsubara, H.; Kazeto, Y.; Ijiri, S.; Hirai, T.; Adachi, S.; Yamauchi, K. Changes in mRNA levels of ovarian steroidogenic enzymes during artificial maturation of Japanese eel Anguilla japonica. Fish. Sci. 2003, 69, 979–988. [Google Scholar] [CrossRef]
- Thirumaran, A.; Wright, J.M. Fatty acid-binding protein (fabp) genes of spotted green pufferfish (Tetraodon nigroviridis): Comparative genomics and spatial transcriptional regulation. Genome 2014, 57, 289–301. [Google Scholar] [CrossRef] [PubMed]
- Bayır, M.; Bayır, A.; Wright, J.M. Divergent spatial regulation of duplicated fatty acid-binding protein (fabp) genes in rainbow trout (Oncorhynchus mykiss). Comp. Biochem. Physiol. D 2015, 14, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Vayda, E.M.; Londraville, L.R.; Cashon, E.R.; Costello, L.; Sidell, D.B. Two distinct types of fatty acid-binding protein are expressed in heart ventricle of Antarctic teleost fishes. Biochem. J. 1998, 330, 375–382. [Google Scholar] [CrossRef] [PubMed]
- Agulleiro, M.J.; André, M.; Morais, S.; Cerda, J.; Babin, P.J. High transcript level of fatty acid-binding protein 11 but not of very low-density lipoprotein receptor is correlated to ovarian follicle atresia in a teleost fish (Solea senegalensis). Biol. Reprod. 2007, 77, 504–516. [Google Scholar] [CrossRef] [PubMed]
- Parmar, M.B.; Venkatachalam, A.B.; Wright, J.M. The evolutionary relationship of the transcriptionally active fabp11a (intronless) and fabp11b genes of medaka with fabp11 genes of other teleost fishes. FEBS J. 2012, 279, 2310–2321. [Google Scholar] [CrossRef] [PubMed]
- Bonen, A.; Chabowski, A.; Luiken, J.J.P.; Glatz, J.F. Mechanisms and regulation of protein-mediated cellular fatty acid uptake: Molecular, biochemical, and physiological evidence. Physiology 2007, 22, 15–28. [Google Scholar] [CrossRef] [PubMed]
- Glatz, J.F. Lipids and lipid binding proteins: A perfect match. Prostaglandins Leukot. Essent. Fat. Acids 2015, 93, 45–49. [Google Scholar] [CrossRef] [PubMed]
- Hiramatsu, N.; Todo, T.; Sullivan, C.V.; Schilling, J.; Reading, B.J.; Matsubara, T.; Ryu, Y.W.; Mizuta, H.; Luo, W.; Nishimiya, O.; et al. Ovarian yolk formation in fishes: Molecular mechanisms underlying formation of lipid droplets and vitellogenin-derived yolk proteins. Gen. Comp. Endocrinol. 2015, 221, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Ryu, Y.W.; Tanaka, R.; Kasahara, A.; Saito, K.; Kanno, K.; Ito, Y.; Hiramatsu, N.; Todo, T.; Sullivan, C.V.; Hara, A. Expression of genes involved in oocyte lipidation in cutthroat trout, Oncorhynchus clarki. Indian J. Sci. Technol. 2011, 4, 203–204. [Google Scholar]
- Yeong Kwon, J.; Prat, F.; Randall, C.R.; Tyler, C. Molecular characterization of putative yolk processing enzymes and their expression during oogenesis and embryogenesis in rainbow trout (Oncorhynchus mykiss). Biol. Reprod. 2001, 65, 1701–1709. [Google Scholar] [CrossRef]
- Hiramatsu, N.; Luo, W.; Reading, B.J.; Sullivan, C.V.; Mizuta, H.; Ryu, Y.W.; Nishimiya, O.; Todo, T.; Hara, A. Multiple ovarian lipoprotein receptors in teleosts. Fish Physiol. Biochem. 2013, 39, 29–32. [Google Scholar] [CrossRef] [PubMed]
