Antimicrobial Peptides Are Expressed during Early Development of Zebrafish (Danio rerio) and Are Inducible by Immune Challenge
Abstract
1. Introduction
2. Results
2.1. Cloning and Identification of Zebrafish Antimicrobial Peptidess
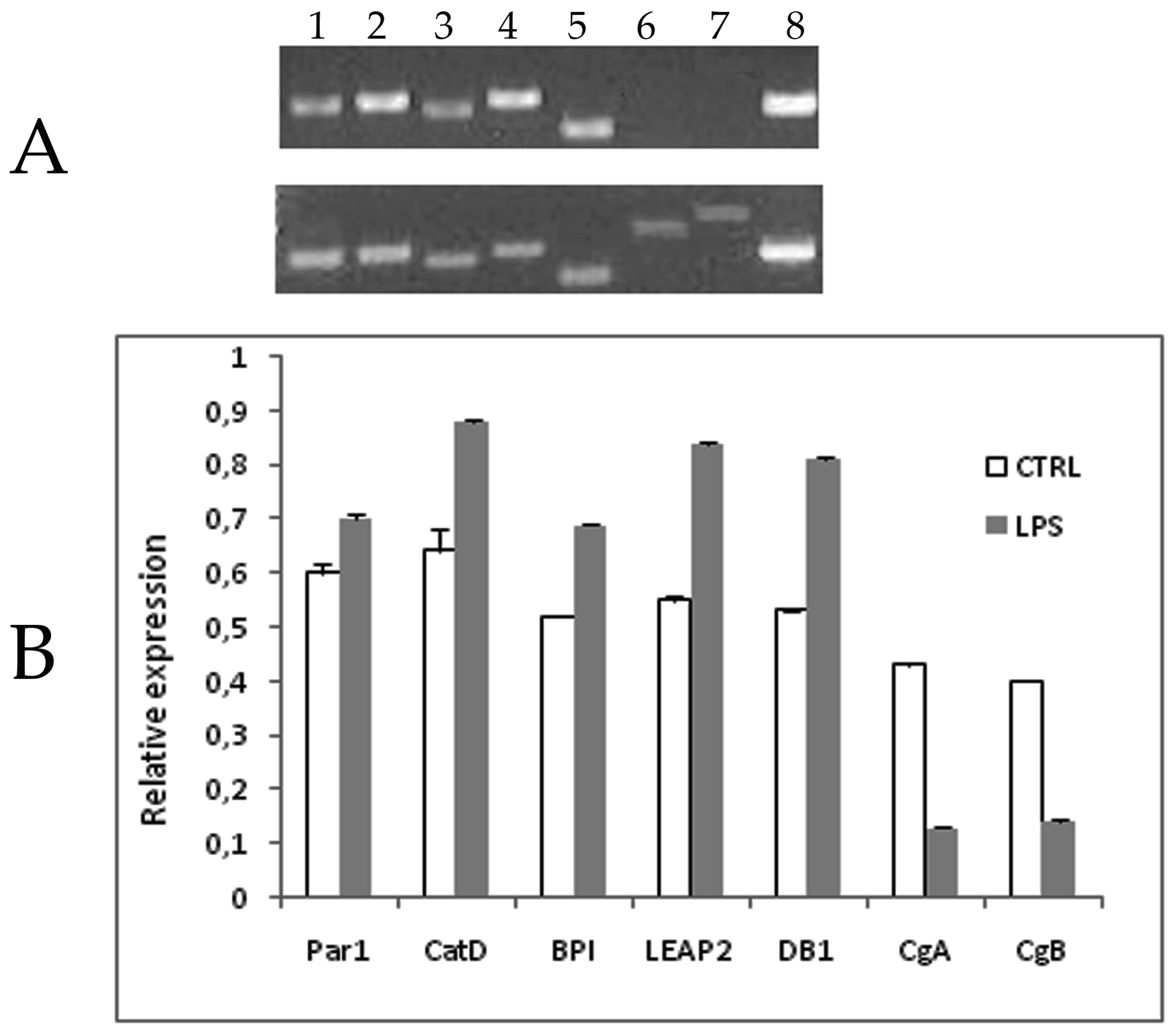
2.2. Ontogeny of AMP Gene Expression in Zebrafish and Response to Experimental Immune Challenge
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Extraction of Total RNA
4.3. RT-PCR Analysis
4.4. cDNA Electrophoresis
4.5. cDNA Purification from Agarose Gel and Cloning of cDNA Fragments
4.6. Bacterial Cultures and Transformation
4.7. Extraction of Plasmid DNA
4.8. Semi-Quantitative Analysis of AMP Expression Levels
4.9. Experimental Immune Challenge of Adult Zebrafish
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Park, C.H.; Valore, E.V.; Waring, A.J.; Ganz, T. Hepcidin, a urinary antimicrobial peptide synthesized in the liver. J. Biol. Chem. 2001, 276, 7806–7810. [Google Scholar] [CrossRef] [PubMed]
- Lauth, X.; Shike, H.; Burns, J.C.; Westerman, M.E.; Ostland, V.E.; Carlberg, J.M.; Van Olst, J.C.; Nizet, V.; Taylor, S.W.; Shimizu, C.; et al. Discovery and characterization of two isoforms of moronecidin, a novel antimicrobial peptide from hybrid striped bass. J. Biol. Chem. 2002, 277, 5030–5039. [Google Scholar] [CrossRef] [PubMed]
- Asmamaw, B. Hepcidin and its roles in fish. J. Zool. Stud. 2016, 3, 1–10. [Google Scholar]
- Katzenback, B.A. Antimicrobial peptides as mediators of innate immunity in teleosts. Biology (Basel) 2015, 4, 607–639. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Li, X.; Wang, Z. APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef] [PubMed]
- Park, C.B.; Lee, J.H.; Park, I.Y.; Kim, M.S.; Kim, S.C. A novel antimicrobial peptide from the loach, Misgurnusanguillicaudatus. FEBS Lett. 1997, 411, 173–178. [Google Scholar] [CrossRef]
- Douglas, S.E.; Patrzykat, A.; Pytyck, J.; Gallant, J.W. Identification, structure and differential expression of novel pleurocidins clustered on the genome of the winter flounder, Pseudopleuronectes americanus (Walbaum). Eur. J. Biochem. 2003, 270, 3720–3730. [Google Scholar] [CrossRef] [PubMed]
- Iijima, N.; Tanimoto, N.; Emoto, Y.; Morita, Y.; Uematsu, K.; Murakami, T.; Nakai, T. Purification and characterization of three isoforms of chrysophsin, a novel antimicrobial peptide in the gills of the red sea bream, Chrysophrys major. Eur. J. Biochem. FEBS 2003, 270, 675–686. [Google Scholar] [CrossRef]
- Shi, J.; Camus, A.C. Hepcidins in amphibians and fishes: Antimicrobial peptides or iron-regulatory hormones? Dev. Comp. Immunol. 2006, 30, 746–755. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.H.; Chen, J.Y.; Kuo, C.M. Three different hepcidins from tilapia, Oreochromis mossambicus: Analysis of their expressions and biological functions. Mol. Immunol. 2007, 44, 1922–1934. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Liu, Z.H.; Tewary, P.; Chen, Q.; de la Rosa, G.; Oppenheim, J.J. Defensin participation in innate and adaptive immunity. Curr. Pharm. Des. 2007, 13, 3131–3139. [Google Scholar] [CrossRef] [PubMed]
