Genome-Wide Identification of the DVR Gene Family and Expression Analysis of GDF8 Genes in Qihe Gibel Carp
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Approval of Animal Research and Animals
2.2. Identification of DVR Family Members in Qihe Gibel Carp
2.3. Evolutionary Analysis of DVR Gene Family
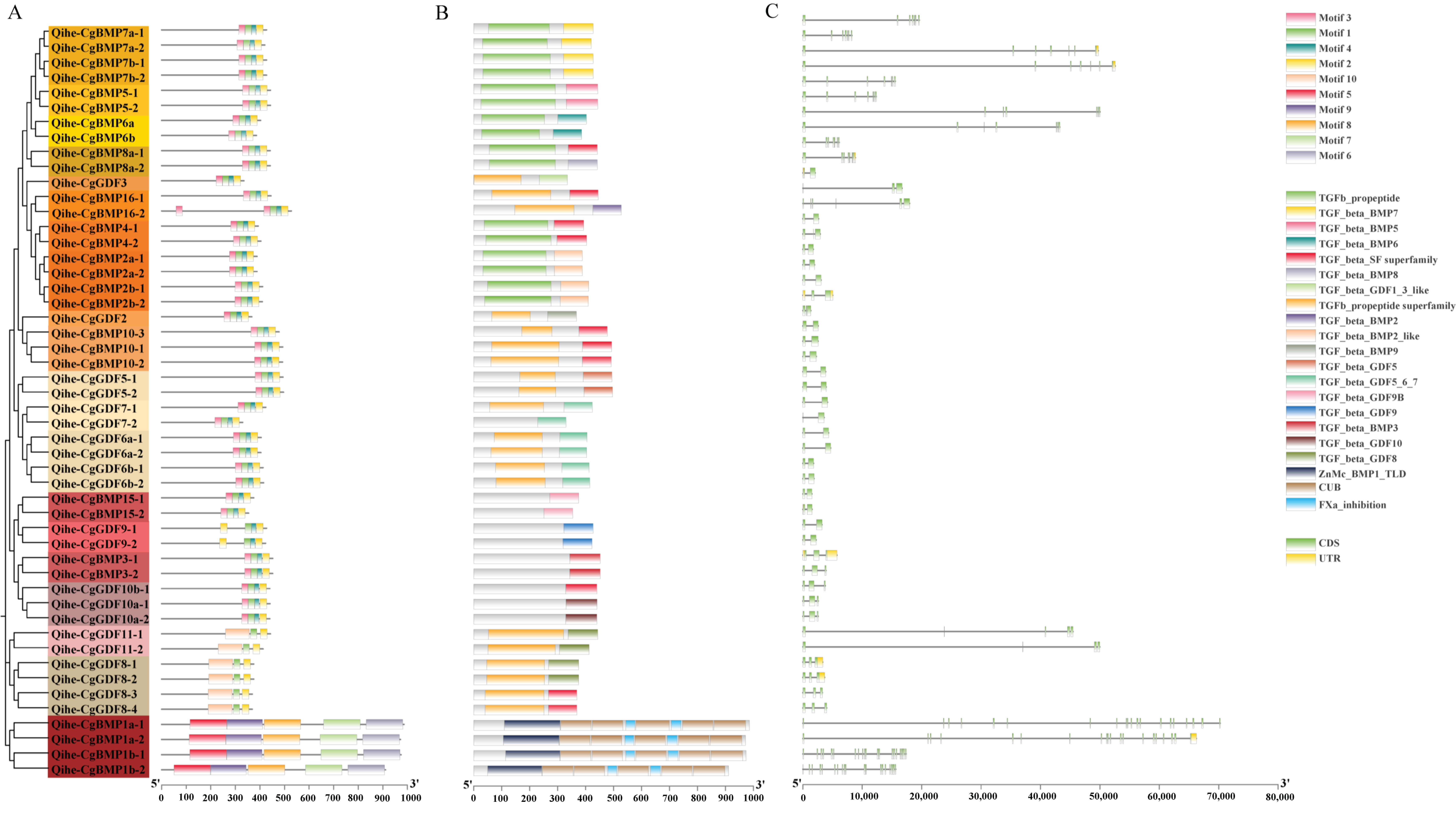
2.4. Analysis of DVR Gene Structure and Conserved Motifs
