Transcriptomic Profiling Reveals Immune-Related Pathway Alterations in Paralichthys olivaceus Infected with Enteromyxum leei
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Approval
2.2. Experimental Fish and Sampling
2.3. Infection Diagnosis of E. leei
2.4. RNA Extraction and Quality Assessment
2.5. Library Preparation and RNA Sequencing
2.6. Differential Expression Analysis
2.7. Functional Enrichment Analysis
2.8. Validation of Candidate Genes by Quantitative Real-Time PCR
2.9. Statistical Analysis
3. Results
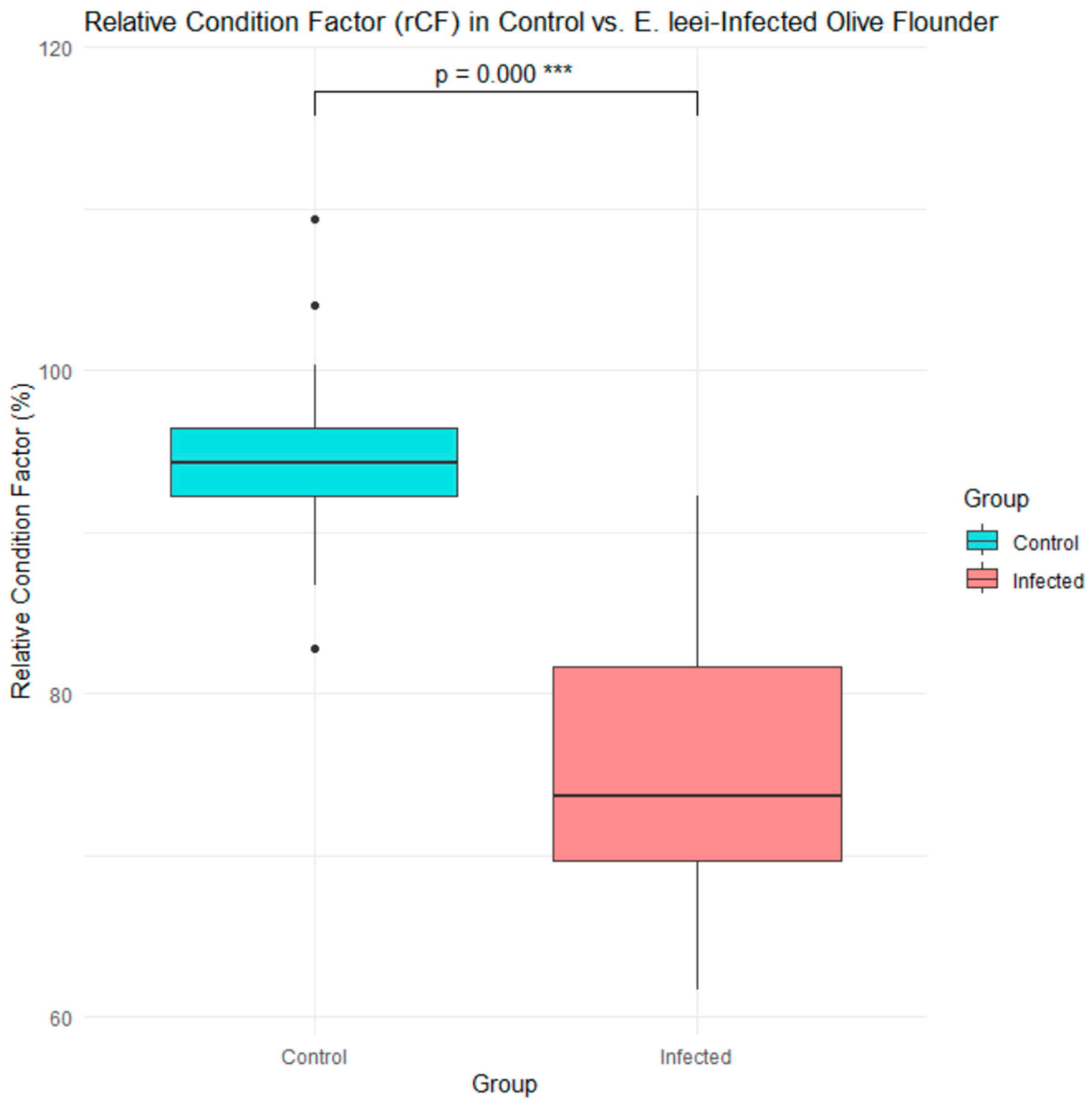
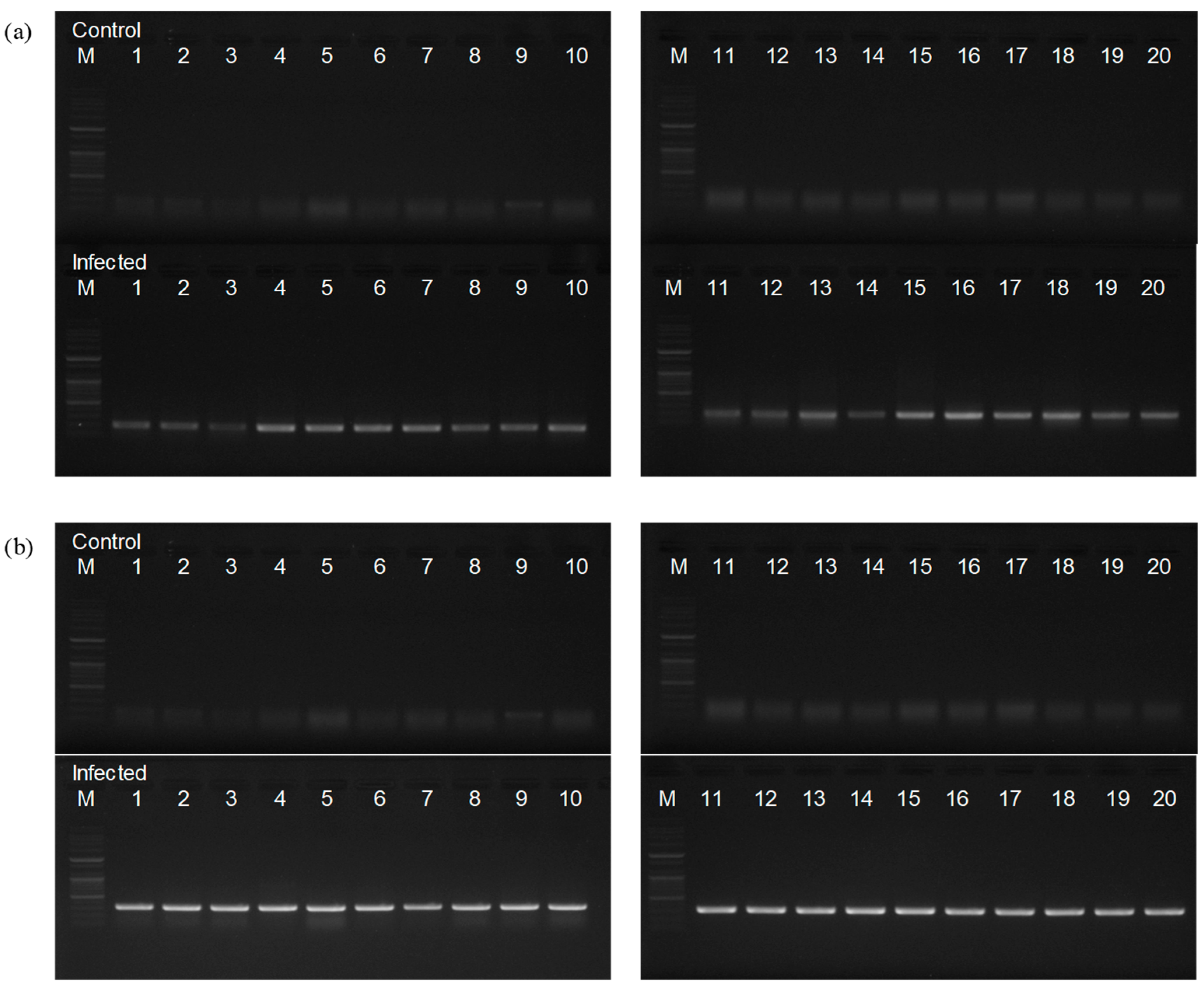
3.1. Phenotypic and Molecular Diagnosis
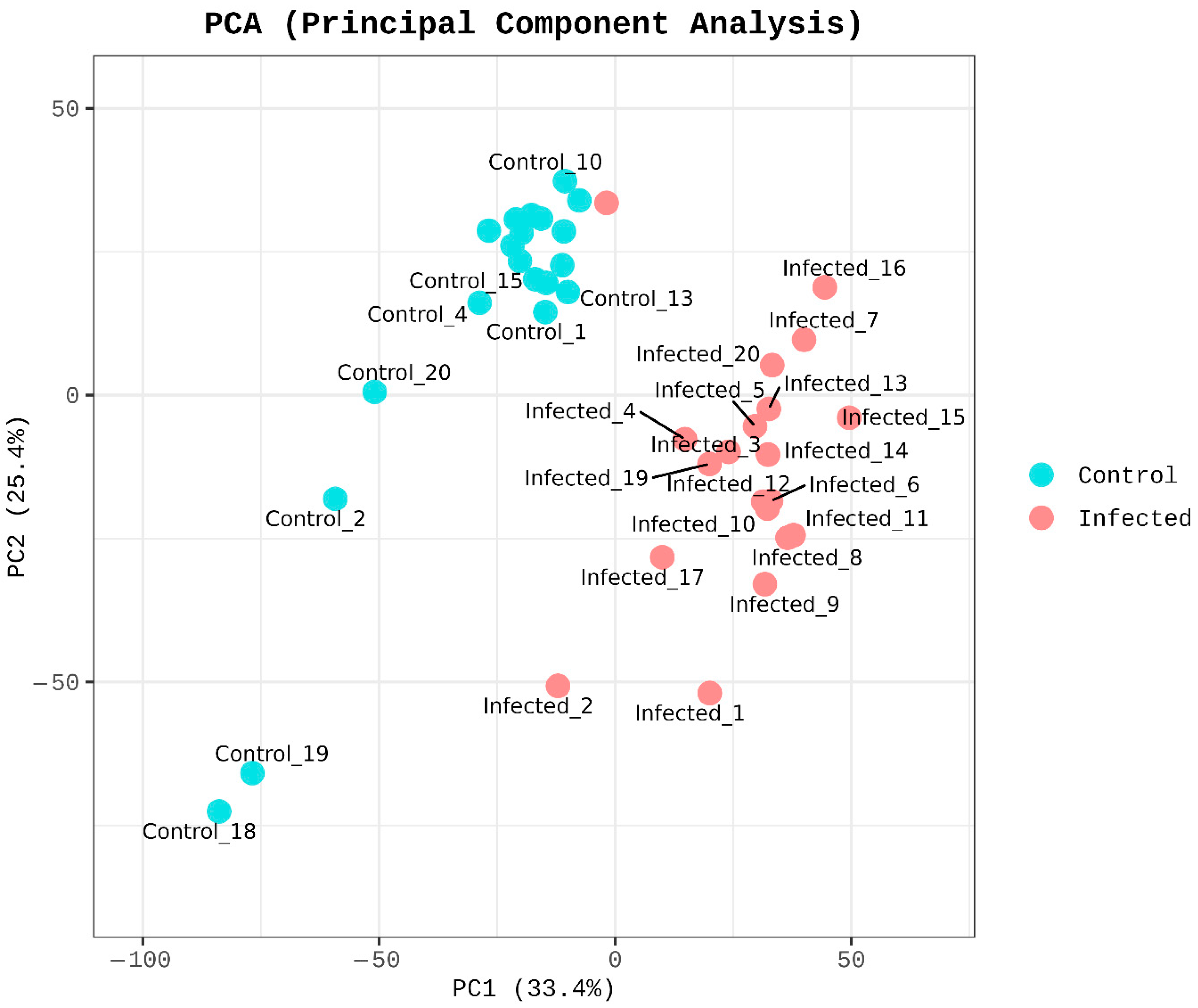
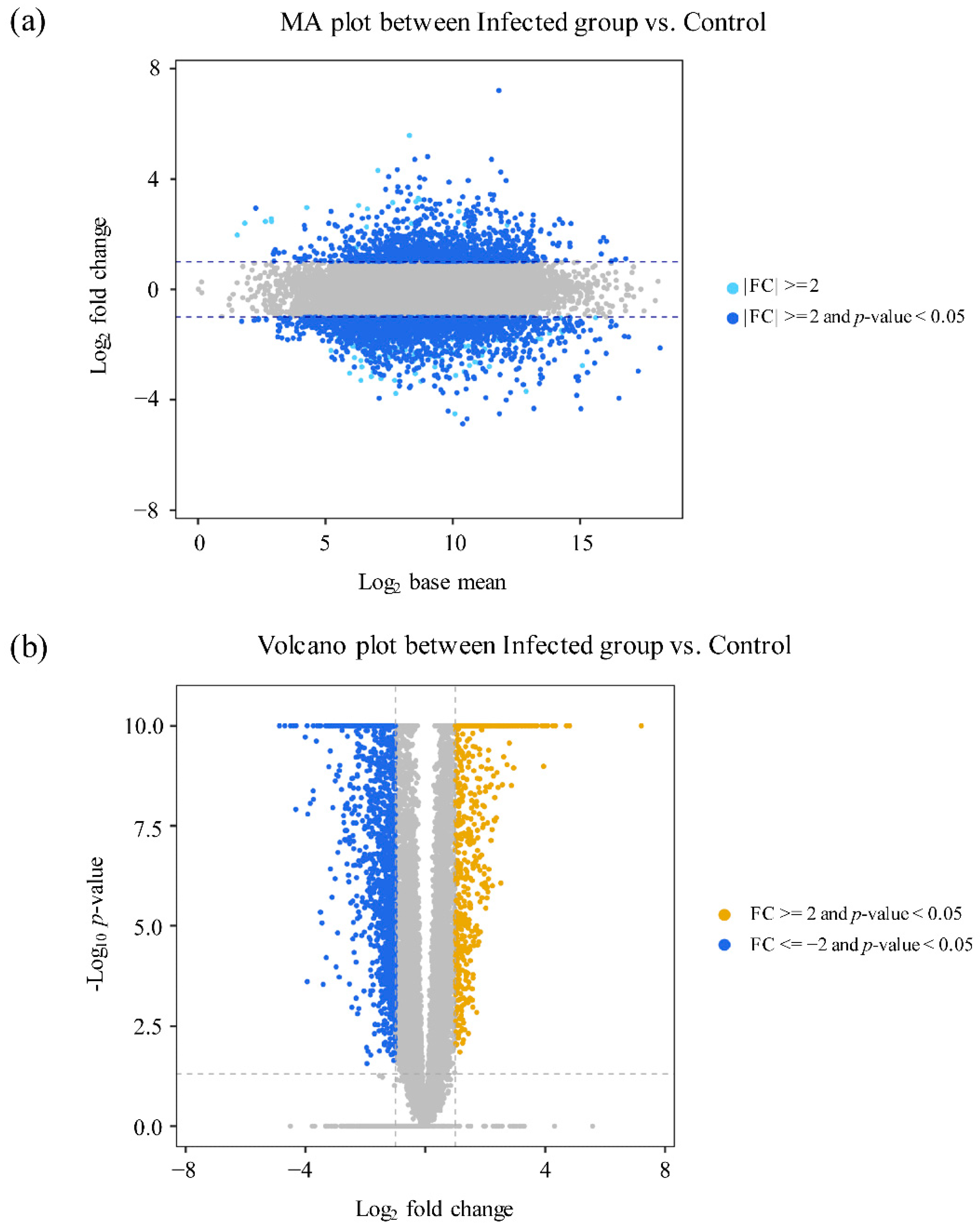
3.2. Transcriptomic Profiling
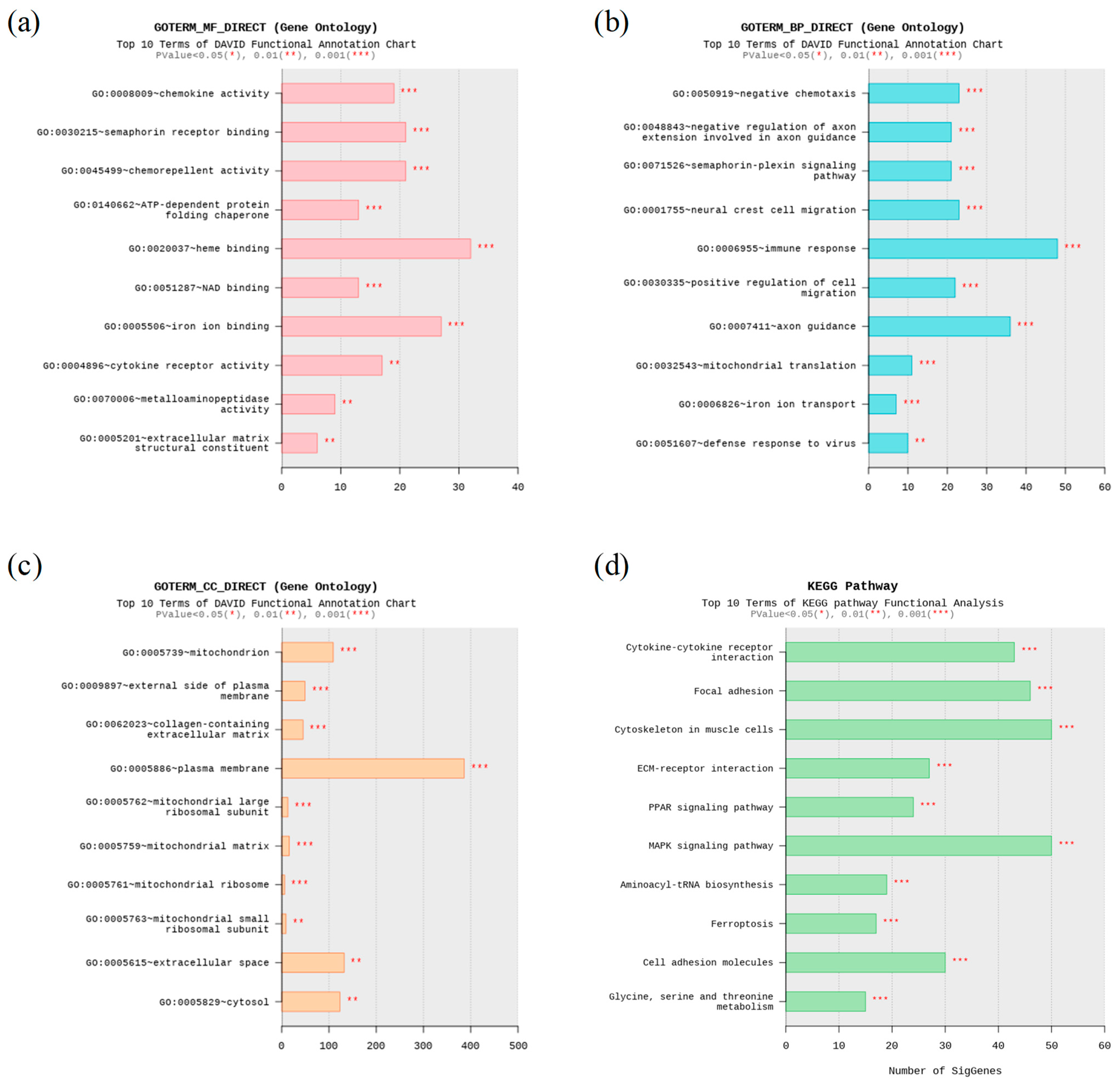
3.3. Functional Enrichment Analyses
3.3.1. GO Enrichment Analysis
3.3.2. KEGG Enrichment Analysis
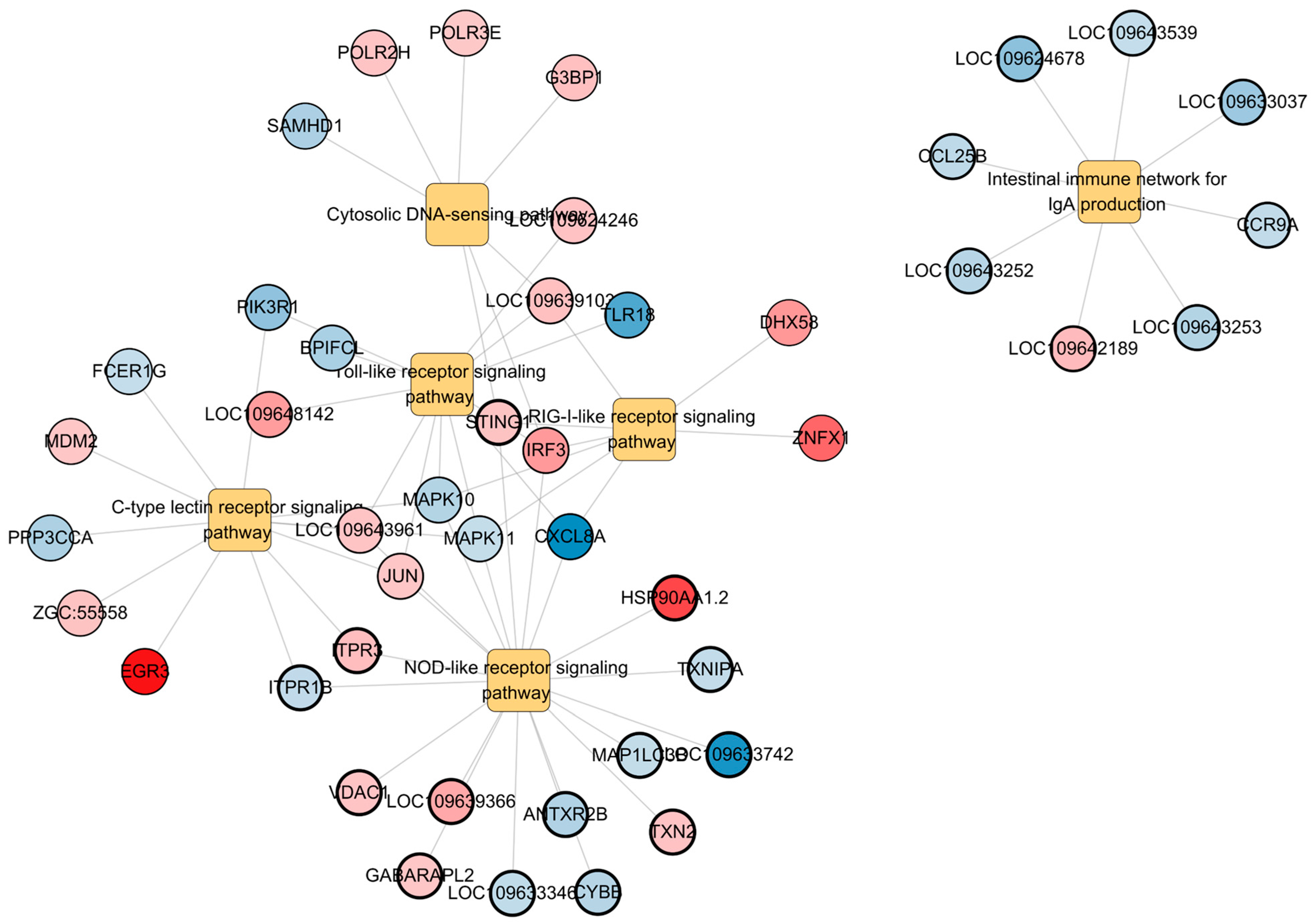
3.4. Immune-Related Pathway Alterations
3.5. Validation of DEGs by qRT-PCR
4. Discussion
4.1. Suppression of Chemokine and Cytokine Signaling
4.2. Activation of Nucleic Acid-Sensing Pathways
4.3. Impairment of Mucosal and Adaptive Immunity
4.4. Functional Roles of Hub Genes and Biomarker Potential
4.5. Network-Level Interactions and Integrative Insights
4.6. Implications and Future Perspectives
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Oh, S.; Lee, S. Fish Welfare-Related Issues and Their Relevance in Land-Based Olive Flounder (Paralichthys olivaceus) Farms in Korea. Animals 2024, 14, 1693. [Google Scholar] [CrossRef]
- Hamidoghli, A.; Won, S.; Lee, S.; Lee, S.; Farris, N.W.; Bai, S.C. Nutrition and feeding of olive flounder Paralichthys olivaceus: A Review. Rev. Fish. Sci. Aquac. 2020, 28, 340–357. [Google Scholar] [CrossRef]
- Raman, R.P.; Prakash, C.; Makesh, M.; Pawar, N. Environmental stress mediated diseases of fish: An overview. Adv. Fish Res. 2013, 5, 141–158. [Google Scholar]
- Rathod, S.V.; Saras, P.; Gondaliya, S.M. Environmental pollution: Threats and challenges for management. In Eco-Restoration of Polluted Environment; CRC Press: Boca Raton, FL, USA, 2024; pp. 1–34. [Google Scholar]
- Yadav, N.K.; Patel, A.B.; Singh, S.K.; Mehta, N.K.; Anand, V.; Lal, J.; Dekari, D.; Devi, N.C. Climate change effects on aquaculture production and its sustainable management through climate-resilient adaptation strategies: A review. Environ. Sci. Pollut. Res. 2024, 31, 31731–31751. [Google Scholar] [CrossRef]
- Christaki, E. Classification of parasitic diseases. In The Surgical Management of Parasitic Diseases; Springer: Berlin/Heidelberg, Germany, 2020; pp. 23–45. [Google Scholar]
- Sitjà-Bobadilla, A.; Estensoro, I.; Palenzuela, O. Enteromyxum leei. In Fish Parasites: A Handbook of Protocols for Their Isolation, Culture and Transmission; CABI GB: Wallingford, UK, 2021; pp. 127–142. [Google Scholar]
- Picard-Sánchez, A. Control of Enteric Parasitic Diseases of Farmed Gilthead Sea Bream: New Insights into Enteromyxum leei (Myxozoa) and Enterospora Nucleophila (Microsporidia) Infections. Ph.D. Thesis, Universidad Politécnica de Valencia, Valencia, Spain, 2021. [Google Scholar]