- Falco, A.; Mas, V.; Tafalla, C.; Perez, L.; Coll, J.M.; Estepa, A. Dual antiviral activity of human alpha-defensin-1 against viral haemorrhagic septicaemia rhabdovirus (VHSV): Inactivation of virus particles and induction of a type I interferon-related response. Antivir. Res. 2007, 76, 111–123. [Google Scholar] [CrossRef] [PubMed]
- Salerno, G.; Parrinello, N.; Roch, P.; Cammarata, M. cDNA sequence and tissue expression of an antimicrobial peptide, dicentracin; a new component of the moronecidin family isolated from head kidney leukocytes of sea bass, Dicentrarchus labrax. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2007, 146, 521–529. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.H.; Park, I.Y.; Kim, H.S.; Lee, W.T.; Kim, M.S.; Kim, S.C. Cathepsin D produces antimicrobial peptide parasin I from histone H2A in the skin mucosa of fish. FASEB J. 2002, 16, 429–431. [Google Scholar] [CrossRef] [PubMed]
- Bulet, P.; Stöcklin, R. Insect antimicrobial peptides: Structures, properties and gene regulation. Protein Pept. Lett. 2005, 12, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Mercier, C.; Koussounadis, A.; Secombes, C. Discovery of multiple beta-defensin-like homologues in teleost fish. Mol. Immunol. 2007, 44, 638–647. [Google Scholar] [CrossRef] [PubMed]
- Krause, A.; Sillard, R.; Kleemeier, B.; Klüver, E.; Maronde, E.; Conejo-García, J.R.; Forssmann, W.G.; Schulz-Knappe, P.; Nehls, M.C.; Wattler, F.; et al. Isolation and biochemical characterization of LEAP-2, a novel blood peptide expressed in the liver. Protein Sci. 2003, 12, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.A.; Zou, J.; Chang, C.I.; Secombes, C.J. Discovery and characterization of two types of liver-expressed antimicrobial peptide 2 (LEAP-2) genes in rainbow trout. Vet. Immunol. Immunopathol. 2004, 101, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Bao, B.; Peatman, E.; Xu, P.; Li, P.; Zeng, H.; He, C.; Liu, Z. The catfish liver-expressed antimicrobial peptide 2 (LEAP-2) gene is expressed in a wide range of tissues and developmentally regulated. Mol. Immunol. 2006, 43, 367–377. [Google Scholar] [CrossRef] [PubMed]
- Elsbach, P. The bactericidal/permeability-increasing protein (BPI) in antibacterial host defense. J. Leukoc. Biol. 1998, 64, 14–18. [Google Scholar] [PubMed]
- Hendy, G.N.; Bevan, S.; Mattei, M.G.; Mouland, A.J. Chromogranin A. Clin. Investig. Med. 1995, 18, 47–65. [Google Scholar]
- Briolat, J.; Wu, S.D.; Mahata, S.K.; Gonthier, B.; Bagnard, D.; Chasserot-Golaz, S.; Helle, K.B.; Aunis, D.; Metz-Boutigue, M.H. New antimicrobial activity for the catecholamine release-inhibitory peptide from chromogranin A. Cell. Mol. Life Sci. 2005, 62, 377–385. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Liu, S.; Hu, L.; Zhang, S. Characterization and bioactivity of hepcidin-2 in zebrafish: Dependence of antibacterial activity upon disulfide bridges. Peptides 2014, 57, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.; Nita-Lazar, M.; González-Montalbán, N.; Wang, J.; Mancini, J.; Ravindran, C.; Ahmed, H.; Vasta, G.R. Manipulating galectin expression in zebrafish (Danio rerio). Methods Mol. Biol. 2015, 1207, 327–341. [Google Scholar] [PubMed]
