Assessing Fish Diversity in the Chishui River Using Environmental DNA (eDNA) Metabarcoding
Abstract
1. Introduction
2. Materials and Methods
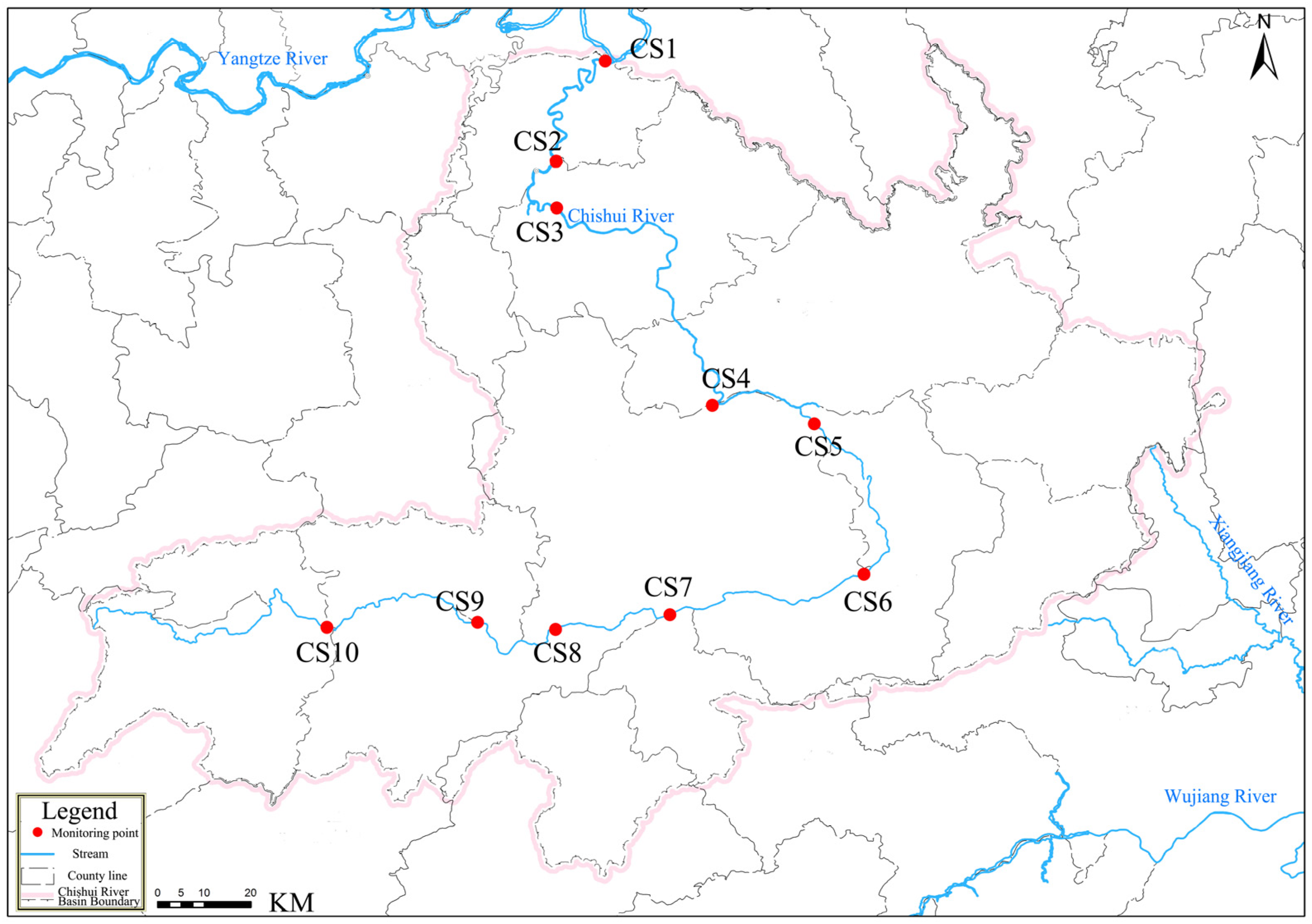
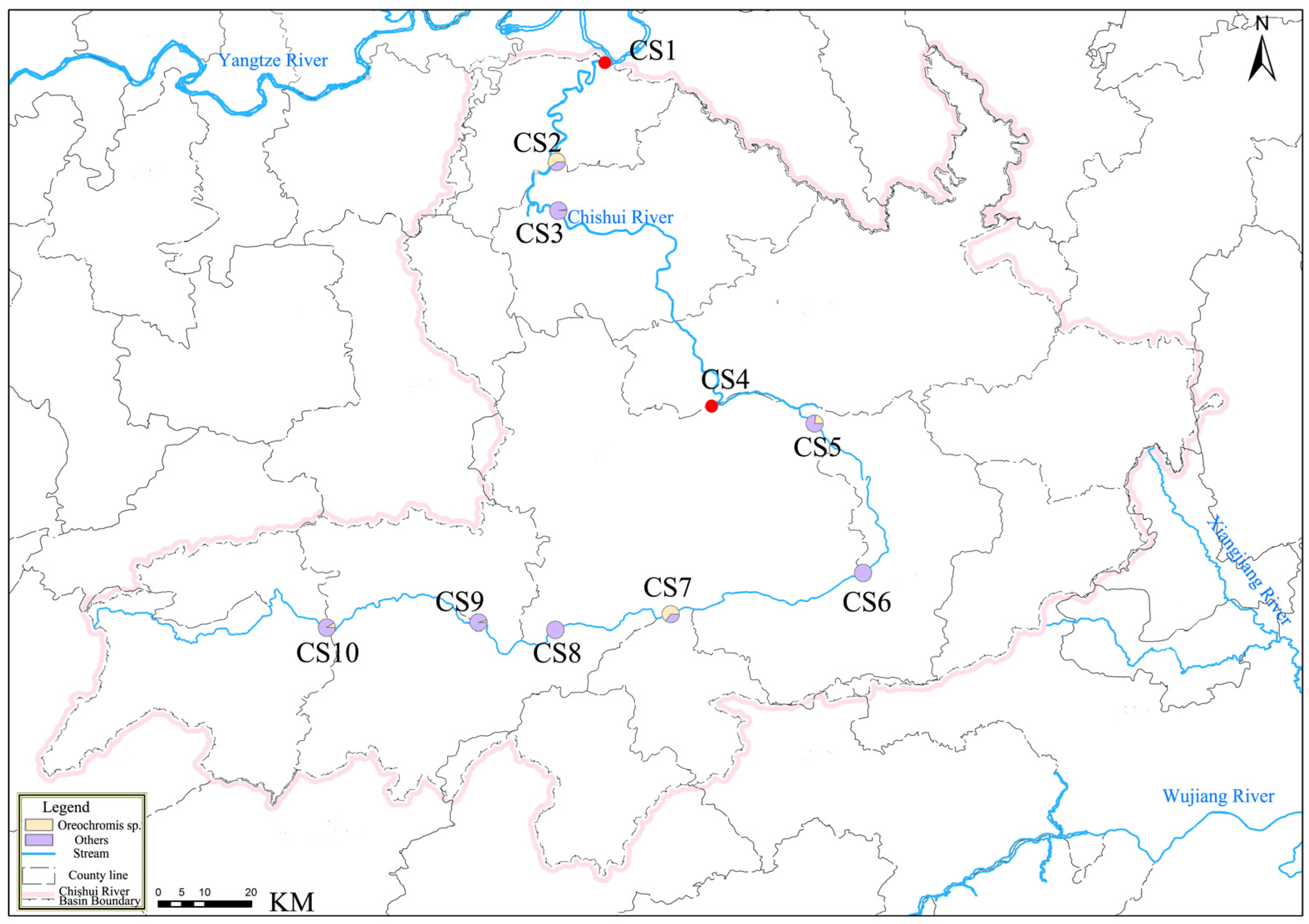
2.1. Study Area and Sample Collection
2.2. DNA Extraction, PCR Amplification, and High-Throughput Sequencing
2.3. Data Analysis
3. Results
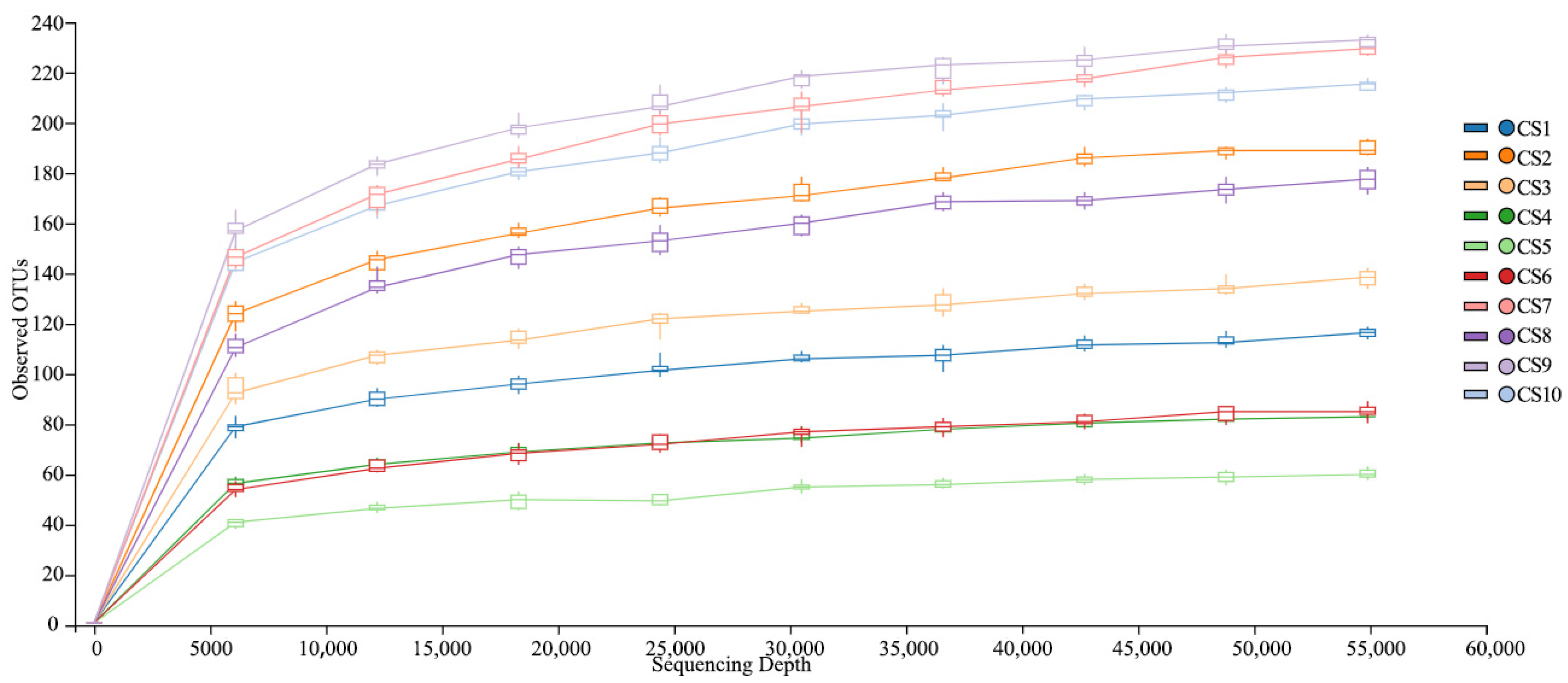
3.1. Sequencing Statistics
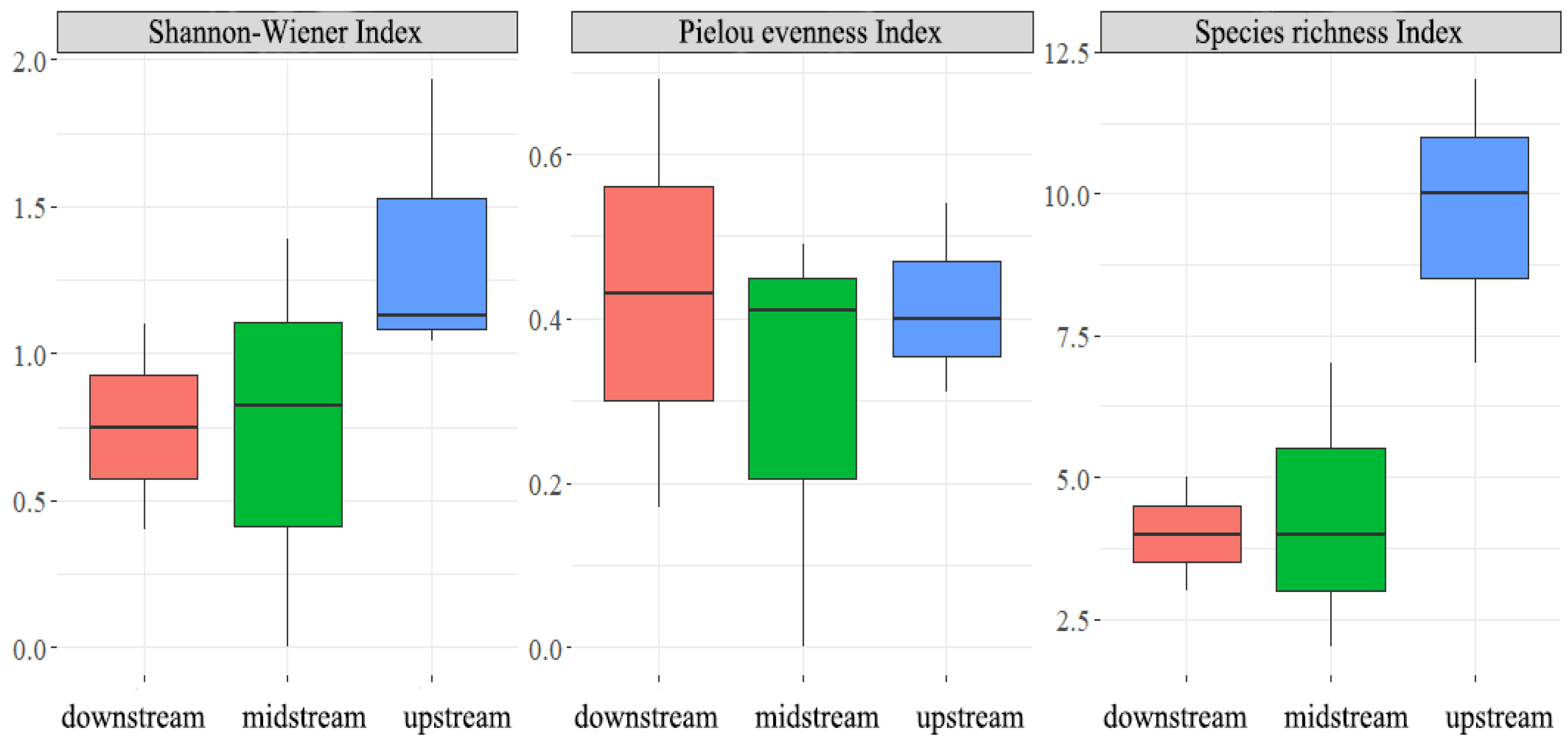
3.2. Alpha Diversity in the Chishui River Fish Community
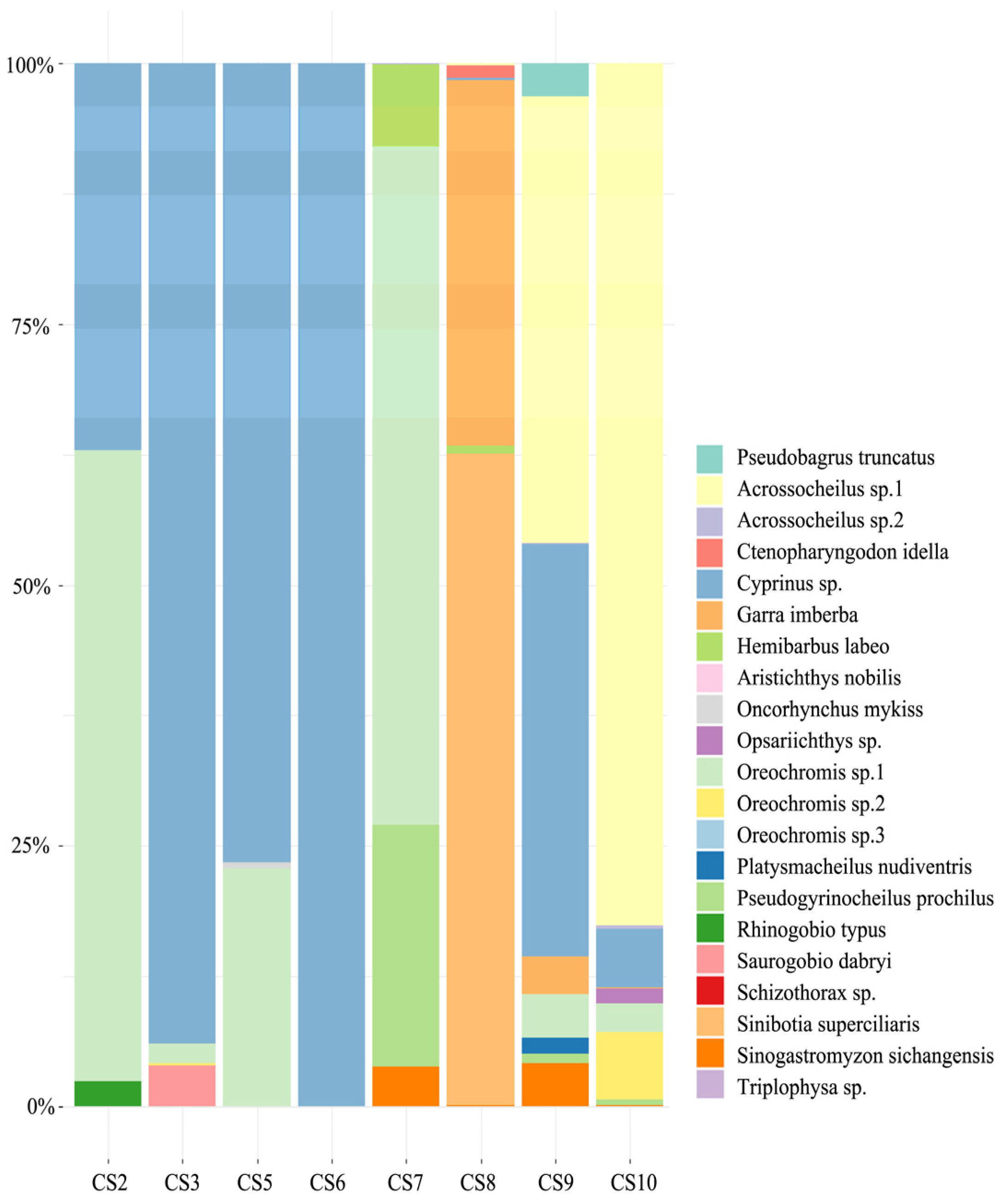
3.3. Fish Community Composition
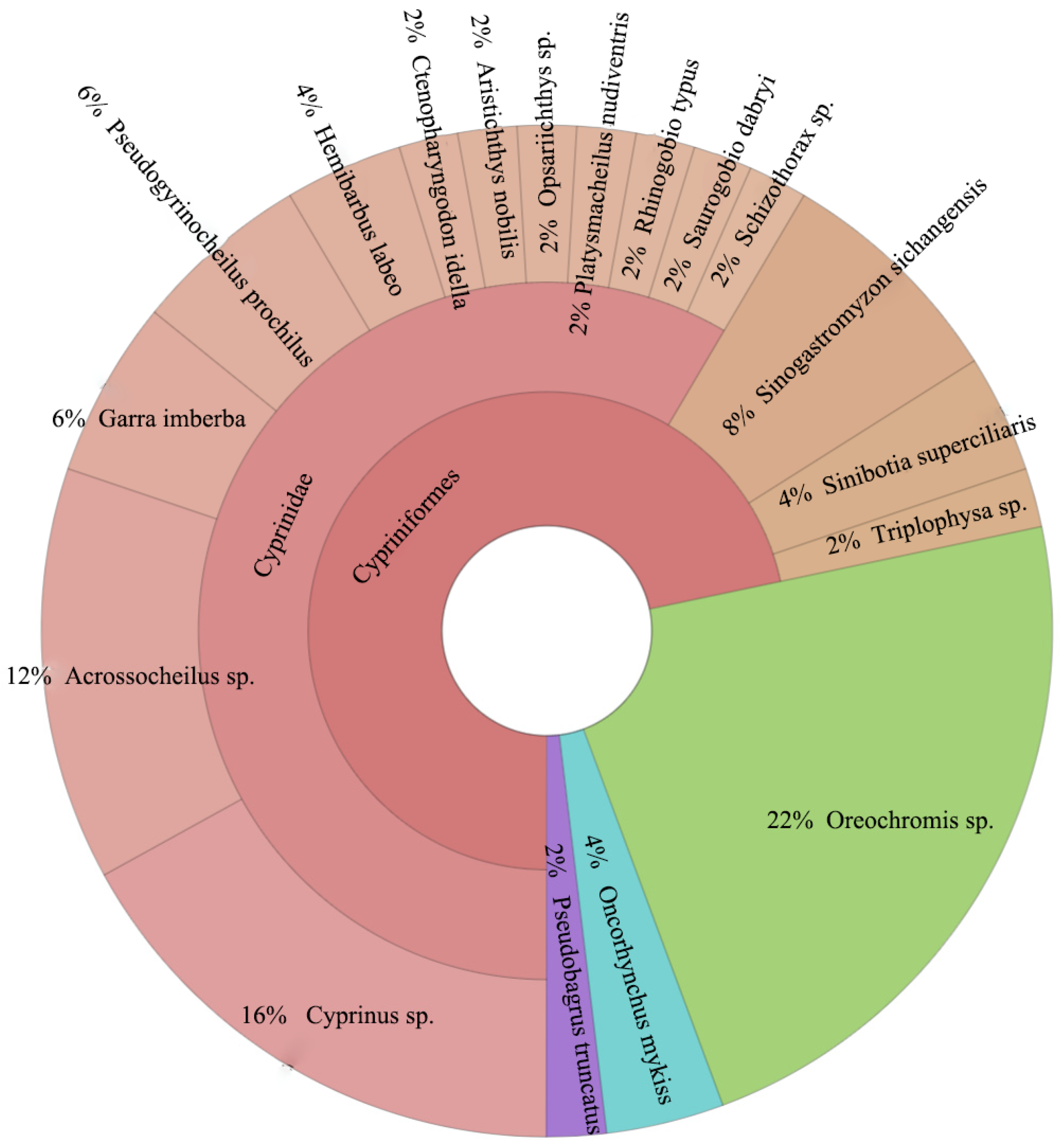
3.4. Fish Species Composition
4. Discussion
4.1. Prospects of Investigating Fish Communities Using Environmental DNA Technology
4.2. Diversity and Spatial Structure of Fish Communities in the Chishui River Based on Environmental DNA
4.3. Potential Risks to the Chishui River Fish Community
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Appendix A
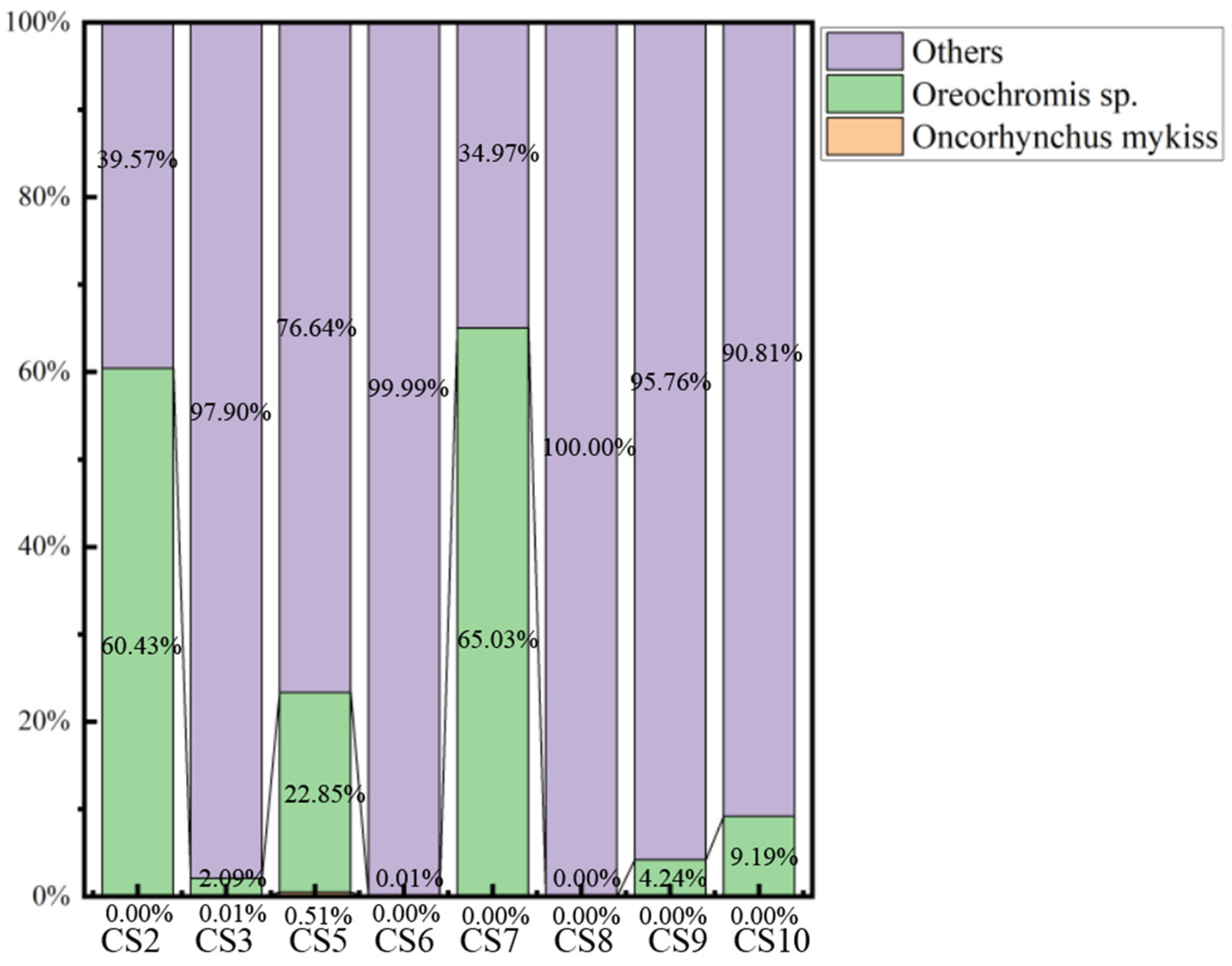
Appendix B
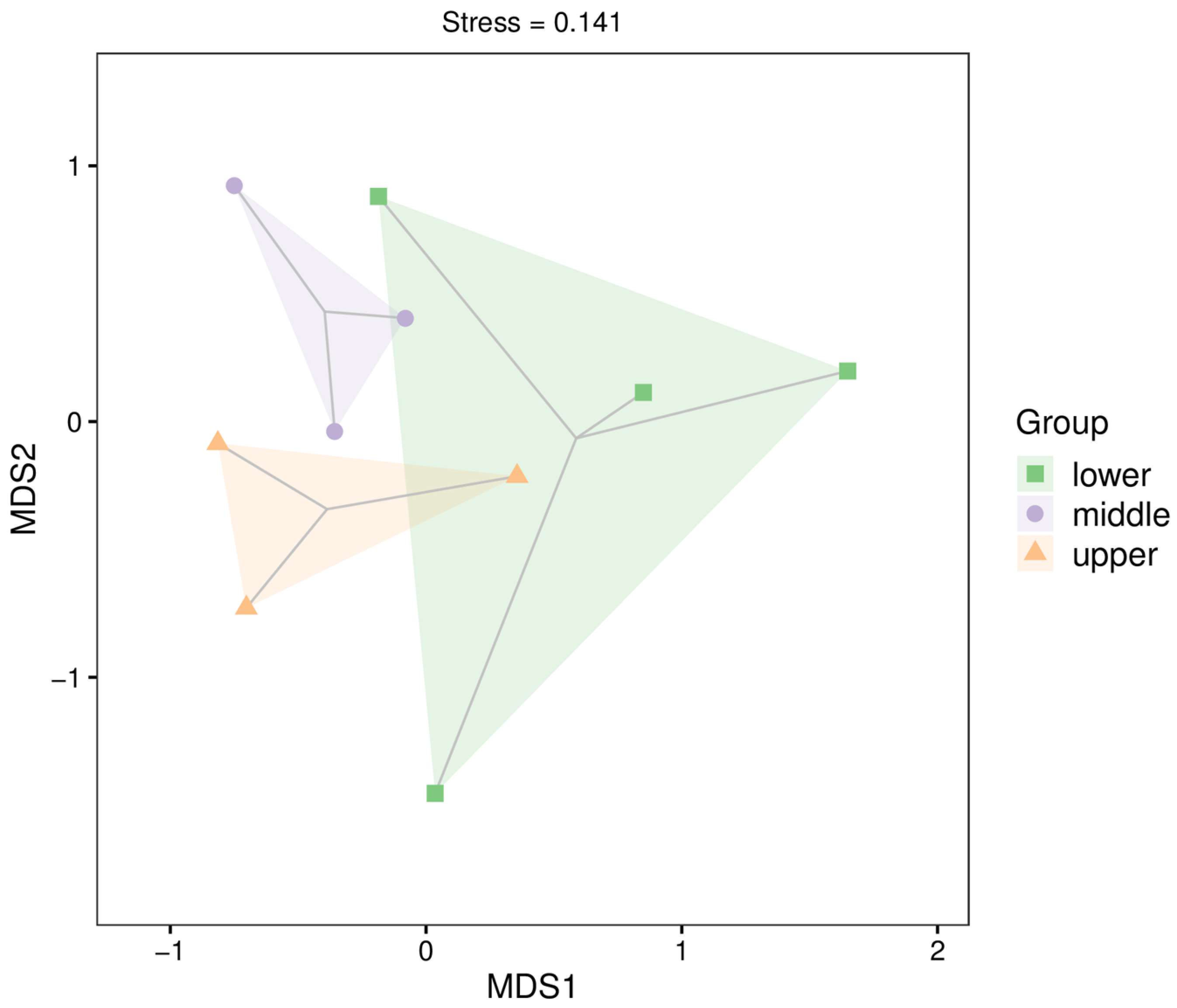
References
- Zhang, Y.; Xiao, D.A.; Yu, T.; Li, G.X.; Wu, Z.Y. Study on Trans-Provincial Lateral Water Environmental Compensation Mechanism in Chishui River Basin. Environ. Prot. Technol. 2019, 25, 23–27. [Google Scholar]
