Microsatellite-Based Evaluation of Genetic-Distance-Driven Crossbreeding in the Endangered Freshwater Fish Pseudopungtungia nigra
Abstract
1. Introduction
2. Materials and Methods
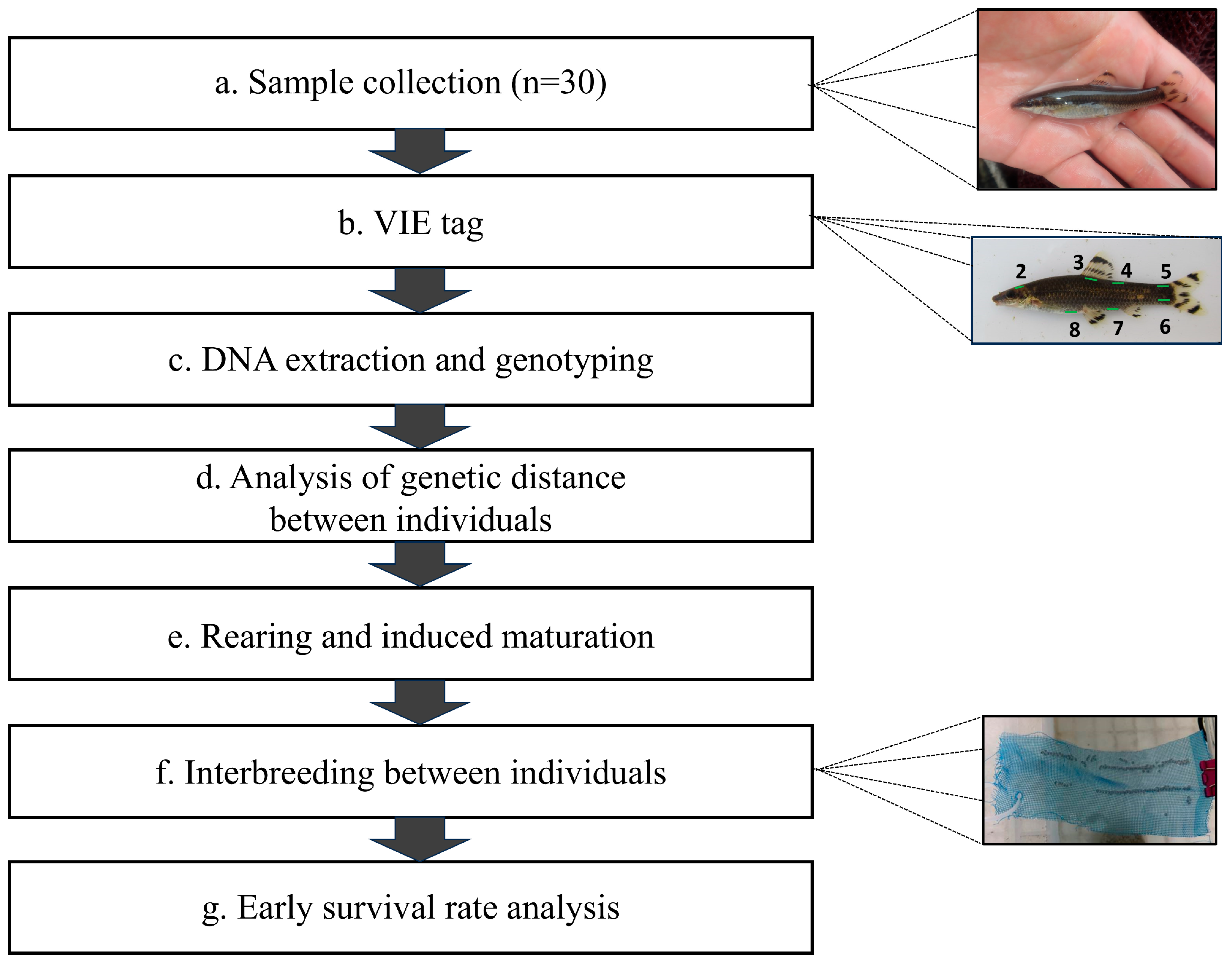
2.1. Fish Collection and Breeding Management for Mating
2.2. VIE Tagging for Broodstock Genotype Verification
2.3. Genomic DNA Extraction from Broodstock and Measuring Genetic Distance
2.4. Production of Crossbred Group Based on Genetic Distance
2.5. Genetic Diversity and Early Survival in the Crossbred Group
2.6. Ethical Approval
3. Results
3.1. Genetic Diversity Analysis Broodstock and Crossbred Group Based on Genetic Distance
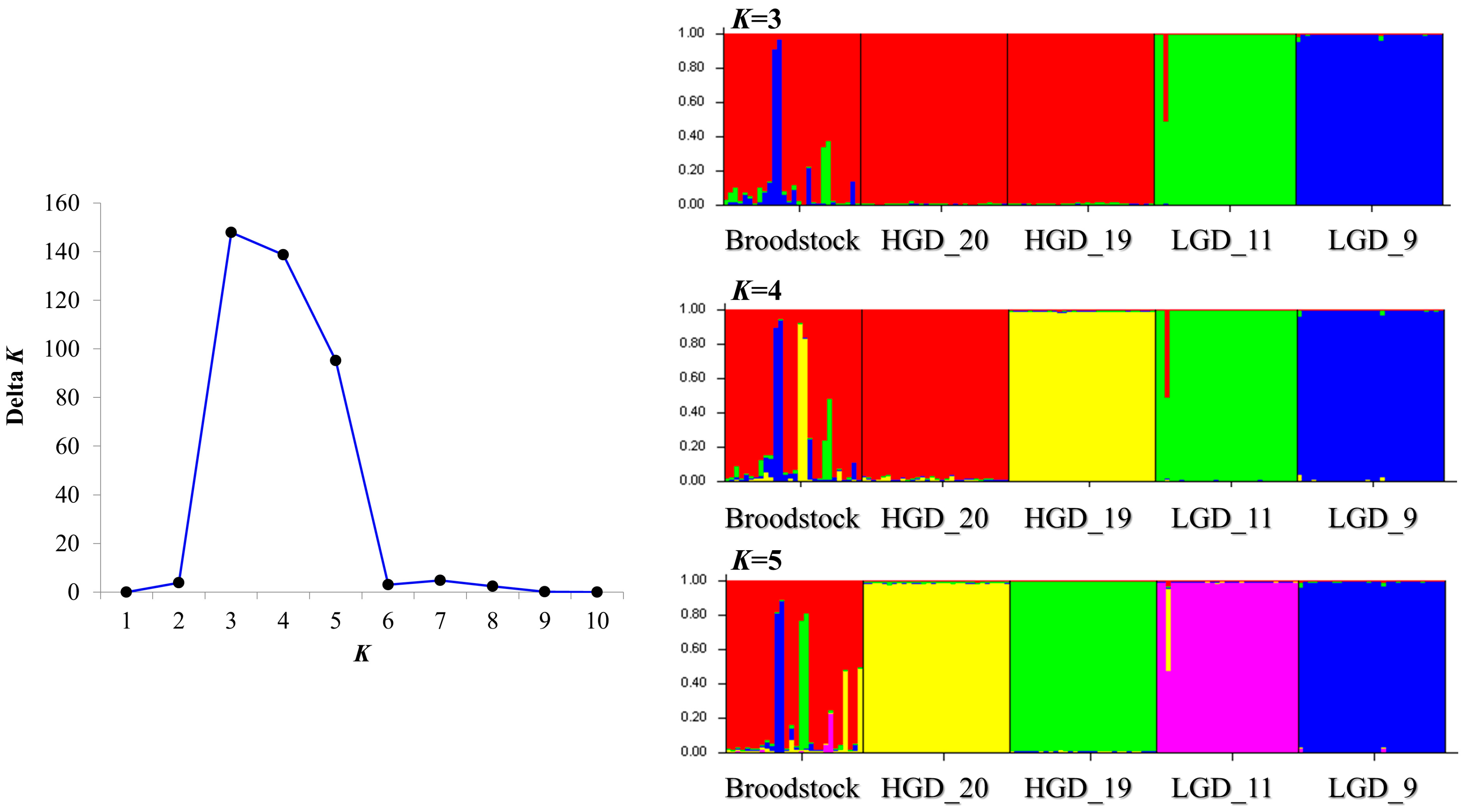
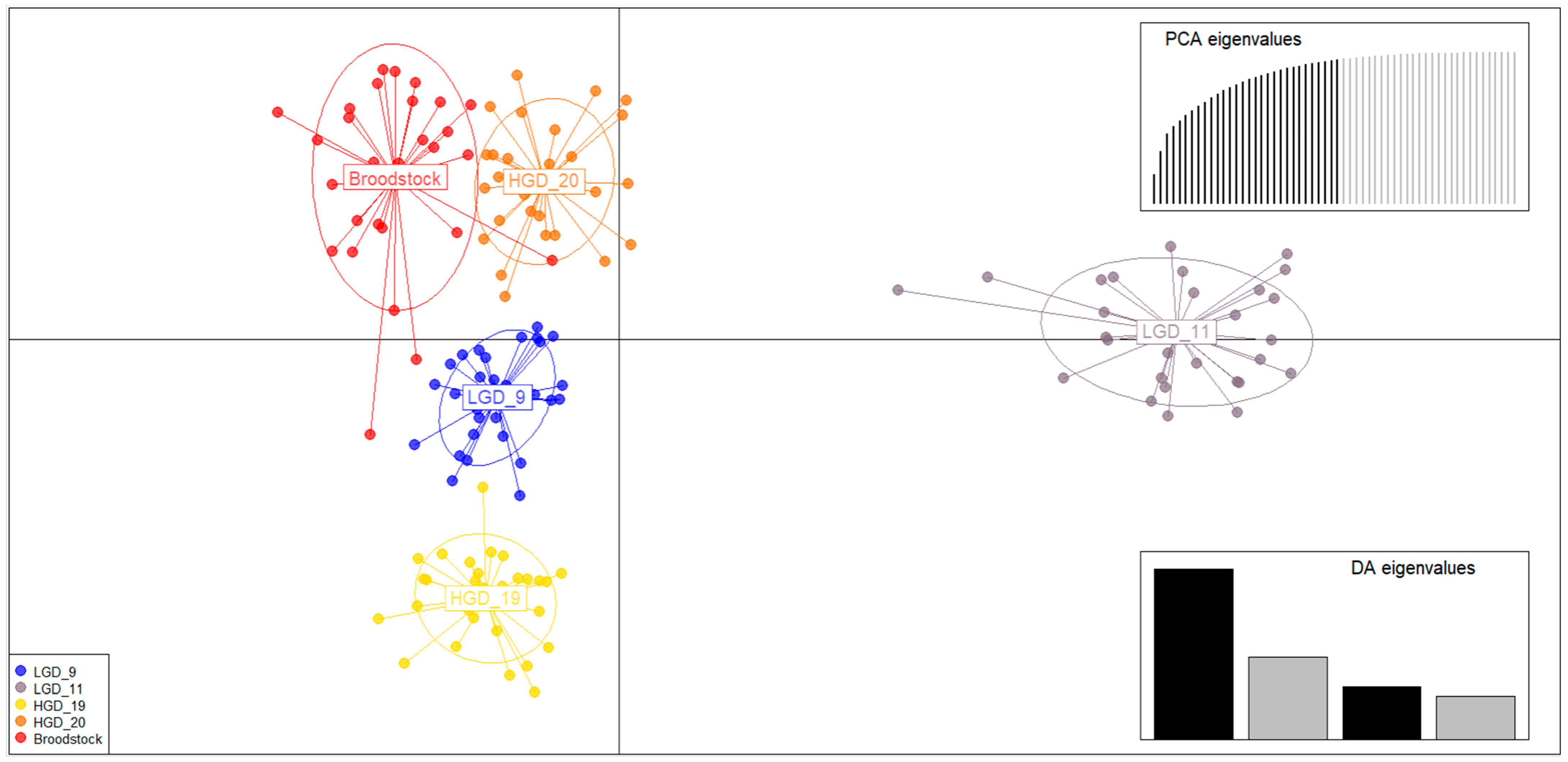
3.2. Analysis of Genetic Structure of Crossbred Groups and Broodstocks
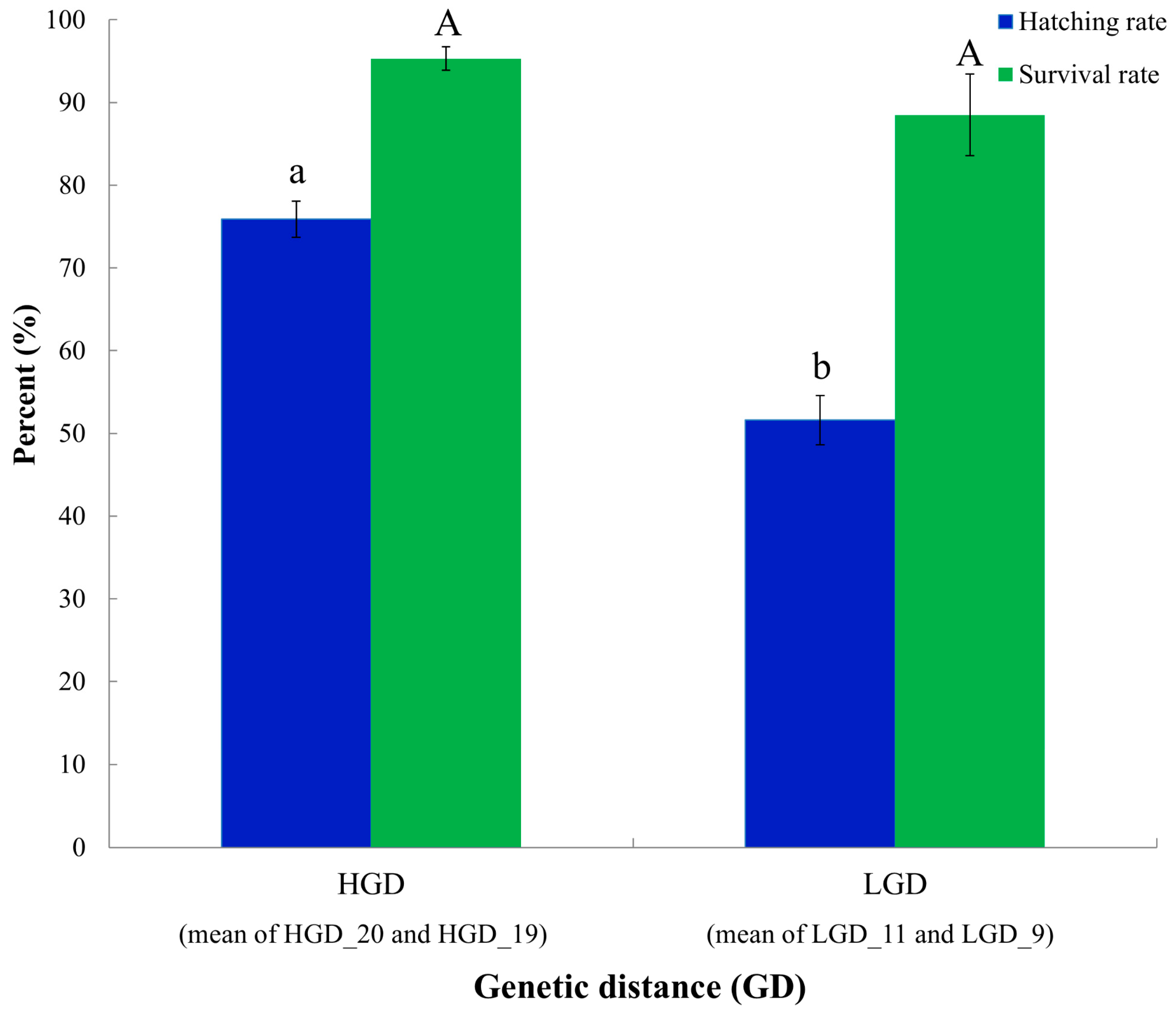
3.3. Early Survival Analysis of the Crossbred Group
4. Discussion
4.1. Genetic Diversity of Crossbred Groups According to Genetic Distance
4.2. Genetic Structure of Crossbred Groups According to Genetic Distance
4.3. Initial Survival Rate by Crossbred Group Produced According to Genetic Distance
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Waples, R.S. The Ne/N ratio in applied conservation. Evol. Appl. 2024, 17, e13695. [Google Scholar] [CrossRef] [PubMed]
- Frankham, R.; Bradshaw, C.J.; Brook, B.W. Genetics in conservation management: Revised recommendations for the 50/500 rules, Red List criteria and population viability analyses. Biol. Conserv. 2014, 170, 56–63. [Google Scholar] [CrossRef]
- Waples, R.S.; Hindar, K.; Karlsson, S.; Hard, J.J. Evaluating the Ryman-Laikre effect for marine stock enhancement and aquaculture. Curr. Zool. 2016, 62, 617–627. [Google Scholar] [CrossRef]
- Mignien, L.; Stoll, S. Reproductive success of stream fish species in relation to high and low flow patterns: The role of life history strategies and species traits. Sci. Total Environ. 2024, 946, 174366. [Google Scholar] [CrossRef]
- Brauer, C.J.; Beheregaray, L.B. Recent and rapid anthropogenic habitat fragmentation increases extinction risk for freshwater biodiversity. Evol. Appl. 2020, 13, 2857–2869. [Google Scholar] [CrossRef]
- Tringali, M.D. Reproductive Success Dynamics Could Limit Precision in Close-Kin Mark–Recapture Abundance Estimation for Atlantic Goliath Grouper (Epinephelus itajara). Fishes 2023, 8, 254. [Google Scholar] [CrossRef]
- Clarke, J.G.; Smith, A.C.; Cullingham, C.I. Genetic rescue often leads to higher fitness as a result of increased heterozygosity across animal taxa. Mol. Ecol. 2024, 33, e17532. [Google Scholar] [CrossRef]