- Divers, S.L.; McQuillan, H.J.; Matsubara, H.; Todo, T.; Lokman, P.M. Effects of reproductive stage and 11-ketotestosterone on LPL mRNA levels in the ovary of the shortfinned eel. J. Lipid Res. 2010, 51, 3250–3258. [Google Scholar] [CrossRef] [PubMed]
- Ibáñez, A.J.; Peinado-Onsurbe, J.; Sánchez, E.; Cerdá-Reverter, J.M.; Prat, F. Lipoprotein lipase (LPL) is highly expressed and active in the ovary of European sea bass (Dicentrarchus labrax L.), during gonadal development. Comp. Biochem. Physiol. A 2008, 150, 347–354. [Google Scholar]
- Carnevali, O.; Cionna, C.; Tosti, L.; Lubzens, E.; Maradonna, F. Role of cathepsins in ovarian follicle growth and maturation. Gen. Comp. Endocrinol. 2006, 146, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Carnevali, O.; Cionna, C.; Tosti, L.; Cerdà, J.; Gioacchini, G. Changes in cathepsin gene expression and relative enzymatic activity during gilthead sea bream oogenesis. Mol. Reprod. Dev. 2008, 75, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Chen, J.; Ding, Y.; Jin, G.; Wu, J.; Huang, H.; Deng, B.; Hua, Z.; Zhou, Y.; Shu, Y.; et al. Genetic variation of PSCA gene is associated with the risk of both diffuse-and intestinal-type gastric cancer in a Chinese population. Int. J. Cancer 2010, 127, 2183–2189. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Noh, J.K.; Kim, H.C.; Park, C.J.; Min, B.H.; Choi, S.J.; Myeong, J.I.; Park, H.J.; Park, C.I. Expressed sequence tags analysis of immune-relevant genes in rock bream Oplegnathus fasciatus peripheral leukocytes stimulated with LPS. J. Fish Pathol. 2009, 22, 353–366. [Google Scholar]
- Wu, M.S.; Chen, C.W.; Lin, C.H.; Tzeng, C.S.; Chang, C.Y. Differential expression profiling of orange-spotted grouper larvae, Epinephelus coioides (Hamilton), that survived a betanodavirus outbreak. J. Fish Dis. 2012, 35, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Zheng, M.; Zheng, J.; Liu, J.; Liu, Y.; Peng, L.; Wang, P.; Zhang, X.; Wang, Q.; Luan, P.; et al. Transcriptomic analyses reveal novel genes with sexually dimorphic expression in yellow catfish (Pelteobagrus fulvidraco) brain. Mar. Biotechnol. 2015, 17, 613–623. [Google Scholar] [CrossRef] [PubMed]
- Erdö, S.L.; László, Á. High specific γ-aminobutyric acid binding to membranes of the human ovary. J. Neurochem. 1984, 42, 1464–1467. [Google Scholar] [CrossRef] [PubMed]
- Cocco, A.; Rönnberg, A.C.; Jin, Z.; André, G.I.; Vossen, L.E.; Bhandage, A.K.; Thörnqvist, P.O.; Birnir, B.; Winberg, S. Characterization of the γ-aminobutyric acid signaling system in the zebrafish (Danio rerio Hamilton) central nervous system by reverse transcription-quantitative polymerase chain reaction. Neuroscience 2017, 343, 300–321. [Google Scholar] [CrossRef] [PubMed]
- Aronesty, E. ea-Utils: “Command-Line Tools for Processing Biological Sequencing Data”. 2011. Available online: http://code.google.com/p/ea-utils (accessed on 6 September 2014).