- Tang, R.; Dai, Y.X.; Liu, H.Z.; Ai, Z.J.; Ouyang, D.D.; Peng, Y.; Tang, Y.; Yu, D. Introduction and Ecological Adaptability Analysis of Exotic Fish Species Rhynchocypris oxycephalus and Barbatula toni in Chishui River. Zool. Res. 2021, 56, 214–228. [Google Scholar] [CrossRef]
- Li, L.; Yuan, W.L.; Liu, F. Status of Fish Resources in the Jiang Segment of Chishui River, Chishui City. Biodivers. Sci. 2015, 24, 1884–1890. [Google Scholar]
- Liu, F.; Liu, D.M.; Yuan, D.C.; Zhang, F.B.; Wang, X.; Zhang, Z.; Qin, Q.; Wang, J.W.; Liu, H.Z. Interannual Variations of Fish Community in Different Sections of Chishui River over the Past Decade. Acta Hydrobiol. Sin. 2020, 44, 122–132. [Google Scholar]
- Fowler, C.M.; Ogburn, M.B.; Aguilar, R.; Heggie, K.; Legett, H.D.; Richie, K.D.; Plough, L.V. Viability of High-Frequency Environmental DNA (eDNA) Sampling as a Fish Enumeration Tool. Ecol. Indic. 2024, 166, 112384. [Google Scholar] [CrossRef]
- Goldberg, C.S.; Pilliod, D.S.; Arkle, R.S.; Waits, L.P. Molecular Detection of Vertebrates in Stream Water: A Demonstration Using Rocky Mountain Tailed Frogs and Idaho Giant Salamanders. PLoS ONE 2011, 6, e22746. [Google Scholar] [CrossRef]
