Research on Morphometric Methods for Larimichthys crocea Based on YOLOv11-CBAM X-Ray Imaging
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Source
2.2. Manual Measurements
2.3. Data Acquisition
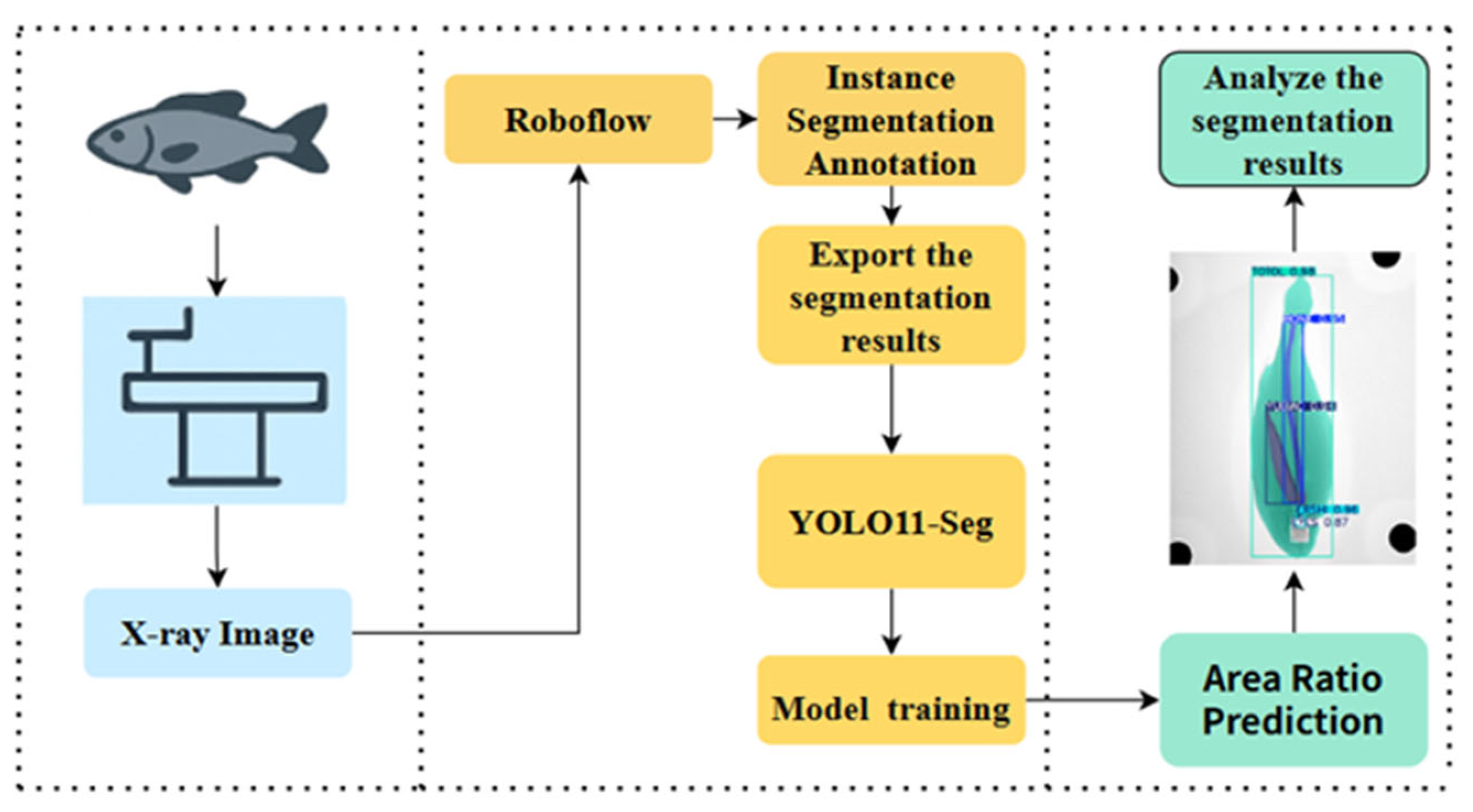
2.4. Dataset Construction
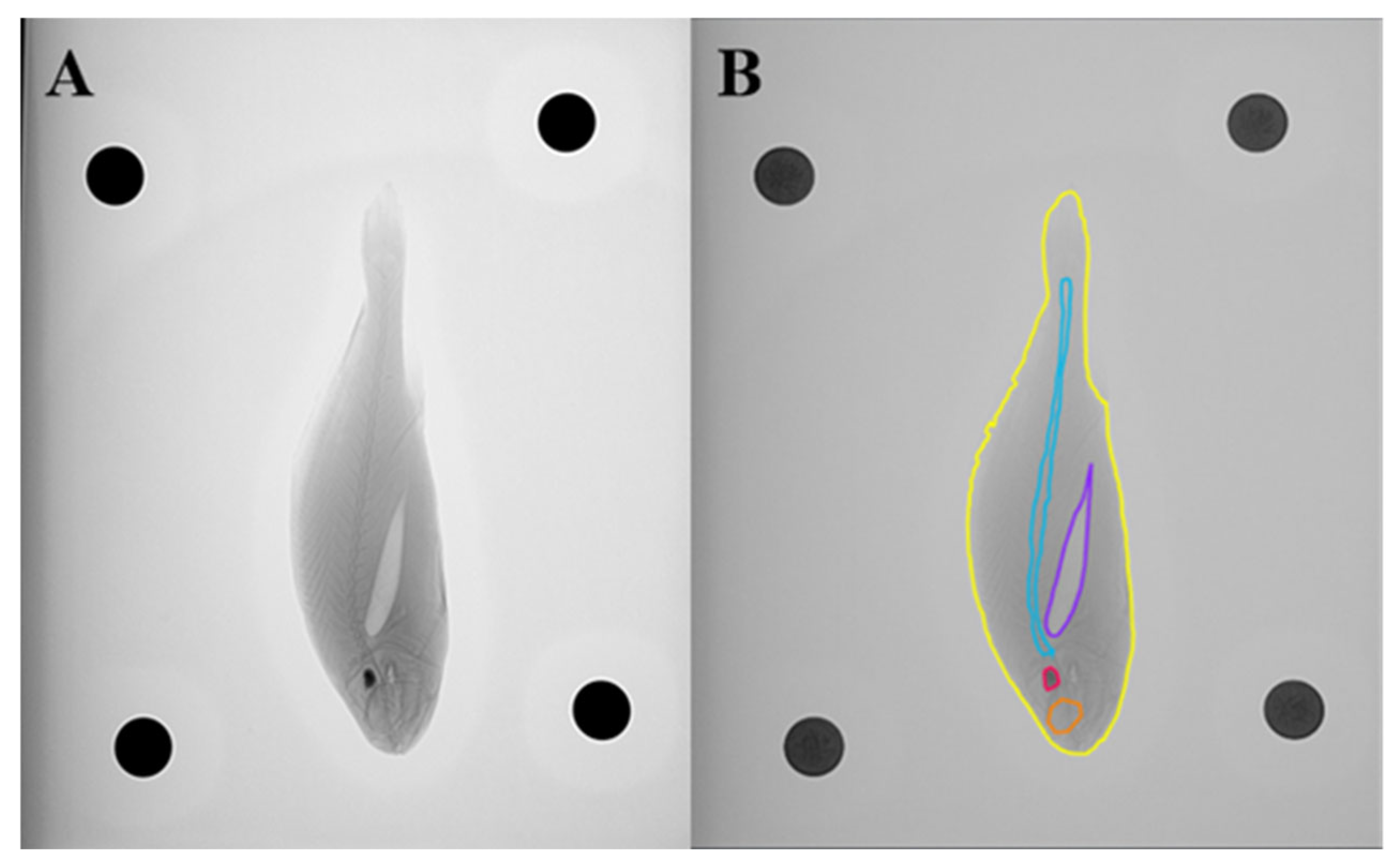
2.5. Instance Segmentation and Area Calculation
2.6. Instance Segmentation Model
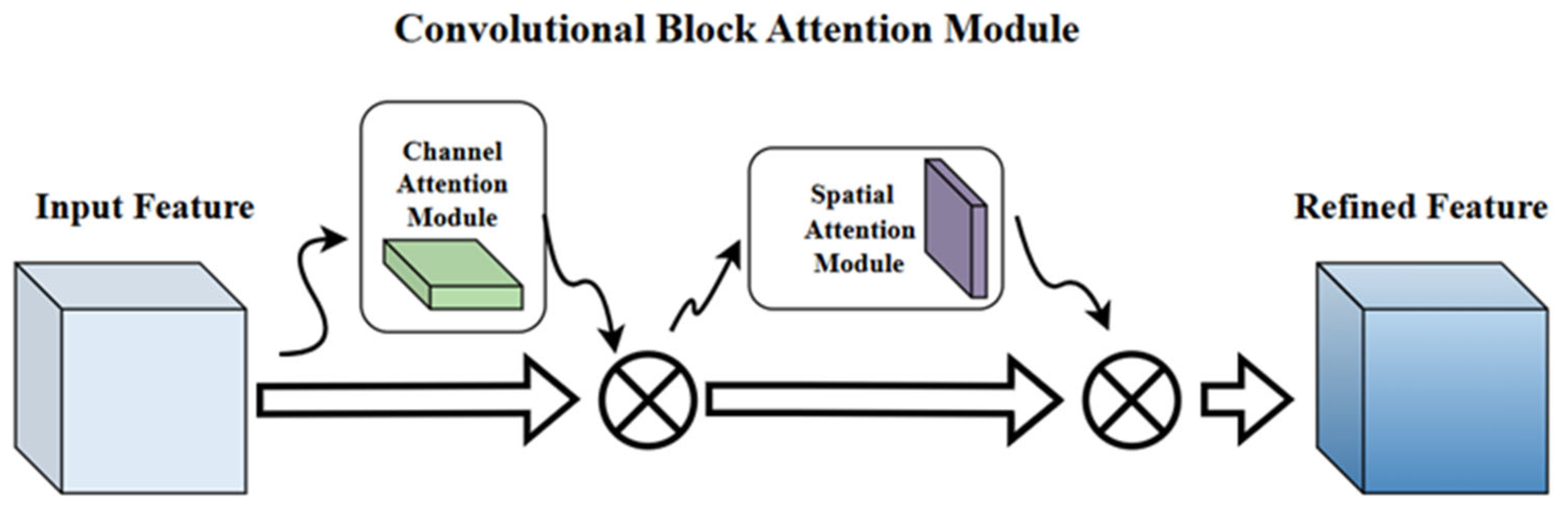
2.7. CBAM-Enhanced Model
2.8. Experimental Environment and Model Performance Evaluation
2.9. Calculation of Morphological Parameters of Large Yellow Croaker
2.9.1. Instance Area Calculation Method
2.9.2. Data Processing
2.9.3. Error Control
- (1)
- Images in which the Hough circle transform failed to stably detect the coin were excluded from area calibration, thereby avoiding errors in the scale factor;
- (2)
- Images for which the segmentation model did not return valid mask information were discarded to prevent false-positive results;
- (3)
- Multiple-circle detection was applied in coin calibration, and the average value was used to reduce single-detection error;
- (4)
- During category statistics, strict consistency was maintained between model output labels and the predefined dictionary to prevent computational deviations caused by label mismatches.