- Lutz, M.L.; Tonkin, Z.; Yen, J.D.L.; Johnson, G.; Ingram, B.A.; Sharley, J.; Lyon, J.; Chapple, D.G.; Sunnucks, P.; Pavlova, A. Using multiple sources during reintroduction of a locally extinct population benefits survival and reproduction of an endangered freshwater fish. Evol. Appl. 2021, 14, 950–964. [Google Scholar] [CrossRef]
- Mamoozadeh, N.R.; Wade, M.J.; Reid, B.N.; Bardwell, E.; Collins, E.E.; Hugentobler, S.A.; Jackson, S.A.; Kline, B.C.; Rothkopf, H.E.; Zhang, A. A practical introduction to effective population size for fisheries management. Trans. Am. Fish. Soc. 2025, 154, 352–371. [Google Scholar] [CrossRef]
- Akers, M.; Quinlan, H.; Johnson, A.; Baker, E.; Welsh, A. Parentage Analysis Reveals Unequal Family Sizes during Hatchery Production. Fishes 2023, 8, 140. [Google Scholar] [CrossRef]
- Lee, S.-H.; Han, G.-H.; Yun, S.-M.; Hwang, D.-S.; Yu, D.-J.; Lee, C.-R.; Kim, I.-S.; Son, Y.-M. Early life history and spawning behavior of Pseudopungtungia nigra. Korean J. Ichthyol. 2004, 16, 309–316. [Google Scholar]
- Kim, K.R.; Kwak, Y.H.; Sung, M.S.; Cho, S.J.; Bang, I.C. Population structure and genetic diversity of the endangered fish black shinner Pseudopungtungia nigra (Cyprinidae) in Korea: A wild and restoration population. Sci. Rep. 2023, 13, 9692. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, A.A.; Miller, A.D.; Weeks, A.R. Genetic mixing for population management: From genetic rescue to provenancing. Evol. Appl. 2021, 14, 634–652. [Google Scholar] [CrossRef] [PubMed]
- Sexton, J.P.; Clemens, M.; Bell, N.; Hall, J.; Fyfe, V.; Hoffmann, A.A. Patterns and effects of gene flow on adaptation across spatial scales: Implications for management. J. Evol. Biol. 2024, 37, 732–745. [Google Scholar] [CrossRef]
- Kardos, M.; Armstrong, E.E.; Fitzpatrick, S.W.; Hauser, S.; Hedrick, P.W.; Miller, J.M.; Tallmon, D.A.; Funk, W.C. The crucial role of genome-wide genetic variation in conservation. Proc. Natl. Acad. Sci. USA 2021, 118, e2104642118. [Google Scholar] [CrossRef]
- Floriani, T.; Lipka, A.E. Rare variants in crops: Theoretical insights and emerging detection strategies. Silico Plants 2025, 7, diaf012. [Google Scholar] [CrossRef]
- Maruyama, T.; Fuerst, P.A. Population bottlenecks and nonequilibrium models in population genetics. II. Number of alleles in a small population that was formed by a recent bottleneck. Genetics 1985, 111, 675–689. [Google Scholar] [CrossRef]
- Luikart, G.; Allendorf, F.W.; Cornuet, J.M.; Sherwin, W.B. Distortion of allele frequency distributions provides a test for recent population bottlenecks. J. Hered. 1998, 89, 238–247. [Google Scholar] [CrossRef]
- Busack, C.; Knudsen, C.M. Using factorial mating designs to increase the effective number of breeders in fish hatcheries. Aquaculture 2007, 273, 24–32. [Google Scholar] [CrossRef]