- Takahara, T.; Minamoto, T.; Yamanaka, H.; Doi, H.; Kawabata, Z. Estimation of Fish Biomass Using Environmental DNA. PLoS ONE 2012, 7, e35868. [Google Scholar] [CrossRef]
- Kelly, R.P.; Port, J.A.; Yamahara, K.M.; Crowder, L.B. Using Environmental DNA to Census Marine Fishes in a Large Mesocosm. PLoS ONE 2014, 9, e86175. [Google Scholar] [CrossRef]
- Sales, N.G.; Wangensteen, O.S.; Carvalho, D.C.; Deiner, K.; Præbel, K.; Coscia, I.; McDevitt, A.D.; Mariani, S. Space-Time Dynamics in Monitoring Neotropical Fish Communities Using eDNA Metabarcoding. Sci. Total Environ. 2021, 754, 142096. [Google Scholar] [CrossRef]
- Chen, Z.; Xu, J.; Yang, F.; Hou, Z.; Ren, K.; Yu, L.; Yang, S.; Li, Z.; Zhang, X. Comprehensive Monitoring and Ecological Risk Assessment of Heavy Metals in Soil and Surface Water of Chishui River Basin in Upper Reaches of the Yangtze River. Water 2023, 15, 2069. [Google Scholar] [CrossRef]
- Liu, Q.; He, D.; Ma, Y.; Wang, H.; Li, Y.; Cheng, Y.; Huang, Y. Sensory Profile and the Contribution of Key Aroma Compounds in Jiang-Flavor Rounded-Base Baijiu Produced in the Chishui River Basin. LWT Food Sci. Technol. 2023, 189, 115474. [Google Scholar] [CrossRef]