2.10. Calibration Error Assessment
2.11. Statistical Analysis and Uncertainty Quantification
3. Results
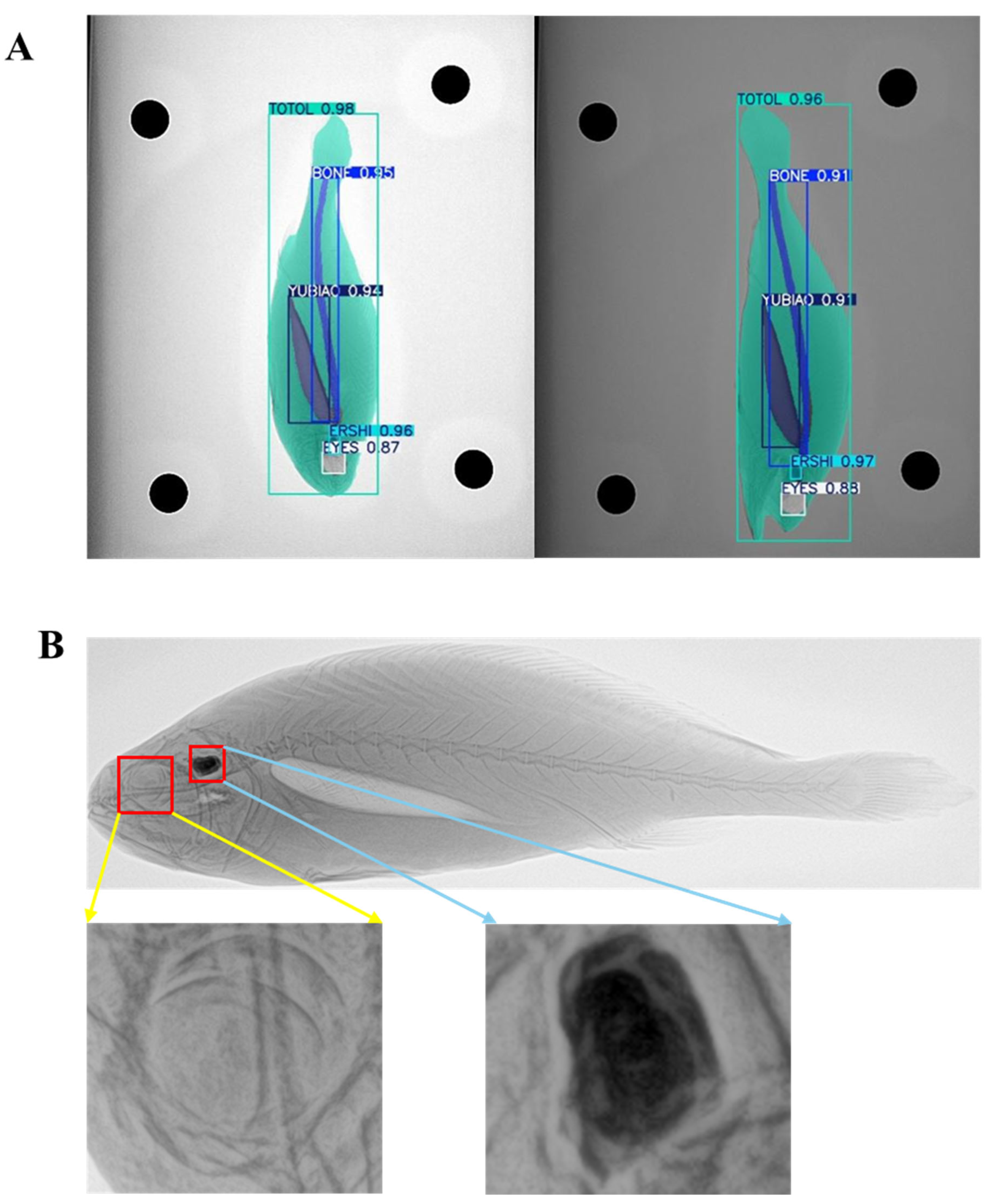
3.1. Performance Evaluation of the Instance Segmentation Model for Large Yellow Croaker
3.2. Analysis of Instance Segmentation Data
4. Discussion
4.1. Biological Interpretation of Internal Morphometric Traits
4.2. Technical Challenges and Uncertainty in Small-Object Segmentation
4.3. Dataset Constraints and Strategies for Scalable Expansion
4.4. Cross-Species Generalization and Future Research Directions
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yuan, J.; Lin, H.; Wu, L.; Zhuang, X.; Ma, J.; Kang, B.; Ding, S. Resource Status and Effect of Long-Term Stock Enhancement of Large Yellow Croaker in China. Front. Mar. Sci. Orig. Res. 2021, 8, 743836. (In English) [Google Scholar] [CrossRef]
- Wu, Y.; Tao, W.; Li, L.; Yang, X.; Chen, S.; Lin, W.; Rong, H. Morphological structure and quality characteristics of cultured Larimichthys crocea in Ningde. J. Fish. China 2019, 43, 1472–1482. [Google Scholar]
- Peng, X.; Qiaozhen, K.; Yongquan, S.; Jiafu, L.; Weiqiang, Z. Protection and utilization status and prospect of large yellow croaker (Larimichthys crocea) germplasm resources. Aquac. Fish. 2022, 46, 674–682. [Google Scholar]
- Vidal, M.; Wolf, N.; Rosenberg, B.; Harris, B.P.; Mathis, A. Perspectives on Individual Animal Identification from Biology and Computer Vision. Integr. Comp. Biol. 2021, 61, 900–916. (In English) [Google Scholar] [CrossRef]
- Schwamborn, R.; Mildenberger, T.K.; Taylor, M.H. Assessing sources of uncertainty in length-based estimates of body growth in populations of fishes and macroinvertebrates with bootstrapped ELEFAN. Ecol. Model. 2019, 393, 37–51. [Google Scholar] [CrossRef]
- Yao, J.-X.; Lin, H.-D.; Wu, L.-S.; Wu, L.-N.; Yuan, J.-G.; Ding, S.-X. Stability of population genetic structure in large yellow croaker (Larimichthys crocea): Insights from temporal, geographical factors, and artificial restocking processes. Ecol. Evol. 2024, 14, e70207. [Google Scholar] [CrossRef]
- Kumar, N.; Marée, R.; Geurts, P.; Muller, M. Recent Advances in Bioimage Analysis Methods for Detecting Skeletal Deformities in Biomedical and Aquaculture Fish Species. Biomolecules 2023, 13, 1797. (In English) [Google Scholar] [CrossRef]
- Song, Y.; Shengmao, Z.; Heng, Z.; Fenghua, T.; Hanye, Z.; Yongchuang, S.; Xuesen, C. Application of X-ray in detection of fish tissue and trace elements. J. Appl. Opt. 2024, 45, 166–176. [Google Scholar] [CrossRef]
- Urazoe, K.; Kuroki, N.; Maenaka, A.; Tsutsumi, H.; Iwabuchi, M.; Fuchuya, K.; Hirose, T.; Numa, M. Automated Fish Bone Detection in X-Ray Images with Convolutional Neural Network and Synthetic Image Generation. IEEJ Trans. Electr. Electron. Eng. 2021, 16, 1510–1517. [Google Scholar] [CrossRef]
- Garcia, R.; Prados, R.; Quintana, J.; Tempelaar, A.; Gracias, N.; Rosen, S.; Vågstøl, H.; Løvall, K. Automatic segmentation of fish using deep learning with application to fish size measurement. ICES J. Mar. Sci. 2019, 77, 1354–1366. [Google Scholar] [CrossRef]
- Mery, D.; Lillo, I.; Loebel, H.; Riffo, V.; Soto, A.; Cipriano, A.; Aguilera, J.M. Automated fish bone detection using X-ray imaging. J. Food Eng. 2011, 105, 485–492. [Google Scholar] [CrossRef]