- Cox, M.P.; Peterson, D.A.; Biggs, P.J. SolexaQA: At-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinform. 2010, 11, 485. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Guo, Y.; Sheng, Q.; Shyr, Y. Advanced heat map and clustering analysis using heatmap3. BioMed Res. Int. 2014. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
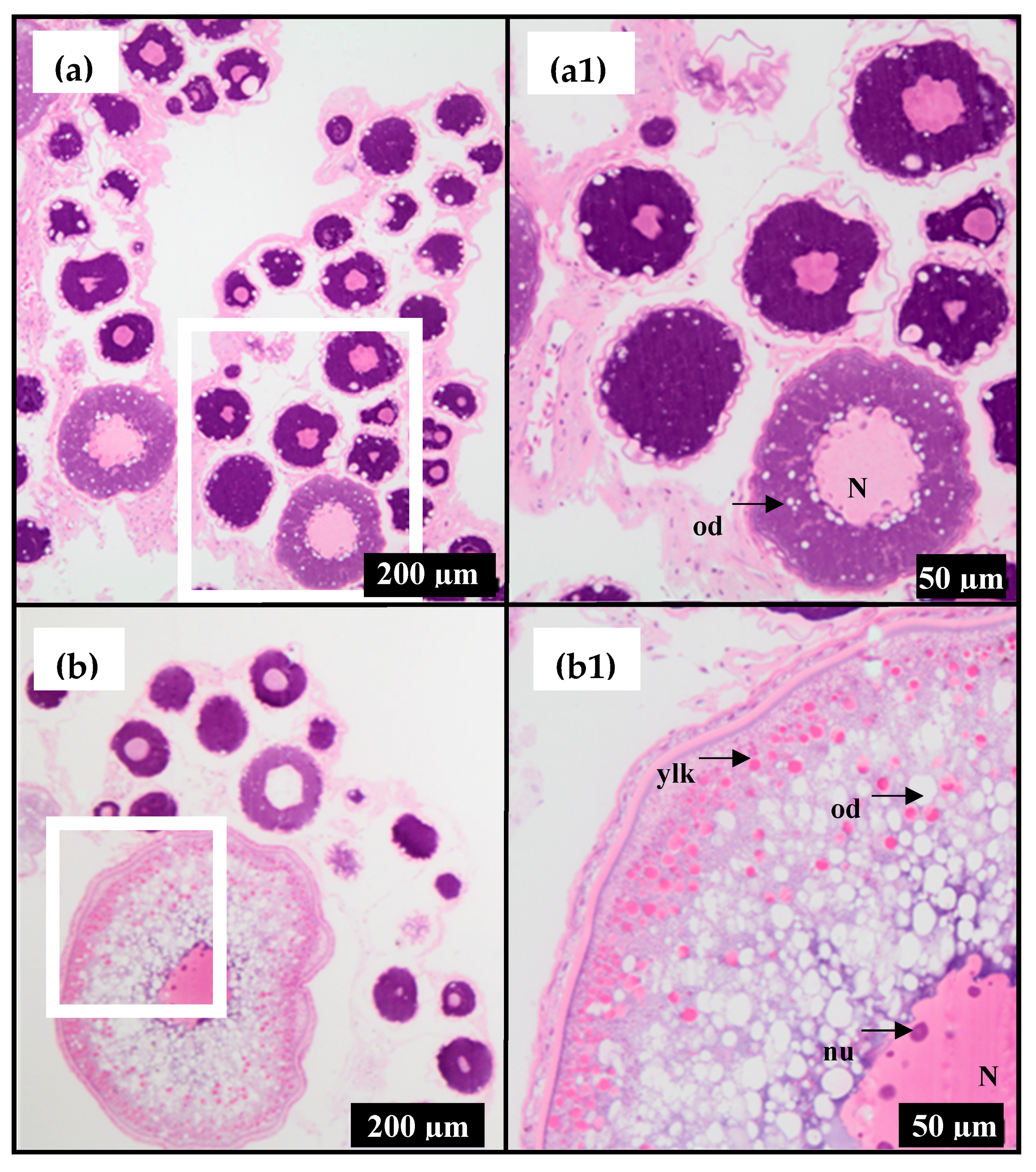
| Fish | Month | Ovarian Stage | RIN * | Oocyte Diameter (mm) | Plasma E2 (ng/mL) |
|---|---|---|---|---|---|
| A | March | Previtellogenic | 5.4 | 0.22 | 0.09 |
| B | March | Previtellogenic | 6.3 | 0.23 | 0.09 |
| C | March | Previtellogenic | 6.9 | 0.22 = 0.22 ± 0.003 | 0.08 = 0.08 ± 0.003 |
| A | May | Early vitellogenic | 9.2 | 0.40 | 0.78 |
| B | May | Early vitellogenic | 8.9 | 0.34 | 0.58 |
| C | May | Early vitellogenic | 9.7 | 0.38 = 0.37 ± 0.017 | 0.53 = 0.63 ± 0.076 |
| Conditions | Transcripts | Components | N50 * | Average Length | Number of Bases |
|---|---|---|---|---|---|
| PE Illumina Q30 and l100 | 146,189 | 120,688 | 1901 | 916.89 | 134,039,555 |
| No. | Contig Name | Hit Description | Gene Name | Assession No. | Species | Identity (%) | E-Value |
|---|---|---|---|---|---|---|---|
| 1 | c35045_g1_i1 | prostate stem cell antigen precursor-like | psca | gb|ADG29182 | Epinephelus coioides | 68 | 5.0E-28 |
| 2 | c42367_g1_i4 | PREDICTED: helicase with zinc finger domain 2 | helz2 | ref|XP_010750041 | Larimichthys crocea | 77 | 0.0E+00 |
| 3 | c39480_g2_i2 | PREDICTED: sodium bicarbonate transporter-like protein 11 | slc4a11 | ref|XP_010740414 | Larimichthys crocea | 93 | 0.0E+00 |
| 4 | c42970_g3_i2 | PREDICTED: sodium- and chloride-dependent GABA transporter 3-like | slc6a11 | ref|XP_006782336 | Neolamprologus brichardi | 74 | 5.0E-144 |
| 5 | c67236_g1_i1 | PREDICTED: sodium- and chloride-dependent betaine transporter-like | slc6a12 | ref|XP_008286219 | Stegastes partitus | 84 | 1.0E-157 |
| 6 | c551_g1_i2 | cytochrome b | myt-cb | gb|AAF06983 | Perca fluviatilis | 92 | 3.0E-64 |
| 7 | c19980_g1_i1 | cytochrome c oxidase subunit I | mt-c01 | ref|YP_163831 | Anguilla australis | 95 | 0.0E+00 |