- Liu, F.; Wang, J.; Cao, W. Long-Term Changes in Fish Assemblage Following the Impoundments of the Three Gorges Reservoir in Hejiang, a Protected Reach of the Upper Yangtze River. Knowl. Manag. Aquat. Ecosyst. 2012, 407, 6. [Google Scholar] [CrossRef]
- Minamoto, T.; Yamanaka, H.; Takahara, T.; Honjo, M.N.; Kawabata, Z. Surveillance of fish species composition using environmental DNA. Limnology 2012, 13, 193–197. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A Versatile Open Source Tool for Metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Ryberg, M.; Kristiansson, E.; Abarenkov, K.; Larsson, K.-H.; Kõljalg, U. Taxonomic Reliability of DNA Sequences in Public Sequence Databases: A Fungal Perspective. PLoS ONE 2006, 1, e59. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and Applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Dixon, P. VEGAN, a Package of R Functions for Community Ecology. J. Veg. Sci. 2003, 14, 927–930. [Google Scholar] [CrossRef]
- Ondov, B.D.; Bergman, N.H.; Phillippy, A.M. Interactive Metagenomic Visualization in a Web Browser. BMC Bioinform. 2011, 12, 385. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Peng, S.; Shen, X.; Lin, Z.; Gong, L.; Zhang, R.; Huang, B. Multiple Scale Impacts of Land Use Intensity on Water Quality in the Chishui River Source Area. Ecol. Indic. 2024, 166, 112396. [Google Scholar] [CrossRef]