2.5. Chromosomal Localization and Gene Duplication Analysis
2.6. RNA Extraction and Expression Analysis of GDF8 Genes
2.7. Data Statistical Analysis
3. Results
3.1. Genome-Wide Identification of DVR Family Members in Qihe Gibel Carp
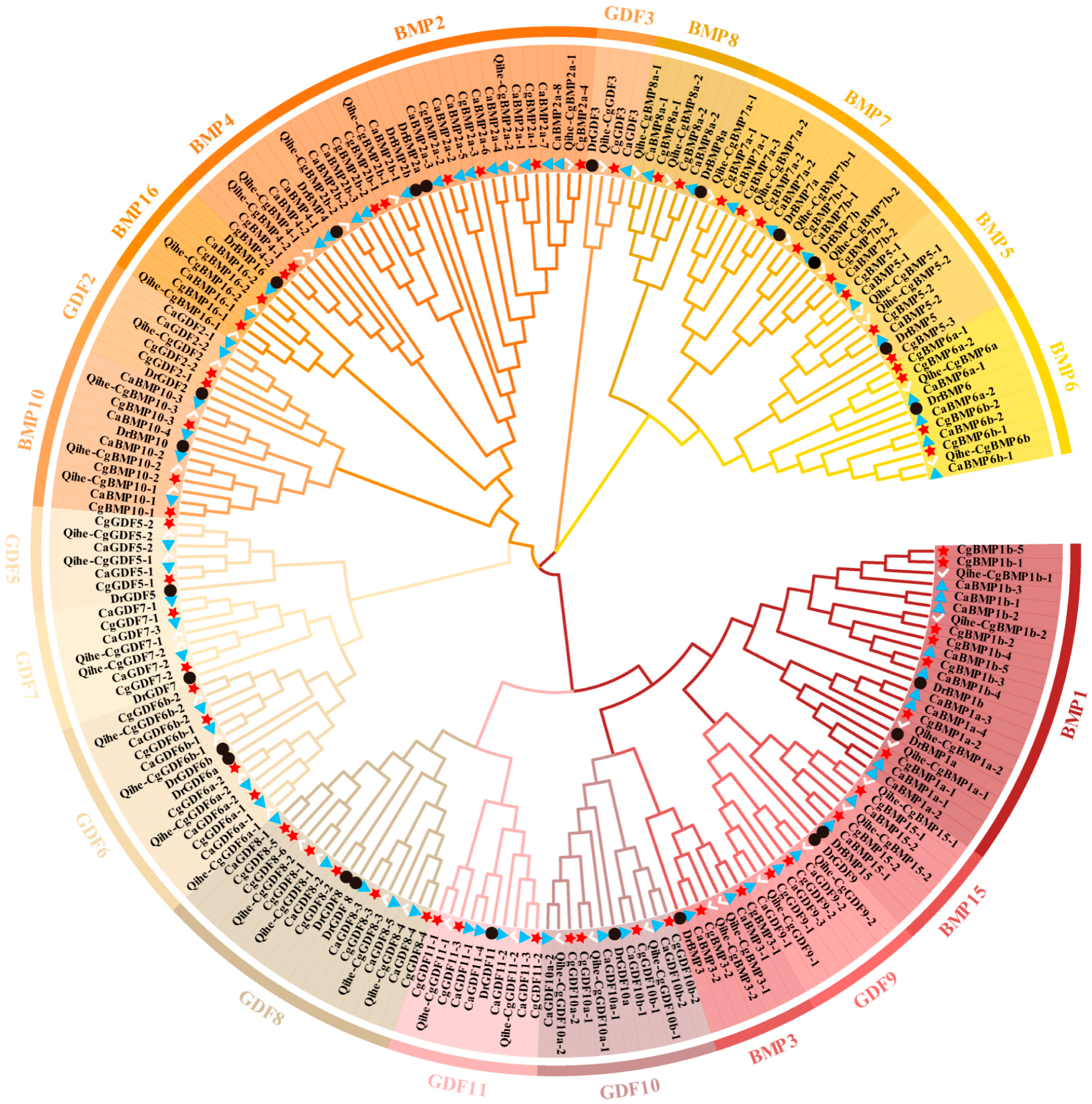
3.2. Phylogenetic Analysis of DVR Gene Family
3.3. Formatting of Mathematical Components
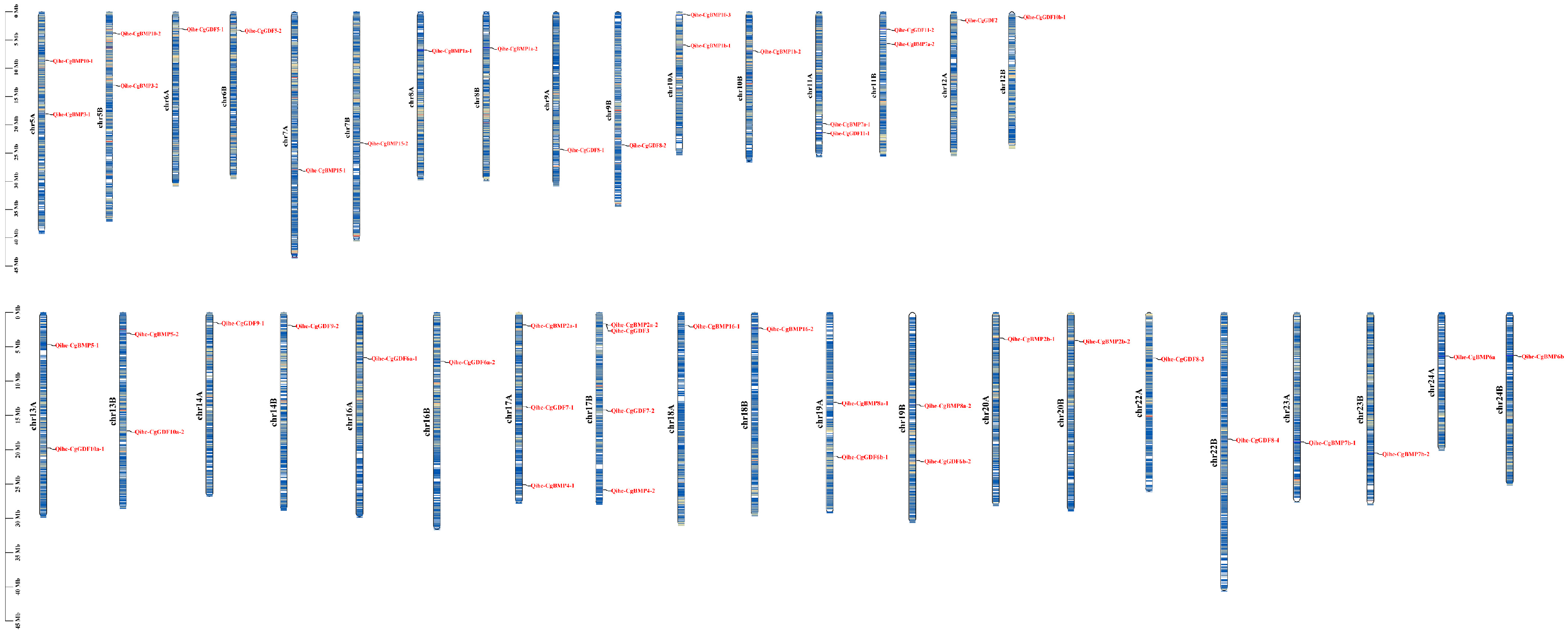