- Brown, K.T.; Southgate, P.C.; Loganimoce, E.M.; Kaure, T.; Stockwell, B.; Lal, M.M. Sandfish generations: Loss of genetic diversity due to hatchery practices in the sea cucumber Holothuria (Metriatyla) scabra. Aquaculture 2024, 578, 740048. [Google Scholar] [CrossRef]
- Frankham, R.; Ballou, J.D.; Eldridge, M.D.; Lacy, R.C.; Ralls, K.; Dudash, M.R.; Fenster, C.B. Predicting the probability of outbreeding depression. Conserv. Biol. 2011, 25, 465–475. [Google Scholar] [CrossRef]
- Edmands, S. Between a rock and a hard place: Evaluating the relative risks of inbreeding and outbreeding for conservation and management. Mol. Ecol. 2007, 16, 463–475. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, J.; Honda, R.; Hosoya, S.; Nochiri, Y.; Matsuzaki, K.; Sugimoto, K.; Nagano, A.J.; Kumagai, A.; Kikuchi, K.; Kurokawa, T. Genome-Assisted Gene-Flow Rescued Genetic Diversity Without Hindering Growth Performance in an Inbred Coho Salmon (Oncorhynchus kisutch) Population Selected for High Growth Phenotype. Mar. Biotechnol. 2025, 27, 38. [Google Scholar] [CrossRef]
- Reid, B.N.; Hofmeier, J.; Crockett, H.; Fitzpatrick, R.; Waters, R.; Fitzpatrick, S.W. Balancing Inbreeding and Outbreeding Risks to Inform Translocations Throughout the Range of an Imperiled Darter. Evol. Appl. 2025, 18, e70088. [Google Scholar] [CrossRef]
- Hedgecock, D.; Davis, J.P. Heterosis for yield and crossbreeding of the Pacific oyster Crassostrea gigas. Aquaculture 2007, 272, S17–S29. [Google Scholar] [CrossRef]
- Lutz, C.G.; Armas-Rosales, A.M.; Saxton, A.M. Genetic effects influencing salinity tolerance in six varieties of tilapia (Oreochromis) and their reciprocal crosses. Aquac. Res. 2010, 41, e770–e780. [Google Scholar] [CrossRef]
- Zheng, H.; Zhang, G.; Guo, X.; Liu, X. Heterosis between two stocks of the bay scallop, Argopecten irradians irradians Lamarck (1819). J. Shellfish Res. 2006, 25, 807–812. [Google Scholar] [CrossRef]
- Linløkken, A.N.; Johnsen, S.I.; Johansen, W. Genetic diversity of hatchery-bred brown trout (Salmo trutta) compared with the wild population: Potential effects of stocking on the indigenous gene pool of a Norwegian reservoir. Diversity 2021, 13, 414. [Google Scholar] [CrossRef]
- Pregler, K.C.; Obedzinski, M.; Gilbert-Horvath, E.A.; White, B.; Carlson, S.M.; Garza, J.C. Assisted gene flow from outcrossing shows the potential for genetic rescue in an endangered salmon population. Conserv. Lett. 2023, 16, e12934. [Google Scholar] [CrossRef]