- Xu, W.; Fang, H.; Yang, S.; Zhang, S.; Shi, Y.; Wu, Z.; Yu, S.; Xiong, X.; Yang, H.; Dai, Y. Research progress on application of deep learning in fish recognition, counting, and tracking: A review. J. Dalian Fish. Univ. 2024, 39, 874–887. [Google Scholar] [CrossRef]
- Kong, J.; Tang, S.; Feng, J.; Mo, L.; Jin, X. AASNet: A Novel Image Instance Segmentation Framework for Fine-Grained Fish Recognition via Linear Correlation Attention and Dynamic Adaptive Focal Loss. Appl. Sci. 2025, 15, 3986. [Google Scholar] [CrossRef]
- Liu, W.; Tan, J.; Lan, G.; Li, A.; Li, D.; Zhao, L.; Yuan, X.; Dong, N. Benchmarking Fish Dataset and Evaluation Metric in Keypoint Detection—Towards Precise Fish Morphological Assessment in Aquaculture Breeding. arXiv 2024, arXiv:2405.12476. [Google Scholar]
- Zhang, J.; Wang, Y. A New Workflow for Instance Segmentation of Fish with YOLO. J. Mar. Sci. Eng. 2024, 12, 1010. [Google Scholar] [CrossRef]
- Woo, S.; Park, J.; Lee, J.-Y.; Kweon, I.S. CBAM: Convolutional Block Attention Module. In Proceedings of the 15th European Conference, Munich, Germany, 8–14 September 2018. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Som, A.; Thopalli, K.; Ramamurthy, K.N.; Venkataraman, V.; Shukla, A.; Turaga, P. Perturbation Robust Representations of Topological Persistence Diagrams. In Computer Vision–ECCV 2018; Ferrari, V., Hebert, M., Sminchisescu, C., Weiss, Y., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 638–659. [Google Scholar]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. (In English) [Google Scholar] [CrossRef]
- Milletarì, F.; Navab, N.; Ahmadi, S.-A. V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation. In Proceedings of the 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; pp. 565–571. [Google Scholar]
- Zaccone, D.; Sengar, M.; Lauriano, E.R.; Pergolizzi, S.; Macri’, F.; Salpietro, L.; Favaloro, A.; Satora, L.; Dabrowski, K.; Zaccone, G. Morphology and innervation of the teleost physostome swim bladders and their functional evolution in non-teleostean lineages. Acta Histochem. 2012, 114, 763–772. [Google Scholar] [CrossRef]
- Kolmann, M.A.; Nagesan, R.S.; Andrews, J.V.; Borstein, S.R.; Figueroa, R.T.; Singer, R.A.; Friedman, M.; López-Fernández, H. DiceCT for fishes: Recommendations for pairing iodine contrast agents with μCT to visualize soft tissues in fishes. J. Fish Biol. 2023, 102, 893–903. [Google Scholar] [CrossRef]
- Kague, E.; Kwon, R.Y.; Busse, B.; Witten, P.E.; Karasik, D. Standardization of bone morphometry and mineral density assessments in zebrafish and other small laboratory fishes using X-ray radiography and micro-computed tomography. J. Bone Min. Res. 2024, 39, 1695–1710. (In English) [Google Scholar] [CrossRef]
- Liao, W.N.; You, M.; Ulhaq, Z.S.; Li, J.; Jiang, Y.; Chen, J.; Tse, W.K.F. Micro-CT analysis reveals the changes in bone mineral density in zebrafish craniofacial skeleton with age. J. Anat. 2023, 242, 544–551. (In English) [Google Scholar] [CrossRef] [PubMed]