| 8 | c11536_g1_i1 | cytochrome c oxidase subunit II | mt-c02 | ref|YP_009058427 | Howella brodiei | 92 | 3.0E-118 |
| 9 | c13004_g1_i1 | cytochrome c oxidase subunit III | mt-c03 | gb|AFQ94066 | Lutjanus lunulatus | 91 | 3.0E-49 |
| 10 | c26582_g1_i1 | PREDICTED: fibrinogen-like protein 1 | fgl1 | ref|XP_010747488 | Larimichthys crocea | 85 | 0.0E+00 |
| 11 | c24576_g1_i1 | PREDICTED: collagen α-1(XV) chain-like | col15a1 | ref|XP_008277067 | Stegastes partitus | 75 | 2.0E-129 |
| 12 | c70752_g1_i1 | PREDICTED: galectin-3-binding protein | lgals3bp | ref|XP_010736908 | Larimichthys crocea | 72 | 9.0E-140 |
| 13 | c68469_g1_i1 | steroid 17-α-hydroxylase/17,20 lyase | cyp17a1 | gb|AEL31248 | Lateolabrax japonicus | 94 | 0.0E+00 |
| 14 | c43269_g4_i4 | toll-like receptor 2 | tlr2 | gb|AEB32453 | Epinephelus coioides | 96 | 1.0E-05 |
| 15 | c33557_g1_i3 | HECT E3 ubiquitin ligase | gb|AER42668 | Epinephelus coioides | 80 | 3.0E-122 | |
| 16 | c61987_g1_i1 | PREDICTED: dentin sialophosphoprotein-like isoform X1 | dspp | ref|XP_010745298 | Larimichthys crocea | 64 | 8.0E-03 |
| 17 | c1460_g1_i1 | PREDICTED: leucine-rich repeat-containing protein 24-like | lrrc24 | ref|XP_008297789 | Stegastes partitus | 95 | 2.0E-96 |
| 18 | c90876_g1_i1 | fatty-acid binding protein H6-isoform | h6-fabp | gb|AAC60352 | Notothenia coriiceps | 93 | 5.0E-86 |
| 19 | c32685_g1_i1 | PREDICTED: fatty-acid desaturase 6 | fads6 | ref|XP_010740675 | Larimichthys crocea | 90 | 0.0E+00 |
| 20 | c43378_g5_i1 | PREDICTED: ubiquitin-like protein ISG15 | igs15 | gb|ADJ57326 | Sciaenops ocellatus | 79 | 8.0E-83 |
| 21 | c43475_g1_i3 | PREDICTED: ERV-FRD provirus ancestral Env polyprotein-like | ervfrd-1 | ref|XP_004920373 | Xenopus (Silurana) tropicalis | 34 | 6.0E-16 |
| 22 | c40949_g1_i2 | PREDICTED: cystine/glutamate transporter | slc7a11 | ref|XP_008290185 | Stegastes partitus | 91 | 0.0E+00 |
| 23 | c35826_g2_i1 | PREDICTED: pleckstrin homology domain-containing family O member 2 | plekho2 | ref|XP_010746837 | Larimichthys crocea | 73 | 0.0E+00 |
| 24 | c83938_g1_i1 | NADH dehydrogenase subunit 2 | nad2 | gb|AFN88677 | Percina shumardi | 88 | 2.0E-28 |