- Feuerstein, C.A.; Kovach, R.P.; Kruse, C.G.; Jaeger, M.E.; Bell, D.A.; Robinson, Z.L.; Whiteley, A.R. Genetic variation and hybridization determine the outcomes of conservation reintroductions. Conserv. Lett. 2024, 17, e13049. [Google Scholar] [CrossRef]
- Wanzenböck, J.; Hopfinger, M.; Wanzenböck, S.; Fuxjäger, L.; Rund, H.; Lamatsch, D.K. First successful hybridization experiment between native European weatherfish (Misgurnus fossilis) and non-native Oriental weatherfish (M. anguillicaudatus) reveals no evidence for postzygotic barriers. NeoBiota 2021, 69, 29–50. [Google Scholar] [CrossRef]
- del Mar Ortega-Villaizan, M.; Noguchi, D.; Taniguchi, N. Minimization of genetic diversity loss of endangered fish species captive broodstocks by means of minimal kinship selective crossbreeding. Aquaculture 2011, 318, 239–243. [Google Scholar] [CrossRef]
- Sriphairoj, K.; Kamonrat, W.; Na-Nakorn, U. Genetic aspect in broodstock management of the critically endangered Mekong giant catfish, Pangasianodon gigas in Thailand. Aquaculture 2007, 264, 36–46. [Google Scholar] [CrossRef]
- Pavlova, A.; Beheregaray, L.B.; Coleman, R.; Gilligan, D.; Harrisson, K.A.; Ingram, B.A.; Kearns, J.; Lamb, A.M.; Lintermans, M.; Lyon, J. Severe consequences of habitat fragmentation on genetic diversity of an endangered Australian freshwater fish: A call for assisted gene flow. Evol. Appl. 2017, 10, 531–550. [Google Scholar] [CrossRef]
- Walsh, M.G.; Winkelman, D.L. Anchor and visible implant elastomer tag retention by hatchery rainbow trout stocked into an Ozark stream. N. Am. J. Fish. Manag. 2004, 24, 1435–1439. [Google Scholar] [CrossRef]
- Bang, I.-C.; Bruggemann, F.; Chen, C.; D’alexis, Q.; Gao, H.; Gélin, P.; Hong, X.; Inoue-Murayama, M.; Kim, K.-R.; Kwak, Y.-H. Microsatellite records for volume 12, issue 2. Conserv. Genet. Resour. 2020, 12, 337–351. [Google Scholar] [CrossRef]
- Nei, M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 1978, 89, 583–590. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Tambasen-Cheong, M.V.P.; Tan-Fermin, J.D.; Garcia, L.M.B.; Baldevarona, R.B. Milt-egg ratio in artificial fertilization of the Asian freshwater catfish, Clarias macrocephalus, injected salmon gonadotropin-releasing hormone analogue and domperidone. Aquat. Living Resour. 1995, 8, 303–307. [Google Scholar] [CrossRef]
- Kalinowski, S.T.; Taper, M.L.; Marshall, T.C. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 2007, 16, 1099–1106. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Raymond, M.; Rousset, F. GENEPOP (version 1.2): Population genetics software for exact tests and ecumenicism. J. Hered. 1995, 86, 248–249. [Google Scholar] [CrossRef]
- Cornuet, J.M.; Luikart, G. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 1996, 144, 2001–2014. [Google Scholar] [CrossRef]