- Picard-Sánchez, A.; Estensoro, I.; Del Pozo, R.; Palenzuela, O.R.; Piazzon, M.C.; Sitjà-Bobadilla, A. Water temperature, time of exposure and population density are key parameters in Enteromyxum leei fish-to-fish experimental transmission. J. Fish Dis. 2020, 43, 491–502. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.P.; Sohn, H.C.; Jin, C.N.; Kang, B.J.; Lee, J. Quantitative investigation of Enteromyxum leei (Myxozoa: Myxosporea) infection and relative condition factor in cultured olive flounder Paralichthys olivaceus (Temminck and Schlegel). J. Fish Dis. 2019, 42, 159–165. [Google Scholar] [CrossRef]
- Shin, S.P.; Jin, C.N.; Sohn, H.; Lee, J. Comparison of oral and anal inoculation of Enteromyxum leei into olive flounder Paralichthys olivaceus. Aquaculture 2022, 561, 738641. [Google Scholar] [CrossRef]
- Fleurance, R.; Sauvegrain, C.; Marques, A.; Le Breton, A.; Guéreaud, C.; Cherel, Y.; Wyers, M. Histopathological changes caused by Enteromyxum leei infection in farmed sea bream Sparus aurata. Dis. Aquat. Org. 2008, 79, 219–228. [Google Scholar] [CrossRef]
- Alvarez-Pellitero, P.; Palenzuela, O.; Sitjà-Bobadilla, A. Histopathology and cellular response in Enteromyxum leei (Myxozoa) infections of Diplodus puntazzo (Teleostei). Parasitol. Int. 2008, 57, 110–120. [Google Scholar] [CrossRef]
- Muñoz, P.; Cuesta, A.; Athanassopoulou, F.; Golomazou, H.; Crespo, S.; Padrós, F.; Sitjà-Bobadilla, A.; Albiñana, G.; Esteban, M.; Alvarez-Pellitero, P. Sharpsnout sea bream (Diplodus puntazzo) humoral immune response against the parasite Enteromyxum leei (Myxozoa). Fish Shellfish Immunol. 2007, 23, 636–645. [Google Scholar] [CrossRef] [PubMed]
- Sitjà-Bobadilla, A.; Schmidt-Posthaus, H.; Wahli, T.; Holland, J.W.; Secombes, C.J. Fish immune responses to Myxozoa. In Myxozoan Evolution, Ecology and Development; Springer: Berlin/Heidelberg, Germany, 2015; pp. 253–280. [Google Scholar]
- Sohn, H.; Jin, C.N.; Kang, B.J.; Shin, S.P.; Lee, J. Infection dynamics of Enteromyxum leei (Myxozoa, Myxosporea) in culture water and its effects on cultured olive flounder, Paralichthys olivaceus (Temminck & Schlegel). J. Fish Dis. 2021, 44, 1475–1479. [Google Scholar] [CrossRef]
- Kim, S.M.; Jun, L.J.; Lee, D.W.; Park, H.K.; Do Jeong, H.; Kim, J.S.; Jeong, J.B. Monitoring of emaciation disease in cultured Paralichthys olivaceus of Jeju island during 2014–2015. Fish. Aquat. Sci. 2018, 21, 17. [Google Scholar] [CrossRef]
- Sohn, H.; Jin, C.N.; Kim, J.; Park, C.U.; Jo, Y.; Jeong, T.; Lee, J. Investigation of Disease Outbreak Trends through Monitoring of Olive Flounder Paralichthys olivaceus in Jeju Island. Smart Media J. 2024, 13, 49–58. [Google Scholar]
- Piazzon, M.C.; Estensoro, I.; Calduch-Giner, J.A.; Del Pozo, R.; Picard-Sánchez, A.; Pérez-Sánchez, J.; Sitjà-Bobadilla, A. Hints on T cell responses in a fish-parasite model: Enteromyxum leei induces differential expression of T cell signature molecules depending on the organ and the infection status. Parasites Vectors 2018, 11, 443. [Google Scholar] [CrossRef] [PubMed]
- Ronza, P.; Robledo, D.; Bermúdez, R.; Losada, A.P.; Pardo, B.G.; Sitjà-Bobadilla, A.; Quiroga, M.I.; Martínez, P. RNA-seq analysis of early enteromyxosis in turbot (Scophthalmus maximus): New insights into parasite invasion and immune evasion strategies. Int. J. Parasitol. 2016, 46, 507–517. [Google Scholar] [CrossRef]
- Toxqui-Rodríguez, S.; Estensoro, I.; Domingo-Bretón, R.; Del Pozo, R.; Pérez-Sánchez, J.; Sipkema, D.; Sitjà-Bobadilla, A.; Piazzon, M.C. Interactions between gilthead seabream intestinal transcriptome and microbiota upon Enteromyxum leei infection: A multi–omic approach. Anim. Microbiome 2025, 7, 22. [Google Scholar] [CrossRef]