- Qu, C.Y. Discussion on Inland Fishery Resources Survey—Methods and Standards of Fish Survey. Henan Fish. 2012, 42, 6–8. [Google Scholar]
- Wang, R.; Lin, L.S.; Li, H.; Zhang, R.; Li, Y. Potential Application of Environmental DNA Method in Fish Diversity Research in Arctic Region. Polar Res. 2021, 33, 148–155. [Google Scholar] [CrossRef]
- Zhou, H.H.; Li, C.; Deng, H.T.; Tian, H.W.; Chen, Y.B.; Gao, X.; Zhu, B.; Chen, D.Q. Analysis of Population Dynamics and Trend of Rare and Endemic Fish Species in the Upper Yangtze River. Freshw. Fish. 2020, 50, 3–14. [Google Scholar] [CrossRef]
- Zhou, R.; Zheng, W.C. Exploration of Fish Species in Main Streams of Dalou Mountains. Chin. Fish. 2018, 11, 102–106. [Google Scholar]
- Li, M.; Wei, T.T.; Shi, B.Y.; Hao, X.Y.; Xu, H.G.; Sun, H.Y. Application of Environmental DNA Technology in Monitoring Diversity of Freshwater Benthic Macroinvertebrates. Biodivers. Sci. 2019, 27, 480–490. [Google Scholar]
- Shpiler, A.R.; Dorfman, B.; Tadmor-Levi, R.; Marcos-Hadad, E.; Perelberg, A.B.; Naor, A.; David, L. Differences in Growth, Pond Survival and CyHV-3 Disease Survival between Genetically CyHV-3 Resistant and Commercial Strains of Common Carp (Cyprinus carpio). Aquaculture 2025, 595, 741584. [Google Scholar] [CrossRef]