- Do, C.; Waples, R.S.; Peel, D.; Macbeth, G.M.; Tillett, B.J.; Ovenden, J.R. NeEstimator v2: Re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol. Ecol. Resour. 2014, 14, 209–214. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Li, Y.L.; Liu, J.X. StructureSelector: A web-based software to select and visualize the optimal number of clusters using multiple methods. Mol. Ecol. Resour. 2018, 18, 176–177. [Google Scholar] [CrossRef] [PubMed]
- Jombart, T. adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef]
- Kim, K.R.; Park, J.W.; Park, K.I.; Lee, H.J. Genetic Diversity and Population Structure in Farmed and Wild Pacific Oysters (Crassostrea gigas): A Comparative Study. Int. J. Mol. Sci. 2025, 26, 4172. [Google Scholar] [CrossRef]
- Storset, A.; Strand, C.; Wetten, M.; Kjøglum, S.; Ramstad, A. Response to selection for resistance against infectious pancreatic necrosis in Atlantic salmon (Salmo salar L.). Aquaculture 2007, 272, S62–S68. [Google Scholar] [CrossRef]
- Robinson, Z.L.; Coombs, J.A.; Hudy, M.; Nislow, K.H.; Letcher, B.H.; Whiteley, A.R. Experimental test of genetic rescue in isolated populations of brook trout. Mol. Ecol. 2017, 26, 4418–4433. [Google Scholar] [CrossRef]
- Waples, R.S. Testing for Hardy–Weinberg proportions: Have we lost the plot? J. Hered. 2015, 106, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Bosco, F.; Castro, D.; Briones, M.R. Neutral and stable equilibria of genetic systems and the hardy-weinberg principle: Limitations of the chi-square test and advantages of auto-correlation functions of allele frequencies. Front. Genet. 2012, 3, 276. [Google Scholar] [CrossRef]
- Christie, M.R.; Marine, M.L.; French, R.A.; Waples, R.S.; Blouin, M.S. Effective size of a wild salmonid population is greatly reduced by hatchery supplementation. Heredity 2012, 109, 254–260. [Google Scholar] [CrossRef]
- Chacón, G.M.; Arias-Pérez, A.; Freire, R.; Martínez, L.; Ojea, J.; Insua, A. Genetic characterization of wild, broodstock and seed samples of Polititapes rhomboides (Bivalvia: Veneridae): Implications for hatchery seed production. Aquac. Rep. 2021, 20, 100658. [Google Scholar] [CrossRef]
- Waples, R.S.; Do, C. ldne: A program for estimating effective population size from data on linkage disequilibrium. Mol. Ecol. Resour. 2008, 8, 753–756. [Google Scholar] [CrossRef] [PubMed]
- Jones, A.T.; Ovenden, J.R.; Wang, Y.G. Improved confidence intervals for the linkage disequilibrium method for estimating effective population size. Heredity 2016, 117, 217–223. [Google Scholar] [CrossRef]