- Hwang, J.Y.; Markkandan, K.; Kwon, M.G.; Seo, J.S.; Yoo, S.-i.; Hwang, S.D.; Son, M.-H.; Park, J. Transcriptome analysis of olive flounder (Paralichthys olivaceus) head kidney infected with moderate and high virulent strains of infectious viral hemorrhagic septicaemia virus (VHSV). Fish Shellfish Immunol. 2018, 76, 293–304. [Google Scholar] [CrossRef]
- Wu, Q.; Ning, X.; Jiang, S.; Sun, L. Transcriptome analysis reveals seven key immune pathways of Japanese flounder (Paralichthys olivaceus) involved in megalocytivirus infection. Fish Shellfish Immunol. 2020, 103, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.I.; Lee, U.H.; Cho, M.; Jung, S.-H.; Min, E.Y.; Park, J.W. Transcriptome analysis based on RNA-seq of common innate immune responses of flounder cells to IHNV, VHSV, and HIRRV. PLoS ONE 2020, 15, e0239925. [Google Scholar] [CrossRef] [PubMed]
- Chinchilla, B.; Encinas, P.; Coll, J.M.; Gomez-Casado, E. Differential immune transcriptome and modulated signalling pathways in rainbow trout infected with viral haemorrhagic septicaemia virus (VHSV) and its derivative non-virion (NV) gene deleted. Vaccines 2020, 8, 58. [Google Scholar] [CrossRef]
- Sun, B.; Li, X.; Ning, X.; Sun, L. Transcriptome analysis of Paralichthys olivaceus erythrocytes reveals profound immune responses induced by edwardsiella tarda infection. Int. J. Mol. Sci. 2020, 21, 3094. [Google Scholar] [CrossRef]
- Ghani, M.U.; Chen, J.; Khosravi, Z.; Wu, Q.; Liu, Y.; Zhou, J.; Zhong, L.; Cui, H. Unveiling the multifaceted role of toll-like receptors in immunity of aquatic animals: Pioneering strategies for disease management. Front. Immunol. 2024, 15, 1378111. [Google Scholar] [CrossRef]
- Sudhagar, A.; Kumar, G.; El-Matbouli, M. Transcriptome analysis based on RNA-Seq in understanding pathogenic mechanisms of diseases and the immune system of fish: A comprehensive review. Int. J. Mol. Sci. 2018, 19, 245. [Google Scholar] [CrossRef]
- Huang, L.; Li, G.; Mo, Z.; Xiao, P.; Li, J.; Huang, J. De novo assembly of the Japanese flounder (Paralichthys olivaceus) spleen transcriptome to identify putative genes involved in immunity. PLoS ONE 2015, 10, e0117642. [Google Scholar]
- Wang, W.; Wang, J.; You, F.; Ma, L.; Yang, X.; Gao, J.; He, Y.; Qi, J.; Yu, H.; Wang, Z. Detection of alternative splice and gene duplication by RNA sequencing in Japanese flounder, Paralichthys olivaceus. G3 Genes Genomes Genet. 2014, 4, 2419–2424. [Google Scholar] [CrossRef]
- Xiu, Y.; Li, Y.; Liu, X.; Li, C. Full-length transcriptome sequencing from multiple immune-related tissues of Paralichthys olivaceus. Fish Shellfish Immunol. 2020, 106, 930–937. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.P.; Sohn, H.C.; Jin, C.N.; Kang, B.J.; Lee, J. Molecular diagnostics for verifying an etiological agent of emaciation disease in cultured olive flounder Paralichthys olivaceus in Korea. Aquaculture 2018, 493, 18–25. [Google Scholar] [CrossRef]
- Estensoro, I.; Redondo, M.J.; Salesa, B.; Kaushik, S.; Pérez-Sánchez, J.; Sitjà-Bobadilla, A. Effect of nutrition and Enteromyxum leei infection on gilthead sea bream Sparus aurata intestinal carbohydrate distribution. Dis. Aquat. Org. 2012, 100, 29–42. [Google Scholar] [CrossRef] [PubMed]