- Zheng, C.Q.; Wu, W.; Wei, J.; Zhuang, H.B.; Sang, G.Q. Analysis of Fish Diversity and Influencing Factors in Dawen River, a Tributary of Lower Yellow River. Water Resour. Prot. 2020, 36, 31–38. [Google Scholar]
- Cen, Z.L.; Hou, X.F.; Zhou, J. Investigation of Exotic Fish Species in Guizhou Province. J. Guizhou Norm. Univ. 2013, 31, 16–19. [Google Scholar] [CrossRef]
- Zhao, L.C.; Wu, Z.Q.; Zhang, M.; Zheng, X.; Li, D.Y.; Liu, B.W. Investigation and Analysis of Wild Tilapia Group Formation in Major Water Systems in Southern Guangxi. J. South Agric. 2019, 50, 397–404. [Google Scholar]
- Wan, C.Y.; Fang, K.; Dong, F.; Zhang, Y.Y.; Wu, J.M.; Mou, X.D.; Zhang, H. Current Status and Ecological Risk Assessment of Exotic Fish Species in the Yangtze River Basin. J. Fish. China 2024. Available online: http://kns.cnki.net/kcms/detail/31.1283.s.20240517.1352.002.html (accessed on 28 May 2025).
- Li, S.; Chen, J.K.; Wang, X.M. Distribution, Invasion Pathways, Mechanisms, and Consequences of Freshwater Fish Invasion Species. Biodivers. Sci. 2016, 24, 672–685. [Google Scholar] [CrossRef]
- Pan, Y.; Cao, W.X.; Xu, L.P.; Yin, S.R. History and Pathways of Fish Invasion in China and Abroad. J. Dalian Fish. Univ. 2006, 21, 72–78. [Google Scholar] [CrossRef]