- Nita-Lazar, M.; Mancini, J.; Feng, C.; González-Montalbán, N.; Ravindran, C.; Jackson, S.; Heras-Sánchez, A.L.; Giomarelli, B.; Ahmed, H.; Haslam, S.M.; et al. The zebrafish galectins Drgal1-L2 and Drgal3-L1 bind in vitro to the infectious hematopoietic necrosis virus (IHNV) glycoprotein and reduce viral adhesion to fish epithelial cells. Dev. Comp. Immunol. 2016, 55, 241–252. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Yoon, H.; Minn, I.; Park, C.B.; Lee, W.T.; Zasloff, M.; Kim, S.C. Pepsin-mediated processing of the cytoplasmic histone H2A to strong antimicrobial peptide Buforin I. J. Immunol. 2000, 165, 3268–3274. [Google Scholar] [CrossRef] [PubMed]
- Birkemo, G.A.; Mantzilas, D.; Lüders, T.; Nes, I.F.; Nissen-Meyer, J. Identification and structural analysis of the antimicrobial domain in hipposin, a 51-mer antimicrobial peptide isolated from Atlantic halibut. Biochim. Biophys. Acta 2004, 1699, 221–227. [Google Scholar] [CrossRef]
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Brogden, K.A.; Guthmiller, J.M.; Salzet, M.; Zasloff, M. The nervous system and innate immunity: The neuropeptide connection. Nat. Immunol. 2005, 6, 558–564. [Google Scholar] [PubMed]
- Skarnes, R.C.; Watson, D.W. Antimicrobial factors of normal tissues and fluids. Bacteriol. Rev. 1957, 21, 273–294. [Google Scholar] [PubMed]
- Sarmasik, A.; Warr, G.; Chen, T.T. Production of transgenic medaka with increased resistance to bacterial pathogens. Mar. Biotechnol. 2002, 4, 310–322. [Google Scholar] [CrossRef] [PubMed]
- Cuesta, A.; Meseguer, J.; Esteban, M.A. The antimicrobial peptide hepcidin exerts an important role in the innate immunity against bacteria in the bony fish gilthead seabream. Mol. Immunol. 2008, 45, 2333–2342. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Claesson, H.E.; Mahshid, Y.; Klein, G.; Klein, E. Leukotriene B4 activates T cells that inhibit B-cell proliferation in EBV-infected cord blood-derived mononuclear cell cultures. Blood 2008, 111, 2693–2703. [Google Scholar] [CrossRef] [PubMed]
- Masso-Silva, J.A.; Diamond, G. Antimicrobial peptides from fish. Pharmaceuticals 2014, 7, 265–310. [Google Scholar] [CrossRef] [PubMed]
- Simon, J.P.; Aunis, D. Biochemistry of the chromogranin A protein family. Biochem. J. 1989, 262, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Metz-Boutigue, M.H.; Kieffer, A.E.; Goumon, Y.; Aunis, D. Innate immunity: Involvement of new neuropeptides. Trends Microbiol. 2003, 11, 585–592. [Google Scholar] [CrossRef] [PubMed]
- Miyakawa, M.; Matsui, T.; Kishikawa, H.; Murayama, R.; Uchiyama, I.; Itoh, T.; Yoshida, T. Salivary chromogranin A as a measure of stress response to noise. Noise Health 2006, 8, 108–113. [Google Scholar] [CrossRef] [PubMed]