- Estensoro, I.; Jung-Schroers, V.; Álvarez-Pellitero, P.; Steinhagen, D.; Sitjà-Bobadilla, A. Effects of Enteromyxum leei (Myxozoa) infection on gilthead sea bream (Sparus aurata) (Teleostei) intestinal mucus: Glycoprotein profile and bacterial adhesion. Parasitol. Res. 2013, 112, 567–576. [Google Scholar] [CrossRef]
- Bjørgen, H.; Li, Y.; Kortner, T.M.; Krogdahl, Å.; Koppang, E.O. Anatomy, immunology, digestive physiology and microbiota of the salmonid intestine: Knowns and unknowns under the impact of an expanding industrialized production. Fish Shellfish Immunol. 2020, 107, 172–186. [Google Scholar] [CrossRef]
- Van der Aa, L.M.; Chadzinska, M.; Tijhaar, E.; Boudinot, P.; Verburg-van Kemenade, B.L. CXCL8 chemokines in teleost fish: Two lineages with distinct expression profiles during early phases of inflammation. PLoS ONE 2010, 5, e12384. [Google Scholar] [CrossRef] [PubMed]
- Sayyaf Dezfuli, B.; Lorenzoni, M.; Carosi, A.; Giari, L.; Bosi, G. Teleost innate immunity, an intricate game between immune cells and parasites of fish organs: Who wins, who loses. Front. Immunol. 2023, 14, 1250835. [Google Scholar] [CrossRef]
- Hu, Y.-H.; Chen, L.; Sun, L. CXCL8 of Scophthalmus maximus: Expression, biological activity and immunoregulatory effect. Dev. Comp. Immunol. 2011, 35, 1032–1039. [Google Scholar] [CrossRef]
- Losada, A.; Bermúdez, R.; Faílde, L.; Di Giancamillo, A.; Domeneghini, C.; Quiroga, M. Effects of Enteromyxum scophthalmi experimental infection on the neuroendocrine system of turbot, Scophthalmus maximus (L.). Fish Shellfish Immunol. 2014, 40, 577–583. [Google Scholar] [CrossRef]
- Davey, G.C.; Calduch-Giner, J.A.; Houeix, B.; Talbot, A.; Sitjà-Bobadilla, A.; Prunet, P.; Pérez-Sánchez, J.; Cairns, M.T. Molecular profiling of the gilthead sea bream (Sparus aurata L.) response to chronic exposure to the myxosporean parasite Enteromyxum leei. Mol. Immunol. 2011, 48, 2102–2112. [Google Scholar] [CrossRef] [PubMed]
- Sitjà-Bobadilla, A.; Calduch-Giner, J.; Saera-Vila, A.; Palenzuela, O.; Álvarez-Pellitero, P.; Pérez-Sánchez, J. Chronic exposure to the parasite Enteromyxum leei (Myxozoa: Myxosporea) modulates the immune response and the expression of growth, redox and immune relevant genes in gilthead sea bream, Sparus aurata L. Fish Shellfish Immunol. 2008, 24, 610–619. [Google Scholar] [CrossRef]
- Holzer, A.S.; Piazzon, M.C.; Barrett, D.; Bartholomew, J.L.; Sitjà-Bobadilla, A. To react or not to react: The dilemma of fish immune systems facing myxozoan infections. Front. Immunol. 2021, 12, 734238. [Google Scholar] [CrossRef]
- Sun, L.; Wu, J.; Du, F.; Chen, X.; Chen, Z.J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 2013, 339, 786–791. [Google Scholar] [CrossRef]
- Mojzesz, M.; Rakus, K.; Chadzinska, M.; Nakagami, K.; Biswas, G.; Sakai, M.; Hikima, J.-i. Cytosolic sensors for pathogenic viral and bacterial nucleic acids in fish. Int. J. Mol. Sci. 2020, 21, 7289. [Google Scholar] [CrossRef]
- Guo, S.; Zeng, M.; Wang, Z.; Zhao, L.; Fan, Y.; Shi, Q.; Song, Z. Characterization and expression profiles of cGAS (cyclic GMP-AMP synthase) and STING (stimulator of interferon) genes in various immune tissues of hybrid yellow catfish under bacterial infections. Aquac. Rep. 2024, 37, 102238. [Google Scholar] [CrossRef]