- D’Ambrosio, J.; Phocas, F.; Haffray, P.; Bestin, A.; Brard-Fudulea, S.; Poncet, C.; Quillet, E.; Dechamp, N.; Fraslin, C.; Charles, M.; et al. Genome-wide estimates of genetic diversity, inbreeding and effective size of experimental and commercial rainbow trout lines undergoing selective breeding. Genet. Sel. Evol. 2019, 51, 26. [Google Scholar] [CrossRef]
- Ryman, N.; Laikre, L. Effects of supportive breeding on the genetically effective population size. Conserv. Biol. 1991, 5, 325–329. [Google Scholar] [CrossRef]
- Xu, Z.; Dou, S.-Z.; Ding, S.-X.; Liu, J.-X. Temporal genetic stability despite decades of overexploitation for large yellow croaker in the East China sea. Front. Mar. Sci. 2022, 9, 861840. [Google Scholar] [CrossRef]
- Fitzpatrick, S.W.; Mittan-Moreau, C.; Miller, M.; Judson, J.M. Genetic rescue remains underused for aiding recovery of federally listed vertebrates in the United States. J. Hered. 2023, 114, 354–366. [Google Scholar] [CrossRef] [PubMed]
- Mathur, S.; Gibbs, H.L. Genomic Evaluation of Assisted Gene Flow Options in an Endangered Rattlesnake. Mol. Ecol. 2025, 34, e70014. [Google Scholar] [CrossRef]
- Fraser, D.J. How well can captive breeding programs conserve biodiversity? A review of salmonids. Evol. Appl. 2008, 1, 535–586. [Google Scholar] [CrossRef]
- Ralls, K.; Sunnucks, P.; Lacy, R.C.; Frankham, R. Genetic rescue: A critique of the evidence supports maximizing genetic diversity rather than minimizing the introduction of putatively harmful genetic variation. Biol. Conserv. 2020, 251, 108784. [Google Scholar] [CrossRef]
- Liang, Y.; Xu, C.; Li, Q. Heterosis and genetic diversity of intraspecific hybrids crosses between two selected lines of the Pacific oyster Crassostrea gigas. Aquaculture 2023, 569, 739369. [Google Scholar] [CrossRef]
- Fernandez Miguez, M.; Presa, P.; Puvanendran, V.; Tveiten, H.; Hansen, O.J.; Perez, M. Gene Expression and Phenotypic Assessment of Egg Quality across Developmental Stages of Atlantic Cod throughout the Spawning Season. Int. J. Mol. Sci. 2024, 25, 7488. [Google Scholar] [CrossRef] [PubMed]
- Ataguba, G.A.; Angela, A. Hybridization and Growth Performance of Progeny from Crosses between Clarias gariepinus and Heterobranchus sp. Aquac. Stud. 2023, 24, AQUAST1154. [Google Scholar] [CrossRef]
- Jiao, Y.; Colvert, B.; Man, Y.; McHenry, M.J.; Kanso, E. Evaluating evasion strategies in zebrafish larvae. Proc. Natl. Acad. Sci. USA 2023, 120, e2218909120. [Google Scholar] [CrossRef]
- Fuiman, L.A.; Cowan, J.H., Jr. Behavior and recruitment success in fish larvae: Repeatability and covariation of survival skills. Ecology 2003, 84, 53–67. [Google Scholar] [CrossRef]