- Liu, Q.G.; Shen, J.Z.; Chen, M.K.; Tong, H.Y.; Li, J.L.; Chen, L.Q. Research Progress on the Miniaturization Issue of Natural Economic Fish Species. J. Shanghai Ocean Univ. 2005, 14, 79–83. [Google Scholar]

| Species | Native/Invasive | Characteristic |
|---|---|---|
| Oreochromis sp. | Invasive | Tropical species |
| Oncorhynchus mykiss | Invasive | Cold-water species, rarely captured by manual fishing |
| Pseudobagrus truncatus | Native | Uncommon; captured annually in low numbers |
| Triplophysa sp. | Native | Rare |
| Sinogastromyzon sichangensis | Native | Seasonally abundant; often used as duck feed |
| Sinibotia superciliaris | Native | Moderately abundant |
| Acrossocheilus sp. | Native | Common species |
| Ctenopharyngodon idella | Native | Rare in natural rivers; likely escapees from aquaculture; scarce in the Chishui River |
| Cyprinus sp. | Native | Rare in natural rivers; likely escapees from aquaculture; scarce in the Chishui River |
| Garra imberba | Native | Rare |
| Hemibarbus | Native | Common species |
| Hypophthalmichthys nobilis | Native | Rare in natural rivers; likely escapees from aquaculture; scarce in the Chishui River |
| Opsariichthys sp. | Native | Uncommon |
| Platysmacheilus nudiventris | Native | Rare |