- Liu, J.; Zhou, J.; Luan, Y.; Li, X.; Meng, X.; Liao, W.; Tang, J.; Wang, Z. cGAS-STING, inflammasomes and pyroptosis: An overview of crosstalk mechanism of activation and regulation. Cell Commun. Signal. 2024, 22, 22. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Chen, Z.J. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci. Signal. 2012, 5, ra20. [Google Scholar] [CrossRef] [PubMed]
- Bowie, A. The STING in the tail for cytosolic DNA–dependent activation of IRF3. Sci. Signal. 2012, 5, pe9. [Google Scholar] [CrossRef]
- Sun, Y.; Cao, Z.; Zhang, P.; Wei, C.; Li, J.; Wu, Y.; Zhou, Y. IFN regulatory factor 3 of golden pompano and its NLS domain are involved in antibacterial innate immunity and regulate the expression of type I interferon (IFNa3). Front. Immunol. 2023, 14, 1128196. [Google Scholar] [CrossRef]
- Holland, J.; Bird, S.; Williamson, B.; Woudstra, C.; Mustafa, A.; Wang, T.; Zou, J.; Blaney, S.; Collet, B.; Secombes, C. Molecular characterization of IRF3 and IRF7 in rainbow trout, Oncorhynchus mykiss: Functional analysis and transcriptional modulation. Mol. Immunol. 2008, 46, 269–285. [Google Scholar] [CrossRef]
- Schonthaler, H.B.; Guinea-Viniegra, J.; Wagner, E.F. Targeting inflammation by modulating the Jun/AP-1 pathway. Ann. Rheum. Dis. 2011, 70, i109–i112. [Google Scholar] [CrossRef]
- Shen, Y.; Gao, J.; Zhang, M.; Li, Y.; Aly, R.S.S.; Wang, L.; Yu, M.; Jiang, H.; Qiao, Z.; Chen, X. Transcriptome to reveal the immunological defenses in mandarin fish (Siniperca chuatsi) skin with LPS and poly (I: C) infection. Fish Shellfish Immunol. 2025, 166, 110652. [Google Scholar] [CrossRef] [PubMed]
- Fang, W.; Chen, Q.; Li, J.; Liu, Y.; Zhao, Z.; Shen, Y.; Mai, K.; Ai, Q. Endoplasmic reticulum stress disturbs lipid homeostasis and augments inflammation in the intestine and isolated intestinal cells of Large yellow croaker (Larimichthys crocea). Front. Immunol. 2021, 12, 738143. [Google Scholar] [CrossRef]
- Cohen-Sánchez, A.; Sánchez-Mairata, A.G.; Valencia, J.M.; Box, A.; Pinya, S.; Tejada, S.; Sureda, A. Immune and oxidative stress response of the fish Xyrichthys novacula infected with the trematode ectoparasite Scaphanocephalus sp. in the Balearic Islands. Fishes 2023, 8, 600. [Google Scholar] [CrossRef]
- Mahdy, O.A.; Abdel-Maogood, S.Z.; Abdelsalam, M.; Salem, M.A. A multidisciplinary study on Clinostomum infections in Nile tilapia: Micro-morphology, oxidative stress, immunology, and histopathology. BMC Vet. Res. 2024, 20, 60. [Google Scholar] [CrossRef]
- Pawłowska, M.; Mila-Kierzenkowska, C.; Szczegielniak, J.; Woźniak, A. Oxidative stress in parasitic diseases—Reactive oxygen species as mediators of interactions between the host and the parasites. Antioxidants 2023, 13, 38. [Google Scholar] [CrossRef]
- Kunkel, E.J.; Campbell, D.J.; Butcher, E.C. Chemokines in lymphocyte trafficking and intestinal immunity. Microcirculation 2003, 10, 313–323. [Google Scholar] [CrossRef]
- Wu, X.; Sun, M.; Yang, Z.; Lu, C.; Wang, Q.; Wang, H.; Deng, C.; Liu, Y.; Yang, Y. The roles of CCR9/CCL25 in inflammation and inflammation-associated diseases. Front. Cell Dev. Biol. 2021, 9, 686548. [Google Scholar] [CrossRef] [PubMed]
- Picard-Sánchez, A.; Estensoro, I.; Perdiguero, P.; Del Pozo, R.; Tafalla, C.; Piazzon, M.C.; Sitjà-Bobadilla, A. Passive immunization delays disease outcome in gilthead sea bream infected with Enteromyxum leei (Myxozoa), despite the moderate changes in IgM and IgT repertoire. Front. Immunol. 2020, 11, 581361. [Google Scholar] [CrossRef]