- Siddique, M.A.M.; Linhart, O.; Krejszeff, S.; Żarski, D.; Pitcher, T.E.; Politis, S.N.; Butts, I.A.E. Paternal identity impacts embryonic development for two species of freshwater fish. Gen. Comp. Endocrinol. 2017, 245, 30–35. [Google Scholar] [CrossRef]
- Yeates, S.E.; Einum, S.; Fleming, I.A.; Megens, H.-J.; Stet, R.J.; Hindar, K.; Holt, W.V.; Van Look, K.J.; Gage, M.J. Atlantic salmon eggs favour sperm in competition that have similar major histocompatibility alleles. Proc. R. Soc. B Biol. Sci. 2009, 276, 559–566. [Google Scholar] [CrossRef] [PubMed]

| Locus Name | Primer Sequence (5′ → 3′) | Motif Repeats | Product Size Range (bp) | Tm (°C) | GenBank Accession No. | Reference |
|---|---|---|---|---|---|---|
| PN4 | F: 6FAM-TCGGTGGATGCGAGGAATAC R: TTCCGCCTGTCAGTCAAGAC | (AC)6 | 190–202 | 58 | MN385013 | Bang et al. [36], Kim et al. [12] |
| PN23 | F: TAMRA-GGCACTCAAGGATAATCTGAAC R: TGTATCCGGCCTCTGTGTAG | (GT)10 | 198–212 | 58 | MN385014 | |
| PN75 | F: HEX-CCTGCATCCATGCCGTATAG R: GCTGTTATAGCGCTGATGATAG | (AC)9 | 199–217 | 58 | MN911279 | |
| PN88 | F: HEX-CAACAGGCTCCACGATTGC R: CTGCCCTCGGAAATAAGATGG | (AC)7 | 174–186 | 58 | MN385015 | |
| PN92 | F: 6FAM-CCGTGCTCATATACAGTCCTC R: CCGCATTGTTCCTCCGATTG | (CA)10 | 242–266 | 58 | MN385016 | |
| PN98 | F: HEX-CAGGATGAGTCCATCGTCTC R: GCTCAGAAGTGACCGACAGA | (GT)10 | 217–301 | 58 | MN385018 | |
| PN123 | F: 6FAM-GGGACACACTTAGCAAGCCT R: AGCCAGTGAGATTGAAAGACCA | (GT)10 | 213–241 | 58 | MN385020 | |
| PN124 | F: 6FAM-GAGACGCACGACTGATGAAG R: GGCTAACAGGGCGATTGATTG | (AC)10 | 246–258 | 58 | MN385021 | |
| PN126 | F: HEX-CCACACTACTGAGACTAAACTG R: TGACAGACCATCTTGCATTCTG | (GT)9 | 176–186 | 58 | MN385022 | |
| PN147 | F: 6FAM-GGCTTATGTGGCTTCGGATAC R: AGGTGAGCCTGAGAGAGAAG | (AC)9 | 122–128 | 58 | MN911282 |
| M1 | M2 | M3 | M4 | M5 | M6 | M7 | M8 | M9 | M10 | M11 | M12 | M13 | M14 | M15 | M16 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F1 | 12 | 15 | 11 | 19 | 15 | 21 | 14 | 18 | 16 | 11 | 16 | 8 | 17 | 14 | 19 | 13 |
| F2 | 14 | 12 | 18 | 24 | 22 | 23 | 18 | 18 | 20 | 13 | 20 | 10 | 20 | 14 | 15 | 12 |
| F3 | 11 | 13 | 11 | 20 | 9 | 14 | 16 | 14 | 17 | 14 | 14 | 8 | 15 | 13 | 15 | 13 |
| F4 | 17 | 20 | 12 | 12 | 17 | 16 | 18 | 12 | 15 | 14 | 13 | 13 | 10 | 14 | 19 | 16 |
| F5 | 14 | 18 | 17 | 21 | 12 | 15 | 18 | 12 | 11 | 13 | 19 | 12 | 17 | 13 | 16 | 9 |
| F6 | 20 | 16 | 14 | 19 | 16 | 11 | 17 | 17 | 12 | 14 | 18 | 16 | 17 | 15 | 20 | 10 |
| F7 | 20 | 12 | 15 | 18 | 13 | 12 | 14 | 11 | 18 | 17 | 17 | 17 | 18 | 12 | 21 | 12 |
| F8 | 21 | 16 | 17 | 17 | 16 | 13 | 16 | 14 | 7 | 14 | 17 | 12 | 15 | 13 | 14 | 13 |
| F9 | 17 | 15 | 19 | 20 | 15 | 15 | 21 | 19 | 17 | 16 | 15 | 18 | 13 | 16 | 19 | 16 |
| F10 | 17 | 17 | 15 | 23 | 17 | 20 | 17 | 19 | 18 | 19 | 18 | 9 | 18 | 18 | 13 | 18 |