- Stauber, M.; Müller, R. Micro-computed tomography: A method for the non-destructive evaluation of the three-dimensional structure of biological specimens. Methods Mol. Biol. 2008, 455, 273–292. (In English) [Google Scholar] [CrossRef]
- Cortesi, F.; Mitchell, L.J.; Tettamanti, V.; Fogg, L.G.; de Busserolles, F.; Cheney, K.L.; Marshall, N.J. Visual system diversity in coral reef fishes. Semin. Cell Dev. Biol. 2020, 106, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Martino, J.C.; Doubleday, Z.A.; Chung, M.-T.; Gillanders, B.M. Experimental support towards a metabolic proxy in fish using otolith carbon isotopes. J. Exp. Biol. 2020, 223, jeb217091. [Google Scholar] [CrossRef]
- Camilieri-Asch, V.; Shaw, J.A.; Mehnert, A.; Yopak, K.E.; Partridge, J.C.; Collin, S.P. diceCT: A Valuable Technique to Study the Nervous System of Fish. eNeuro 2020, 7. (In English) [Google Scholar] [CrossRef]
- Tanner, S.E.; Reis-Santos, P.; Cabral, H.N. Otolith chemistry in stock delineation: A brief overview, current challenges and future prospects. Fish. Res. 2016, 173, 206–213. [Google Scholar] [CrossRef]
- Schulz-Mirbach, T.; Olbinado, M.; Rack, A.; Mittone, A.; Bravin, A.; Melzer, R.R.; Ladich, F.; Heß, M. In-situ visualization of sound-induced otolith motion using hard X-ray phase contrast imaging. Sci. Rep. 2018, 8, 3121. (In English) [Google Scholar] [CrossRef] [PubMed]
- Maiditsch, I.P.; Ladich, F.; Heß, M.; Schlepütz, C.M.; Schulz-Mirbach, T. Revealing sound-induced motion patterns in fish hearing structures in 4D: A standing wave tube-like setup designed for high-resolution time-resolved tomography. J. Exp. Biol. 2022, 225, jeb243614. (In English) [Google Scholar] [CrossRef] [PubMed]
- Gollas-Galván, T.; Avila-Villa, L.A.; Martínez-Porchas, M.; Hernandez-Lopez, J. Rickettsia-like organisms from cultured aquatic organisms, with emphasis on necrotizing hepatopancreatitis bacterium affecting penaeid shrimp: An overview on an emergent concern. Rev. Aquac. 2014, 6, 256–269. [Google Scholar] [CrossRef]
- Vasconcelos-Filho, J.E.; Thomsen, F.S.L.; Stosic, B.; Antonino, A.C.D.; Duarte, D.A.; Heck, R.J.; Lessa, R.P.T.; Santana, F.M.; Ferreira, B.P.; Duarte-Neto, P.J. Peeling the Otolith of Fish: Optimal Parameterization for Micro-CT Scanning. Front. Mar. Sci. Orig. Res. 2019, 6, 728. (In English) [Google Scholar] [CrossRef]
- Barrett, J.F.; Keat, N. Artifacts in CT: Recognition and avoidance. Radiographics 2004, 24, 1679–1691. (In English) [Google Scholar] [CrossRef]
- Wang, C.; Zhang, Q.; Wang, X.; Zhou, L.; Li, Q.; Xia, Z.; Ma, B.; Shi, Y.-Q. Light-Field Image Multiple Reversible Robust Watermarking Against Geometric Attacks. IEEE Trans. Dependable Secur. Comput. 2025, 22, 5861–5875. [Google Scholar] [CrossRef]
- Cerveri, P.; Forlani, C.; Borghese, N.A.; Ferrigno, G. Distortion correction for x-ray image intensifiers: Local unwarping polynomials and RBF neural networks. Med. Phys. 2002, 29, 1759–1771. [Google Scholar] [CrossRef] [PubMed]