| Pseudogyrinocheilus prochilus | Native | Uncommon; widely distributed in upper tributaries of the Pearl River Basin, China |
| Rhinogobio typus | Native | Highly abundant |
| Saurogobio dabryi | Native | Highly abundant |
| Schizothorax | Native | Uncommon |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, J.; Zhang, J.; Pan, C.; Xu, S.; Wu, Y.; Lv, W.; Hong, M.; Hu, Y.; Wang, Y. Assessing Fish Diversity in the Chishui River Using Environmental DNA (eDNA) Metabarcoding. Fishes 2025, 10, 279. https://doi.org/10.3390/fishes10060279
Gao J, Zhang J, Pan C, Xu S, Wu Y, Lv W, Hong M, Hu Y, Wang Y. Assessing Fish Diversity in the Chishui River Using Environmental DNA (eDNA) Metabarcoding. Fishes. 2025; 10(6):279. https://doi.org/10.3390/fishes10060279
Chicago/Turabian StyleGao, Jing, Jing Zhang, Chengrong Pan, Sheng Xu, Yajie Wu, Wei Lv, Min Hong, Yuxin Hu, and Yingru Wang. 2025. "Assessing Fish Diversity in the Chishui River Using Environmental DNA (eDNA) Metabarcoding" Fishes 10, no. 6: 279. https://doi.org/10.3390/fishes10060279
APA StyleGao, J., Zhang, J., Pan, C., Xu, S., Wu, Y., Lv, W., Hong, M., Hu, Y., & Wang, Y. (2025). Assessing Fish Diversity in the Chishui River Using Environmental DNA (eDNA) Metabarcoding. Fishes, 10(6), 279. https://doi.org/10.3390/fishes10060279