- Turquier, V.R.; Vaudry, H.; Je Gou, S.; Anouar, Y. Frog chromogranin A messenger ribonucleic acid encodes three highly conserved peptides. Coordinate regulation of proopiomelanocortin and chromogranin a gene expression in the pars intermedia of the pituitary during background colour adaptation. Endocrinology 1999, 140, 4104–4112. [Google Scholar] [CrossRef] [PubMed]
- Ait-Ali, D.; Turquier, V.; Alexandre, D.; Grumolato, L.; Jegou, S.; Vaudry, H.; Anouar, Y. Molecular characterization of frog chromogranin B reveals conservation of selective sequences encoding potential novel regulatory peptides. FEBS Lett. 2002, 511, 127–132. [Google Scholar] [CrossRef]
- Strub, J.-M.; Garcia-Stablone, P.; Lonning, K.; Taupenot, L.; Hubert, P.; Van Dorsselaers, A.; Aunis, D.; Metz-Boutigue, M.C. Processing of chromogranin B in bovine adrenal medulla-Identification of secretolytin, the endogenous C-terminal fragment of residues 614–626 with antibacterial activity. Eur. J. Biochem. (FEBS) 1995, 229, 356–368. [Google Scholar] [CrossRef]
- Park, I.Y.; Park, C.B.; Kim, M.S.; Kim, S.C. Parasin I, an antimicrobial peptide derived from histone H2A in the catfish, Parasilurus asotus. FEBS Lett. 1998, 437, 258–262. [Google Scholar] [CrossRef]
- Diamond, G.M.; Beckloff, N.; Weinberg, A.; Kisich, K.O. The Roles of Antimicrobial Peptides in Innate Host Defense. Curr. Pharm. Des. 2009, 15, 2377–2392. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, J.M.; Molle, G.; Kemp, G.D.; Smith, V.J. Isolation and characterisation of oncorhyncin II, a histone H1-derived antimicrobial peptide from skin secretions of rainbow trout, Oncorhynchu smykiss. Dev. Comp. Immunol. 2004, 28, 127–138. [Google Scholar] [CrossRef]
- Birkemo, G.A.; Lüders, T.; Andersen, Ø.; Nes, I.F.; Nissen-Meyer, J. Hipposin, a histone-derived antimicrobial peptide in Atlantic halibut (Hippoglossus hippoglossus L.). Biochim. Biophys. Acta 2003, 1646, 207–215. [Google Scholar] [CrossRef]
- Tall, A. Plasma lipid transfer proteins. Annu. Rev. Biochem. 1995, 64, 235–257. [Google Scholar] [CrossRef] [PubMed]
- Ooi, C.E.; Weiss, J.; Doerfler, M.E.; Elsbach, P. Endotoxin-neutralizing properties of the 25 kD N-terminal fragment and a newly isolated 30 kD C-terminal fragment of the 55–60 kD bactericidal/permeability-increasing protein of human neutrophils. J. Exp. Med. 1991, 174, 649–655. [Google Scholar] [CrossRef] [PubMed]
- Horwitz, A.H.; Leigh, S.D.; Abrahamson, S.; Gazzano-Santoro, H.; Liu, P.S.; Williams, R.E.; Carroll, S.F.; Theofan, G. Expression and characterization of cysteine-modified variants of an amino-terminal fragment of bactericidal/permeability-increasing protein. Protein Expr. Purif. 1996, 8, 28–40. [Google Scholar] [CrossRef] [PubMed]
- Weiss, J.; Franson, C.; Schmeidler, K.; Elsbach, P. Reversible envelope effects during and after killing of Escherichia coli Wby a highly-purified rabbit polymorpho-nuclear leukocyte fraction. Biochim. Biophys. Acta 1976, 436, 154–169. [Google Scholar] [CrossRef]