- Walker, A.; Metcalfe, P.; Liney, G.; Batumalai, V.; Dundas, K.; Glide-Hurst, C.; Delaney, G.P.; Boxer, M.; Yap, M.L.; Dowling, J.; et al. MRI geometric distortion: Impact on tangential whole-breast IMRT. J. Appl. Clin. Med. Phys. 2016, 17, 7–19. (In English) [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, C.; Lu, M.; Yang, J.; Gui, J.; Zhang, S. From Simple to Complex Scenes: Learning Robust Feature Representations for Accurate Human Parsing. IEEE Trans. Pattern Anal. Mach. Intell. 2024, 46, 5449–5462. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Ren, X.; Zhu, B.; Tang, T.; Tan, X.; Gui, Y.; Yao, Q. An Adaptive Attention Fusion Mechanism Convolutional Network for Object Detection in Remote Sensing Images. Remote Sens. 2022, 14, 516. [Google Scholar] [CrossRef]
- Lee, Y.-W.; Kim, B.-G. Attention-based scale sequence network for small object detection. Heliyon 2024, 10, e32931. [Google Scholar] [CrossRef]
- Mia, M.S.; Voban, A.A.B.; Arnob, A.B.H.; Naim, A.; Ahmed, M.K.; Islam, M.S. DANet: Enhancing Small Object Detection through an Efficient Deformable Attention Network. In Proceedings of the 2023 International Conference on the Cognitive Computing and Complex Data (ICCD), Huai’an, China, 21–22 October 2023; pp. 51–62. [Google Scholar]
- Klever, J.; de Motte, A.; Meyer-Lindenberg, A.; Brühschwein, A. Evaluation and Comparison of Self-Made and Commercial Calibration Markers for Radiographic Magnification Correction in Veterinary Digital Radiography. Vet. Comp. Orthop. Traumatol. 2021, 35, 010–017. (In English) [Google Scholar] [CrossRef]
- Weinhardt, V.; Shkarin, R.; Wernet, T.; Wittbrodt, J.; Baumbach, T.; Loosli, F. Quantitative morphometric analysis of adult teleost fish by X-ray computed tomography. Sci. Rep. 2018, 8, 16531. [Google Scholar] [CrossRef]
- Hur, M.; Gistelinck, C.A.; Huber, P.; Lee, J.; Thompson, M.H.; Monstad-Rios, A.T.; Watson, C.J.; McMenamin, S.K.; Willaert, A.; Parichy, D.M.; et al. MicroCT-Based Phenomics in the Zebrafish Skeleton Reveals Virtues of Deep Phenotyping in a Distributed Organ System. Zebrafish 2018, 15, 77–78. (In English) [Google Scholar] [CrossRef]
- Andrialovanirina, N.; Poloni, L.; Laffont, R.; Caillault, É.P.; Couette, S.; Mahé, K. 3D meshes dataset of sagittal otoliths from red mullet in the Mediterranean Sea. Sci. Data 2024, 11, 813. [Google Scholar] [CrossRef]
- Fernandes, A.F.A.; Turra, E.M.; de Alvarenga, É.R.; Passafaro, T.L.; Lopes, F.B.; Alves, G.F.; Singh, V.; Rosa, G.J. Deep Learning image segmentation for extraction of fish body measurements and prediction of body weight and carcass traits in Nile tilapia. Comput. Electron. Agric. 2020, 170, 105274. [Google Scholar] [CrossRef]
- Nguyen, S.V.; Lanni, D.; Xu, Y.; Michaelson, J.S.; McMenamin, S.K. Dynamics of the Zebrafish Skeleton in Three Dimensions During Juvenile and Adult Development. Front. Physiol. Orig. Res. 2022, 13, 875866. (In English) [Google Scholar] [CrossRef]
- Watson, C.J.; Monstad-Rios, A.T.; Bhimani, R.M.; Gistelinck, C.; Willaert, A.; Coucke, P.; Hsu, Y.-H.; Kwon, R.Y. Phenomics-Based Quantification of CRISPR-Induced Mosaicism in Zebrafish. Cell Syst. 2020, 10, 275–286.e5. [Google Scholar] [CrossRef]
- Bucklow, C.V.; Genner, M.J.; Turner, G.F.; Maclaine, J.; Benson, R.; Verd, B. A whole-body micro-CT scan library that captures the skeletal diversity of Lake Malawi cichlid fishes. Sci. Data 2024, 11, 984. [Google Scholar] [CrossRef]