| 25 | c80759_g1_i1 | ATP synthase F0 subunit 6 | mt-atp6 | ref|YP_007317144 | Pseudopentaceros richardsoni | 92 | 8.0E-34 |
| 26 | c19734_g1_i1 | PREDICTED: uncharacterized protein LOC102082036 | ref|XP_005478141 | Oreochromis niloticus | 78 | 6.0E-97 | |
| 27 | c43363_g4_i1 | PREDICTED: uncharacterized protein LOC100536992 | ref|XP_009301840 | Danio rerio | 39 | 3.0E-52 | |
| 28 | c43475_g1_i1 | PREDICTED: uncharacterized protein LOC101948489 | ref|XP_008168575 | Chrysemys picta bellii | 46 | 0.0E+00 | |
| 29 | c43269_g4_i2 | NO HIT | |||||
| 30 | c12262_g1_i1 | NO HIT | |||||
| 31 | c31915_g1_i1 | NO HIT | |||||
| 32 | c29645_g1_i1 | NO HIT | |||||
| 33 | c57351_g1_i1 | NO HIT | |||||
| 34 | c101392_g1_i1 | NO HIT | |||||
| 35 | c42887_g2_i1 | NO HIT |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wylie, M.J.; Symonds, J.E.; Setiawan, A.N.; Irvine, G.W.; Liu, H.; Elizur, A.; Lokman, P.M. Transcriptomic Changes during Previtellogenic and Vitellogenic Stages of Ovarian Development in Wreckfish (Hāpuku), Polyprion oxygeneios (Perciformes). Fishes 2019, 4, 16. https://doi.org/10.3390/fishes4010016
Wylie MJ, Symonds JE, Setiawan AN, Irvine GW, Liu H, Elizur A, Lokman PM. Transcriptomic Changes during Previtellogenic and Vitellogenic Stages of Ovarian Development in Wreckfish (Hāpuku), Polyprion oxygeneios (Perciformes). Fishes. 2019; 4(1):16. https://doi.org/10.3390/fishes4010016
Chicago/Turabian StyleWylie, Matthew J, Jane E Symonds, Alvin N Setiawan, Glen W Irvine, Hui Liu, Abigail Elizur, and P Mark Lokman. 2019. "Transcriptomic Changes during Previtellogenic and Vitellogenic Stages of Ovarian Development in Wreckfish (Hāpuku), Polyprion oxygeneios (Perciformes)" Fishes 4, no. 1: 16. https://doi.org/10.3390/fishes4010016
APA StyleWylie, M. J., Symonds, J. E., Setiawan, A. N., Irvine, G. W., Liu, H., Elizur, A., & Lokman, P. M. (2019). Transcriptomic Changes during Previtellogenic and Vitellogenic Stages of Ovarian Development in Wreckfish (Hāpuku), Polyprion oxygeneios (Perciformes). Fishes, 4(1), 16. https://doi.org/10.3390/fishes4010016