| F11 | 16 | 11 | 14 | 20 | 10 | 15 | 20 | 15 | 16 | 12 | 16 | 10 | 13 | 13 | 17 | 11 |
| F12 | 22 | 21 | 16 | 15 | 14 | 16 | 21 | 19 | 17 | 18 | 18 | 17 | 22 | 20 | 18 | 14 |
| Max | 22 | 21 | 19 | 24 | 22 | 23 | 21 | 19 | 20 | 19 | 20 | 18 | 22 | 20 | 21 | 18 |
| Min | 11 | 11 | 11 | 12 | 9 | 11 | 14 | 11 | 7 | 11 | 13 | 8 | 10 | 12 | 13 | 9 |
| Group Name | Genetic Distance Index | No. of Individuals | NA | HE | HO | FIS | PHWE |
|---|---|---|---|---|---|---|---|
| Broodstock | - | 28 | 5.8 ± 1.8 | 0.739 ± 0.120 | 0.697 ± 0.158 | 0.058 | 0.221 |
| HGD | 19 | 30 | 2.7 ± 0.3 | 0.549 ± 0.050 | 0.787 ± 0.082 | −0.445 | 0.000 *** |
| 20 | 30 | 2.9 ± 0.3 | 0.547 ± 0.071 | 0.800 ± 0.108 | −0.448 | 0.000 *** | |
| LGD | 11 | 29 | 2.9 ± 0.3 | 0.538 ± 0.035 | 0.638 ± 0.060 | −0.227 | 0.000 *** |
| 9 | 30 | 2.6 ± 0.4 | 0.469 ± 0.071 | 0.573 ± 0.094 | −0.190 | 0.000 *** |
| Group Name | N | Wilcoxon Sign-Rank Test | Ne | (95% CI) | |||
|---|---|---|---|---|---|---|---|
| PIAM | PTPM | PSMM | Mode-Shift | ||||
| Broodstock | 28 | 0.000 *** | 0.000 *** | 0.001 ** | SHIFTED | ∞ | (96–∞) |
| HGD_20 | 30 | 0.000 *** | 0.000 *** | 0.000 *** | SHIFTED | ∞ | (∞–∞) |
| HGD_19 | 30 | 0.000 *** | 0.000 *** | 0.000 *** | SHIFTED | ∞ | (∞–∞) |
| LGD_11 | 29 | 0.000 *** | 0.000 *** | 0.000 *** | SHIFTED | 114 | (7–∞) |
| LGD_9 | 30 | 0.000 *** | 0.000 *** | 0.000 *** | SHIFTED | 65 | (15–∞) |
| Broodstock | HGD_20 | HGD_19 | LGD_11 | LGD_9 | |
|---|---|---|---|---|---|
| Broodstock | - | 0.000 | 0.000 | 0.000 | 0.000 |
| HGD_20 | 0.105 | - | 0.000 | 0.000 | 0.000 |
| HGD_19 | 0.152 | 0.224 | - | 0.000 | 0.000 |
| LGD_11 | 0.201 | 0.316 | 0.321 | - | 0.000 |
| LGD_9 | 0.170 | 0.288 | 0.298 | 0.371 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, K.-R.; Bang, I.-C. Microsatellite-Based Evaluation of Genetic-Distance-Driven Crossbreeding in the Endangered Freshwater Fish Pseudopungtungia nigra. Fishes 2025, 10, 603. https://doi.org/10.3390/fishes10120603
Kim K-R, Bang I-C. Microsatellite-Based Evaluation of Genetic-Distance-Driven Crossbreeding in the Endangered Freshwater Fish Pseudopungtungia nigra. Fishes. 2025; 10(12):603. https://doi.org/10.3390/fishes10120603
Chicago/Turabian StyleKim, Kang-Rae, and In-Chul Bang. 2025. "Microsatellite-Based Evaluation of Genetic-Distance-Driven Crossbreeding in the Endangered Freshwater Fish Pseudopungtungia nigra" Fishes 10, no. 12: 603. https://doi.org/10.3390/fishes10120603
APA StyleKim, K.-R., & Bang, I.-C. (2025). Microsatellite-Based Evaluation of Genetic-Distance-Driven Crossbreeding in the Endangered Freshwater Fish Pseudopungtungia nigra. Fishes, 10(12), 603. https://doi.org/10.3390/fishes10120603