- Solstad, T.; Stenvik, J.; Jørgensen, T.Ø. mRNA expression patterns of the BPI/LBP molecule in the Atlantic cod (Gadusmorhua L.). Fish Shellfish Immunol. 2007, 23, 260–271. [Google Scholar] [CrossRef] [PubMed]
- Romano, N.; Picchietti, S.; Taverne-Thiele, J.J.; Tavern, N.; Abelli, L.; Mastrolia, L.; Verburg-van Kemenade, B.M.L.; Rombout, J.H.W.M. Distribution of macrophages during fish development: An immunohistochemical study in carp (Cyprinuscarpio L.). Anat. Embryol. 1998, 198, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Ganz, T. Defensins: Antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 2003, 3, 710–720. [Google Scholar] [CrossRef] [PubMed]
- Lynn, D.J.; Lloyd, A.T.; Fares, M.A.; O’Farrelly, C. Evidence of positively selected sites in mammalian alpha-defensins. Mol. Biol. Evol. 2004, 21, 819–827. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Hughes, A.L.; Ando, J.; Matsuda, Y.; Cheng, J.F.; Skinner-Noble, D.; Zhang, G. A genome-wide screen identifies a single beta-defensin gene cluster in the chicken: Implications for the origin and evolution of mammalian defensins. BMC Genom. 2004, 5, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Selsted, M.E.; Ouellette, A.J. Mammalian defensins in the antimicrobial immune response. Nat. Immunol. 2005, 6, 551–557. [Google Scholar] [CrossRef] [PubMed]
- Diamond, G.; Kaiser, V.; Rhodes, J.; Russell, J.P.; Bevins, C.L. Transcriptional regulation of beta-defensin gene expression in tracheal epithelial cells. Infect. Immun. 2000, 68, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.H.; Lockwood, C.M. A comprehensive method to purify three major ANCA antigens: Proteinase 3, myeloperoxidase and bactericidal/permeability-increasing protein from human neutrophil granule acid extract. J. Immunol. Methods 1996, 197, 121–130. [Google Scholar] [CrossRef]
- Falco, A.; Chico, V.; Marroquí, L.; Perez, L.; Coll, J.M.; Estepa, A. Expression and antiviral activity of a beta-defensin-like peptide identified in the rainbow trout (Oncorhynchus mykiss) EST sequences. Mol. Immunol. 2008, 45, 757–765. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Neely, M.; Pfeifer, J.; Caparon, M.G. Streptococcus-zebrafish model of bacterial pathogenesis. Infect. Immun. 2002, 70, 3904–3914. [Google Scholar] [CrossRef] [PubMed]







| Target | Product Size (bp) | Forward (5′–3′) | Reverse (5′–3′) |
|---|---|---|---|
| zfParI | 350 | CGGCAAAGCTAGAGCTAAGG | CAAGAAGACCGAGAAGGCTG |
| zfDB1 * | 242 | GAAAACTGGAGCTCCTGATC | ATGCTCTGGGGCTTCATGTT |
| zfLEAP2 | 360 | CAGTGCCTACTGCCAGAACA | GGATTGGCATGACAGTTTTG |
| zfCATD | 362 | GGAGGATGTGAAGCCATTGT | AACAGAGTGGGCTTCGCTAA |
| zfBPI | 325 | CATCTCAGCAAAGACGACGA | TATGGATTTCAGGGGCATGT |
| zfChA | 463 | CATCTCCTGGGAGTTTCAGC | TTTCGTCGGTTCCTCAGATC |
| zfChB | 504 | TTCCCTCACGTTTACCCTTG | TCTCCTGTCTTTGTTATCCGG |
| zfACT | 380 | GACAACGGCTCCGGTATG | TCTCTGTTGGTTTGGGATT |
| Gene | Gene Product (aa/kDa) | Similar Polypeptide (aa) | % Identity | Organism | Accession n. |
|---|---|---|---|---|---|
| CgA | 246/27,806 | 34 | Bos taurus | AAB21297 | |