- Estensoro, I.; Calduch-Giner, J.A.; Kaushik, S.; Pérez-Sánchez, J.; Sitjà-Bobadilla, A. Modulation of the IgM gene expression and IgM immunoreactive cell distribution by the nutritional background in gilthead sea bream (Sparus aurata) challenged with Enteromyxum leei (Myxozoa). Fish Shellfish Immunol. 2012, 33, 401–410. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Yu, B.; Chen, J.; Wu, H.; Xu, Y.; Yang, B.; Lu, Q. Thymic stromal lymphopoietin epigenetically upregulates Fc receptor γ subunit–related receptors on antigen-presenting cells and induces TH2/TH17 polarization through dectin-2. J. Allergy Clin. Immunol. 2019, 144, 1025–1035.e7. [Google Scholar] [CrossRef]
- Di Genova, B.M.; Tonelli, R.R. Infection strategies of intestinal parasite pathogens and host cell responses. Front. Microbiol. 2016, 7, 256. [Google Scholar] [CrossRef] [PubMed]
- Brinchmann, M.F.; Patel, D.M.; Pinto, N.; Iversen, M.H. Functional aspects of fish mucosal lectins—Interaction with non-self. Molecules 2018, 23, 1119. [Google Scholar] [CrossRef]
- Pérez, T.; Balcázar, J.; Ruiz-Zarzuela, I.; Halaihel, N.; Vendrell, D.; de Blas, I.; Múzquiz, J. Host–microbiota interactions within the fish intestinal ecosystem. Mucosal Immunol. 2010, 3, 355–360. [Google Scholar] [CrossRef]
- Yu, Y.; Wang, Q.; Huang, Z.; Ding, L.; Xu, Z. Immunoglobulins, mucosal immunity and vaccination in teleost fish. Front. Immunol. 2020, 11, 567941. [Google Scholar] [CrossRef]
- Aghaallaei, N.; Agarwal, R.; Benjaminsen, J.; Lust, K.; Bajoghli, B.; Wittbrodt, J.; Feijoo, C.G. Antigen-presenting cells and T cells interact in a specific area of the intestinal mucosa defined by the ccl25-ccr9 axis in medaka. Front. Immunol. 2022, 13, 812899. [Google Scholar] [CrossRef]
- Salinas, I.; Fernández-Montero, Á.; Ding, Y.; Sunyer, J.O. Mucosal immunoglobulins of teleost fish: A decade of advances. Dev. Comp. Immunol. 2021, 121, 104079. [Google Scholar] [CrossRef]
- Salinas, I. The mucosal immune system of teleost fish. Biology 2015, 4, 525–539. [Google Scholar] [CrossRef]
- Russo, R.C.; Garcia, C.C.; Teixeira, M.M.; Amaral, F.A. The CXCL8/IL-8 chemokine family and its receptors in inflammatory diseases. Expert Rev. Clin. Immunol. 2014, 10, 593–619. [Google Scholar] [CrossRef]
- He, X.; Zhu, Z.; Johnson, C.; Stoops, J.; Eaker, A.E.; Bowen, W.; DeFrances, M.C. PIK3IP1, a negative regulator of PI3K, suppresses the development of hepatocellular carcinoma. Cancer Res. 2008, 68, 5591–5598. [Google Scholar] [CrossRef]
- Zheng, W.; Xu, X.-w.; E, Z.; Liu, Y.; Chen, S. Genome-wide identification of the MAPK gene family in turbot and its involvement in abiotic and biotic stress responses. Front. Mar. Sci. 2022, 9, 1005401. [Google Scholar] [CrossRef]
- Kojima, K.; Ichijo, H.; Naguro, I. Molecular functions of ASK family in diseases caused by stress-induced inflammation and apoptosis. J. Biochem. 2021, 169, 395–407. [Google Scholar] [CrossRef] [PubMed]
- Blanco, E.; Camps, C.; Bahal, S.; Kerai, M.D.; Ferla, M.P.; Rochussen, A.M.; Handel, A.E.; Golwala, Z.M.; Spiridou Goncalves, H.; Kricke, S. Dominant negative variants in ITPR3 impair T cell Ca2+ dynamics causing combined immunodeficiency. J. Exp. Med. 2024, 222, e20220979. [Google Scholar] [CrossRef]