- Kumar, N.; Di Biagio, C.; Dellacqua, Z.; Raman, R.; Martini, A.; Boglione, C.; Muller, M.; Geurts, P.; Marée, R. Empirical Evaluation of Deep Learning Approaches for Landmark Detection in Fish Bioimages. In Computer Vision–ECCV 2022 Workshops; Springer: Cham, Switzerland, 2023; pp. 470–486. [Google Scholar]
- He, Z.; Jiang, Y.; Wang, X.; Xie, Y.; Cheng, Y.; Mei, J. Machine learning for extracting morphological phenotypic traits and estimating weight in largemouth bass. Aquac. Fish. 2025, in press. [Google Scholar] [CrossRef]
- Sakashita, M.; Sato, M.; Kondo, S. Comparative morphological examination of vertebral bodies of teleost fish using high-resolution micro-CT scans. J. Morphol. 2019, 280, 778–795. (In English) [Google Scholar] [CrossRef]
- Scadeng, M.; McKenzie, C.; He, W.; Bartsch, H.; Dubowitz, D.J.; Stec, D.; Leger, J.S. Morphology of the Amazonian Teleost Genus Arapaima Using Advanced 3D Imaging. Front. Physiol. Orig. Res. 2020, 11, 260. (In English) [Google Scholar] [CrossRef]
- Hasegawa, T.; Kondo, K.; Senou, H. Transferable Deep Learning Model for the Identification of Fish Species for Various Fishing Grounds. J. Mar. Sci. Eng. 2024, 12, 415. [Google Scholar] [CrossRef]
- Siri, D.; Vellaturi, G.; Ibrahim, S.H.S.; Molugu, S.; Desanamukula, V.S.; Kocherla, R.; Vatambeti, R. Enhanced deep learning models for automatic fish species identification in underwater imagery. Heliyon 2024, 10, e35217. [Google Scholar] [CrossRef] [PubMed]
- Gignac, P.M.; Kley, N.J.; Clarke, J.A.; Colbert, M.W.; Morhardt, A.C.; Cerio, D.; Cost, I.N.; Cox, P.G.; Daza, J.D.; Early, C.M.; et al. Diffusible iodine-based contrast-enhanced computed tomography (diceCT): An emerging tool for rapid, high-resolution, 3-D imaging of metazoan soft tissues. J. Anat. 2016, 228, 889–909. (In English) [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Vanselow, D.J.; Yakovlev, M.A.; Katz, S.R.; Lin, A.Y.; Clark, D.P.; Vargas, P.; Xin, X.; Copper, J.E.; Canfield, V.A.; et al. Computational 3D histological phenotyping of whole zebrafish by X-ray histotomography. eLife 2019, 8, e44898. [Google Scholar] [CrossRef] [PubMed]
- Fisher, S.; Jagadeeswaran, P.; Halpern, M.E. Radiographic analysis of zebrafish skeletal defects. Dev. Biol. 2003, 264, 64–76. [Google Scholar] [CrossRef]
- Zhang, Y.; Yu, J.; Wang, Y.; Tang, S.; Li, H.; Xin, Z.; Wang, C.; Zhao, Z. Small object detection based on hierarchical attention mechanism and multi-scale separable detection. IET Image Process. 2023, 17, 3986–3999. [Google Scholar] [CrossRef]
- Ni, J.; Zhu, S.; Tang, G.; Ke, C.; Wang, T. A Small-Object Detection Model Based on Improved YOLOv8s for UAV Image Scenarios. Remote Sens. 2024, 16, 2465. [Google Scholar] [CrossRef]
- Liu, M.; Wu, Y.; Li, R.; Lin, C. LFN-YOLO: Precision underwater small object detection via a lightweight reparameterized approach. Front. Mar. Sci. Orig. Res. 2025, 11, 1513740. [Google Scholar] [CrossRef]
- Fu, G.; Yuna, Y. Phenotyping and phenomics in aquaculture breeding. Aquac. Fish. 2022, 7, 140–146. (In English) [Google Scholar] [CrossRef]




| ID | Body Length (mm) | Body Height (mm) | Body Thickness (mm) | Total Length (mm) | Weight (g) |
|---|---|---|---|---|---|
| 1 | 182 | 58 | 29.6 | 210 | 125 |
| 2 | 225 | 75 | 40 | 256 | 241 |
| 3 | 210 | 75.6 | 36 | 232 | 198 |