| 31 | Rana ridibunda | AAD38522 | |||
| 31 | Homo sapiens | EDW81506 | |||
| CgB | 677/81,950 | 37 | Homo sapiens | NP001810 | |
| 22 | Bos taurus | 18142994 | |||
| 20 | Rana ridibunda | AAL76931 | |||
| Par I | 154/16,741 | unamed protein product | 100 | Tetraodon negroviridis | CAF98588 |
| histone H2A | 99 | Oncorhynchus mykiss | CAA25528 | ||
| histone H2A | 96 | Xenopus tropicalis | CAJ82985 | ||
| similar to histone H2A | 93 | Canis familiaris | XP545430 | ||
| similar to histone H2A | 86 | Rattus norvegicus | XP001062486 | ||
| CatD | 398/43,187.51 | cathepsin D prepo | 86 | Silarus asotus | AAM62283 |
| cathepsin D precursor | 85 | Clupea harengus | AAG27733 | ||
| cathepsin D | 83 | Oncorhynchus mykiss | AAC60301 | ||
| cathepsin D1 | 80 | Takifugu rubripes | NP001072052 | ||
| unanamed protein prod. | 77 | Tetraodon negroviridis | CAF91576 | ||
| cathepsin D precursor | 75 | Chionodraco hamatus | CAA07719 | ||
| 67 | Homo sapiens | NP001900 | |||
| BPI | 487/54,463 | unnamed protein product | 73 | Tetraodon negroviridis | CAF90160 |
| phospholipid transfer protein | 70 | Xenopus tropicalis | AAI36237 | ||
| similar to phospholipid | 64 | Gallus gallus | XP425722 | ||
| phospholipid | 60 | Homo sapiens | EAW75784 | ||
| pbi | 23 | Cyprinus carpio | BAC56095 | ||
| Om2 | 23 | Oncorhynchus mykiss | |||
| LEAP-2 | 92/10,212 | leap2 | 62 | Ictalurus punctatus | AAX45791 |
| leap2 | 62 | Ictalurus furcatus | AAX45792 | ||
| leap2 isoform A | 61 | Oncorhynchus mykiss | AAR11766 | ||
| unnamed protein product | 59 | Tetraodon negroviridis | CAF87882 | ||
| peptide2B | 47 | Oncorhynchus mykiss | AAR11767 | ||
| 43 | Homo sapiens | NP443203 | |||
| DB1 | 67/7263 | unnamed protein product | 85 | Tetraodon negroviridis | CAG02912 |
| beta defensin like peptides 1 | 78 | Oncorhynchus mykiss | CAK54951 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caccia, E.; Agnello, M.; Ceci, M.; Strickler Dinglasan, P.; Vasta, G.R.; Romano, N. Antimicrobial Peptides Are Expressed during Early Development of Zebrafish (Danio rerio) and Are Inducible by Immune Challenge. Fishes 2017, 2, 20. https://doi.org/10.3390/fishes2040020
Caccia E, Agnello M, Ceci M, Strickler Dinglasan P, Vasta GR, Romano N. Antimicrobial Peptides Are Expressed during Early Development of Zebrafish (Danio rerio) and Are Inducible by Immune Challenge. Fishes. 2017; 2(4):20. https://doi.org/10.3390/fishes2040020
Chicago/Turabian StyleCaccia, Elisabetta, Maria Agnello, Marcello Ceci, Patricia Strickler Dinglasan, Gerardo R. Vasta, and Nicla Romano. 2017. "Antimicrobial Peptides Are Expressed during Early Development of Zebrafish (Danio rerio) and Are Inducible by Immune Challenge" Fishes 2, no. 4: 20. https://doi.org/10.3390/fishes2040020
APA StyleCaccia, E., Agnello, M., Ceci, M., Strickler Dinglasan, P., Vasta, G. R., & Romano, N. (2017). Antimicrobial Peptides Are Expressed during Early Development of Zebrafish (Danio rerio) and Are Inducible by Immune Challenge. Fishes, 2(4), 20. https://doi.org/10.3390/fishes2040020