- Thackray, L.B.; Duan, E.; Lazear, H.M.; Kambal, A.; Schreiber, R.D.; Diamond, M.S.; Virgin, H.W. Critical role for interferon regulatory factor 3 (IRF-3) and IRF-7 in type I interferon-mediated control of murine norovirus replication. J. Virol. 2012, 86, 13515–13523. [Google Scholar] [CrossRef]
- Wang, Y.-D.; Wang, Y.-H.; Hui, C.-F.; Chen, J.-Y. Transcriptome analysis of the effect of Vibrio alginolyticus infection on the innate immunity-related TLR5-mediated induction of cytokines in Epinephelus lanceolatus. Fish Shellfish Immunol. 2016, 52, 31–43. [Google Scholar] [CrossRef] [PubMed]
- Trung, N.B.; Nan, F.-H.; Lee, M.-C.; Loh, J.-Y.; Gong, H.-Y.; Lu, M.-W.; Hang, H.T.; Lin, Y.-L.; Lee, P.-T. Fish-specific TLR18 in Nile tilapia (Oreochromis niloticus) recruits MyD88 and TRIF to induce expression of effectors in NF-κB and IFN pathways in melanomacrophages. Fish Shellfish Immunol. 2021, 119, 587–601. [Google Scholar] [CrossRef]
- Zhu, L.-y.; Nie, L.; Zhu, G.; Xiang, L.-x.; Shao, J.-z. Advances in research of fish immune-relevant genes: A comparative overview of innate and adaptive immunity in teleosts. Dev. Comp. Immunol. 2013, 39, 39–62. [Google Scholar] [CrossRef] [PubMed]
- Muangrerk, C.; Uchuwittayakul, A.; Srisapoome, P. Identification, expression and antimicrobial functional analysis of interleukin-8 (IL-8) in response to Streptococcus iniae and Flavobacterium covae in Asian seabass (Lates calcarifer Bloch, 1790). Animals 2024, 14, 475. [Google Scholar] [CrossRef]
- Ashour, D.S. Toll-like receptor signaling in parasitic infections. Expert Rev. Clin. Immunol. 2015, 11, 771–780. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, R.; Tiwari, M.; Chaudhary, A.; Sharan Thakur, R.; Pande, V.; Das, J. Chemokines: A key driver for inflammation in protozoan infection. Int. Rev. Immunol. 2024, 43, 211–228. [Google Scholar] [CrossRef] [PubMed]
- Ronza, P.; Robledo, D.; Bermúdez, R.; Losada, A.P.; Pardo, B.G.; Martínez, P.; Quiroga, M.I. Integrating genomic and morphological approaches in fish pathology research: The case of turbot (Scophthalmus maximus) enteromyxosis. Front. Genet. 2019, 10, 26. [Google Scholar]
- Estensoro, I.; Benedito-Palos, L.; Palenzuela, O.; Kaushik, S.; Sitjà-Bobadilla, A.; Pérez-Sánchez, J. The nutritional background of the host alters the disease course in a fish–myxosporean system. Vet. Parasitol. 2011, 175, 141–150. [Google Scholar] [CrossRef]



| Primer | Sequence (5′–3′) | Source | Target Size (bp) |
|---|---|---|---|
| EL-Shin-FW | CGGTGACGCCAATCCGTG | Shin, et al. (2018) [32] | 192 |
| EL-Shin-RV | GACGGTATCTGATCGTCTTCGA | ||
| EL-New-FW | CGCCAATCCGTGTTGGTTTT | This study | 318 |
| EL-New-RV | CAAATTAAGCCGCAGGCTCC |
| Gene Symbol | Accession No. | Primer Sequence (5′–3′) | Product Size (bp) |
|---|---|---|---|
| cxcl8a | XM_020100336.2 * | FW: TGGCCATTCCTGATGGAACC | 105 |
| RV: TCCACGCTTCCTATGTGACG | |||
| pik3r1 | XM_020082243.2 | FW: AGACGCCGAATGGTACTGG | 116 |
| RV: GTCTCCGTGCATTTTCGTCG | |||
| mapk10 | XM_069513303.1 | FW: ATCTGCACTCAGCTGGCATT | 249 |
| RV: GGATTTTGTGGCGCACCATT | |||
| itpr1b | XM_069512494.1 | FW: CGCCGCTGAGATAGACACAT | 200 |
| RV: CGATCTCGAACCTCGACAGG | |||
| mapk11 | XM_069519302.1 | FW: CTCATACCGGGAACTCAGGC | 210 |
| RV: CCGCGGAGGAGCTGATAAAT | |||
| jun | XM_069522324.1 | FW: CTGTCTCGGCTCCGAACTAC | 179 |
| RV: CATGTCGATCGGGGAGAGTG | |||
| sting1 | XM_069531174.1 | FW: CTACTTGCGATTGGTGCTGC | 210 |
| RV: CCTGCCCTGTCAATCTCGTT | |||
| itpr3 | XM_069527461.1 | FW: CTCAAAGGAGGCGATGTGGT | 263 |