| 4 | 224 | 67 | 34 | 253 | 208 |
| 5 | 223 | 72 | 39 | 250 | 220 |
| 6 | 260 | 83 | 44 | 285 | 333 |
| 7 | 242 | 74 | 39 | 276 | 244 |
| 8 | 218 | 65 | 36 | 239 | 196 |
| 9 | 238 | 74 | 41 | 268 | 272 |
| 10 | 209 | 66 | 38 | 233 | 186 |
| Model | mAP50 | mAP50–95 | Recall | Precision |
|---|---|---|---|---|
| YOLOv11 | 0.983 | 0.752 | 0.977 | 0.979 |
| YOLOv11-CBAM | 0.985 | 0.754 | 1.000 | 0.986 |
| Mask R-CNN | 0.955 | 0.835 | 0.952 | 0.941 |
| RetinaNet | 0.942 | 0.812 | 0.933 | 0.925 |
| Faster R-CNN | 0.928 | 0.785 | 0.918 | 0.909 |
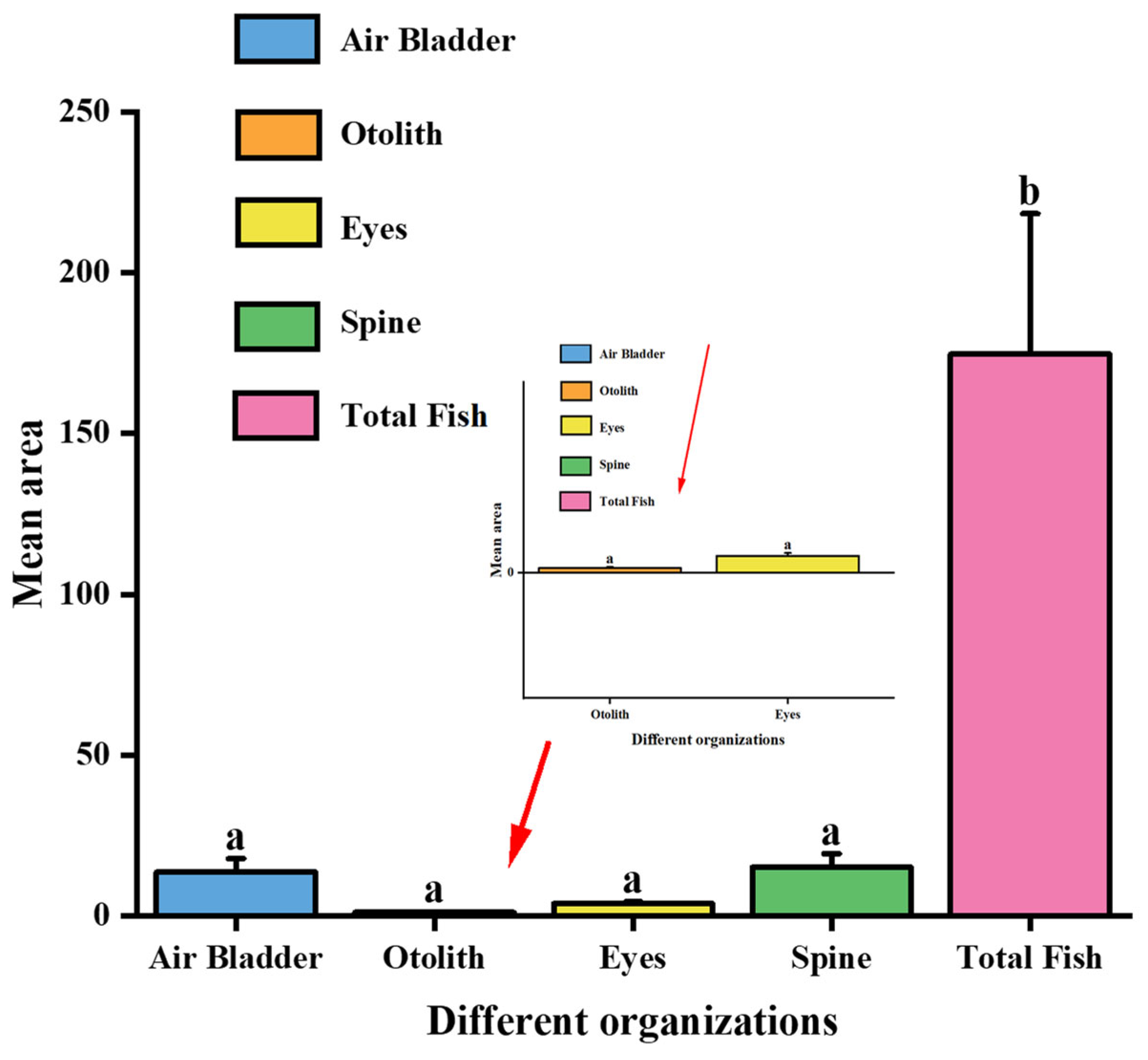
| No. | Air Bladder Area (cm2) | Otolith Area (cm2) | Eye Area (cm2) | Spine Area (cm2) | Whole-Fish Area (cm2) |
|---|---|---|---|---|---|
| 1 | 14.545 | 1.035 | 2.950 | 12.940 | 166.635 |
| 2 | 21.695 | 1.483 | 5.143 | 21.845 | 279.350 |
| 3 | 9.430 | 0.688 | 4.070 | 11.165 | 140.378 |
| 4 | 19.813 | 1.195 | 3.323 | 17.923 | 216.228 |
| 5 | 13.788 | 0.810 | 3.560 | 13.763 | 171.135 |
| 6 | 10.278 | 0.793 | 3.653 | 11.143 | 144.090 |
| 7 | 10.098 | 0.943 | 3.000 | 11.408 | 140.490 |
| 8 | 13.438 | 1.345 | 3.615 | 21.363 | 173.890 |
| 9 | 9.350 | 0.888 | 3.983 | 10.065 | 142.115 |
| 10 | 13.603 | 0.998 | 3.870 | 16.135 | 166.105 |
| Category | Air Bladder (cm2) | Otolith (cm2) | Eye (cm2) | Spine (cm2) | Total Fish (cm2) |
|---|---|---|---|---|---|
| Mean area | 13.509 | 1.015 | 3.716 | 14.959 | 174.784 |
| Standard deviation | 4.267 | 0.254 | 0.628 | 4.336 | 43.679 |
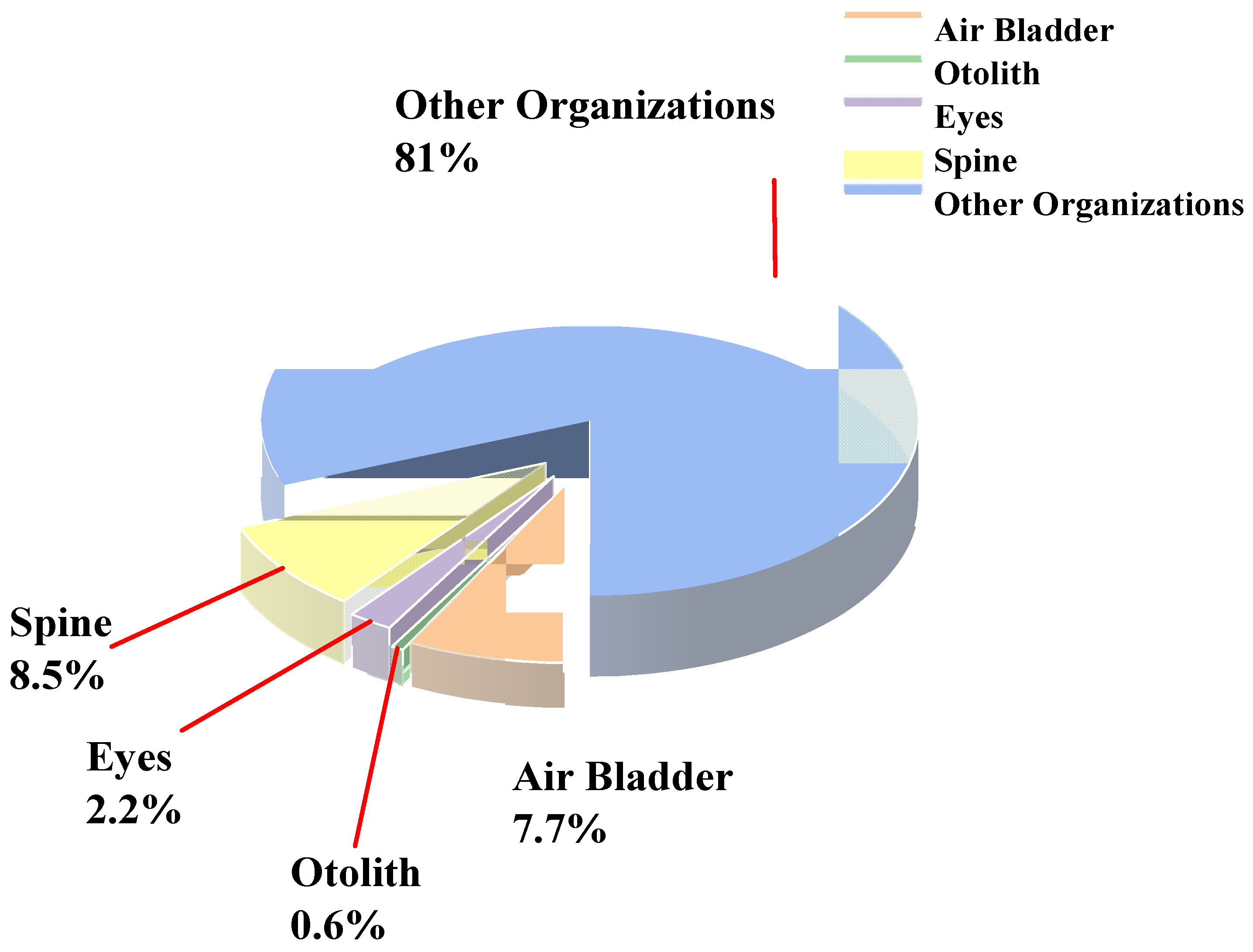
| No | Air Bladder (%) | Otolith (%) | Eye (%) | Spine (%) |
|---|---|---|---|---|
| 1 | 8.73% | 0.62% | 1.77% | 7.77% |
| 2 | 7.77% | 0.53% | 1.84% | 7.82% |
| 3 | 6.72% | 0.49% | 2.90% | 7.95% |
| 4 | 9.16% | 0.55% | 1.54% | 8.29% |
| 5 | 8.06% | 0.47% | 2.08% | 8.04% |
| 6 | 7.13% | 0.55% | 2.53% | 7.73% |
| 7 | 7.19% | 0.67% | 2.14% | 8.12% |
| 8 | 7.73% | 0.77% | 2.08% | 12.29% |
| 9 | 6.58% | 0.62% | 2.80% | 7.08% |
| 10 | 8.19% | 0.60% | 2.33% | 9.71% |
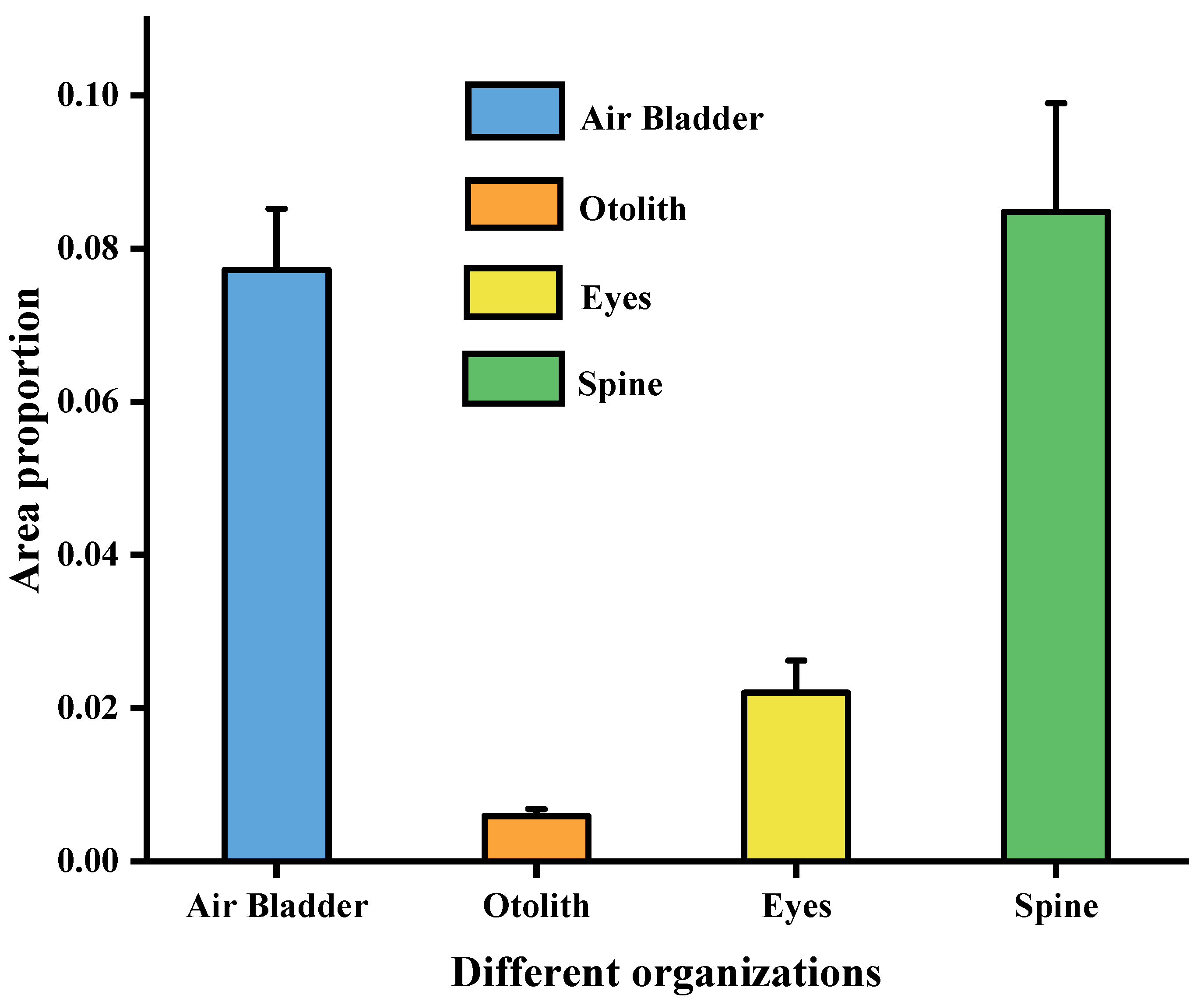
| Structure | Air Bladder | Otolith | Eye | Spine |
|---|---|---|---|---|
| Mean ratio (%) | 7.72% | 0.59% | 2.20% | 8.48% |
| Standard deviation (%) | 0.80% | 0.09% | 0.42% | 1.42% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, Y.; Qiao, G.; Zhang, S.; Wu, C.; Wu, Z.; Cheng, T.; Zheng, H. Research on Morphometric Methods for Larimichthys crocea Based on YOLOv11-CBAM X-Ray Imaging. Fishes 2025, 10, 641. https://doi.org/10.3390/fishes10120641
Yao Y, Qiao G, Zhang S, Wu C, Wu Z, Cheng T, Zheng H. Research on Morphometric Methods for Larimichthys crocea Based on YOLOv11-CBAM X-Ray Imaging. Fishes. 2025; 10(12):641. https://doi.org/10.3390/fishes10120641
Chicago/Turabian StyleYao, Yatong, Guangde Qiao, Shengmao Zhang, Chong Wu, Zuli Wu, Tianfei Cheng, and Hanfeng Zheng. 2025. "Research on Morphometric Methods for Larimichthys crocea Based on YOLOv11-CBAM X-Ray Imaging" Fishes 10, no. 12: 641. https://doi.org/10.3390/fishes10120641
APA StyleYao, Y., Qiao, G., Zhang, S., Wu, C., Wu, Z., Cheng, T., & Zheng, H. (2025). Research on Morphometric Methods for Larimichthys crocea Based on YOLOv11-CBAM X-Ray Imaging. Fishes, 10(12), 641. https://doi.org/10.3390/fishes10120641