| RV: CTACCGTCCATCTCAGCAGC | |||
| irf3 | OR047553.1 | FW: CAGTGGACGAATCCGGAACA | 247 |
| RV: CCGGCCAGTGAAACACTTTG | |||
| gapdh (housekeeping) | AB029337.1 | FW: CATCAAATGGGGCGATGCTG | 216 |
| RV: CAGTTGGTTGTGCAGGAAGC |
| Sample | Raw Reads | Clean Reads | Clean Bases | Q20 (%) | Mapping Ratio (%) | Number of Transcripts | Mean Length of Transcripts (bp) |
|---|---|---|---|---|---|---|---|
| Control_Pi | 78,139,897 | 74,898,452 | 10,813,483,192 | 99.51 | 94.73 | 20,559 | 17,428 |
| Infected_Pi | 79,405,829 | 75,623,078 | 10,890,747,787 | 99.50 | 77.73 | 20,152 | 17,269 |
| Pathway | Gene Symbol | Log2FC | p-Value |
|---|---|---|---|
| Toll-like receptor signaling pathway | cxcl8a * | −7.89 | 0.0036 |
| tlr18 | −6.12 | 0.0036 | |
| pik3r1 * | −4.00 | 0.0036 | |
| bpifcl | −3.03 | 0.0036 | |
| mapk10 * | −2.66 | 0.0036 | |
| mapk11 * | −2.02 | 0.0036 | |
| Jun * | 2.09 | 0.0036 | |
| LOC109643961 | 2.21 | 0.0036 | |
| LOC109624246 | 2.25 | 0.0036 | |
| LOC109639103 | 2.32 | 0.0036 | |
| LOC109648142 | 3.78 | 0.0036 | |
| irf3 * | 3.93 | 0.0036 | |
| NOD-like receptor signaling pathway | sting1 * | 2.21 | 0.0000 |
| antxr2b | −2.79 | 0.0000 | |
| itpr1b * | −2.19 | 0.0000 | |
| hsp90aa1.2 | 7.28 | 0.0000 | |
| LOC109633346 | −2.16 | 0.0000 | |
| LOC109633742 | −7.33 | 0.0000 | |
| vdac1 | 2.21 | 0.0000 | |
| LOC109639366 | 3.37 | 0.0000 | |
| map1lc3b | −2.06 | 0.0000 | |
| txnipa | −2.09 | 0.0000 | |
| gabarapl2 | 2.09 | 0.0000 | |
| txn2 | 2.24 | 0.0000 | |
| itpr3 * | 2.46 | 0.0000 | |
| cybb | −2.57 | 0.0000 | |
| Intestinal immune network for IgA production | LOC109624678 | −4.09 | 0.0002 |
| ccl25b | −2.31 | 0.0002 | |
| LOC109633037 | −3.55 | 0.0002 | |
| LOC109642189 | 2.58 | 0.0002 | |
| LOC109643252 | −2.52 | 0.0002 | |
| LOC109643253 | −2.67 | 0.0002 | |
| LOC109643539 | −2.04 | 0.0002 | |
| ccr9a | −2.15 | 0.0002 | |
| C-type lectin receptor signaling pathway | egr3 | 9.22 | 0.0094 |
| ppp3cca | −2.93 | 0.0094 | |
| fcer1g | −2.05 | 0.0094 | |
| zgc:55558 | 2.21 | 0.0094 | |
| mdm2 | 2.07 | 0.0094 | |
| RIG-I-like receptor signaling pathway | dhx58 | 4.03 | 0.0213 |
| znfx1 | 6.01 | 0.0213 | |
| Cytosolic DNA-sensing pathway | g3bp1 | 2.33 | 0.0117 |
| samhd1 | −3.02 | 0.0117 | |
| polr2h | 2.17 | 0.0117 | |
| polr3e | 2.00 | 0.0117 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, H.; Kim, T.-M.; Oh, H.-M.; Lim, H.-K.; Cho, J.-H. Transcriptomic Profiling Reveals Immune-Related Pathway Alterations in Paralichthys olivaceus Infected with Enteromyxum leei. Fishes 2025, 10, 601. https://doi.org/10.3390/fishes10120601
Lee H, Kim T-M, Oh H-M, Lim H-K, Cho J-H. Transcriptomic Profiling Reveals Immune-Related Pathway Alterations in Paralichthys olivaceus Infected with Enteromyxum leei. Fishes. 2025; 10(12):601. https://doi.org/10.3390/fishes10120601
Chicago/Turabian StyleLee, Hyobin, Tae-Min Kim, Hye-Min Oh, Han-Kyu Lim, and Jeong-Hyeon Cho. 2025. "Transcriptomic Profiling Reveals Immune-Related Pathway Alterations in Paralichthys olivaceus Infected with Enteromyxum leei" Fishes 10, no. 12: 601. https://doi.org/10.3390/fishes10120601
APA StyleLee, H., Kim, T.-M., Oh, H.-M., Lim, H.-K., & Cho, J.-H. (2025). Transcriptomic Profiling Reveals Immune-Related Pathway Alterations in Paralichthys olivaceus Infected with Enteromyxum leei. Fishes, 10(12), 601. https://doi.org/10.3390/fishes10120601