3.4. Chromosomal Localization and Gene Replication of DVR Gene Family
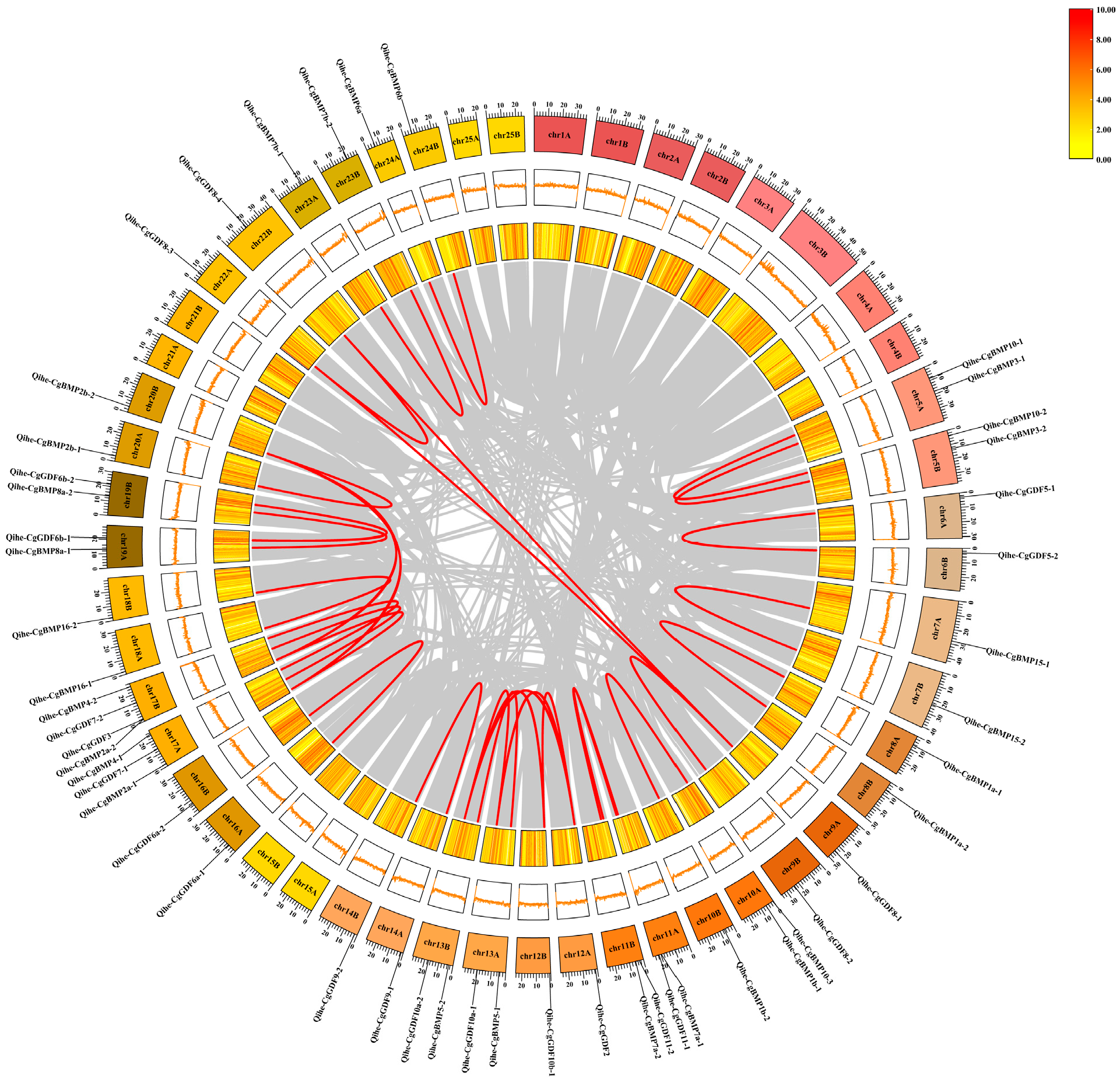
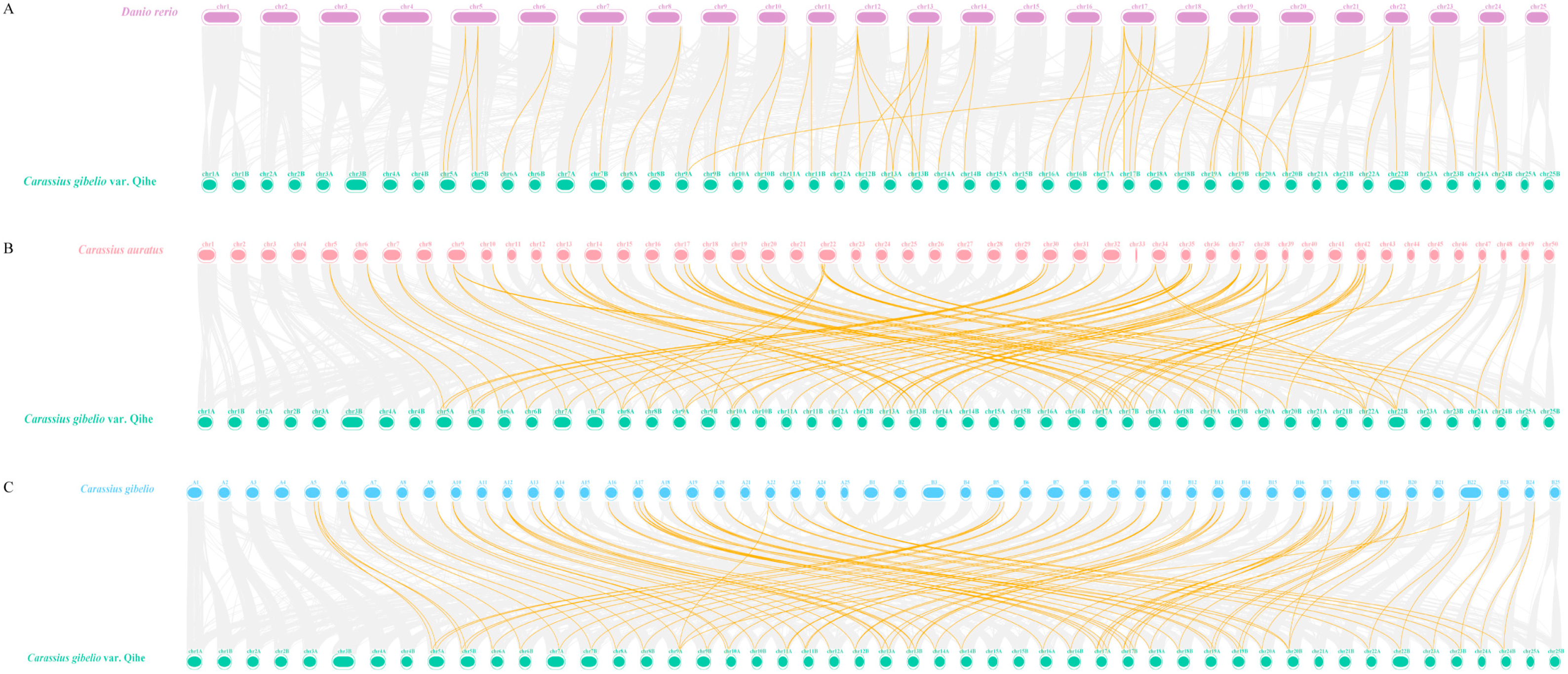
3.5. Synteny Analysis of DVR Gene Family in Qihe Gibel Carp
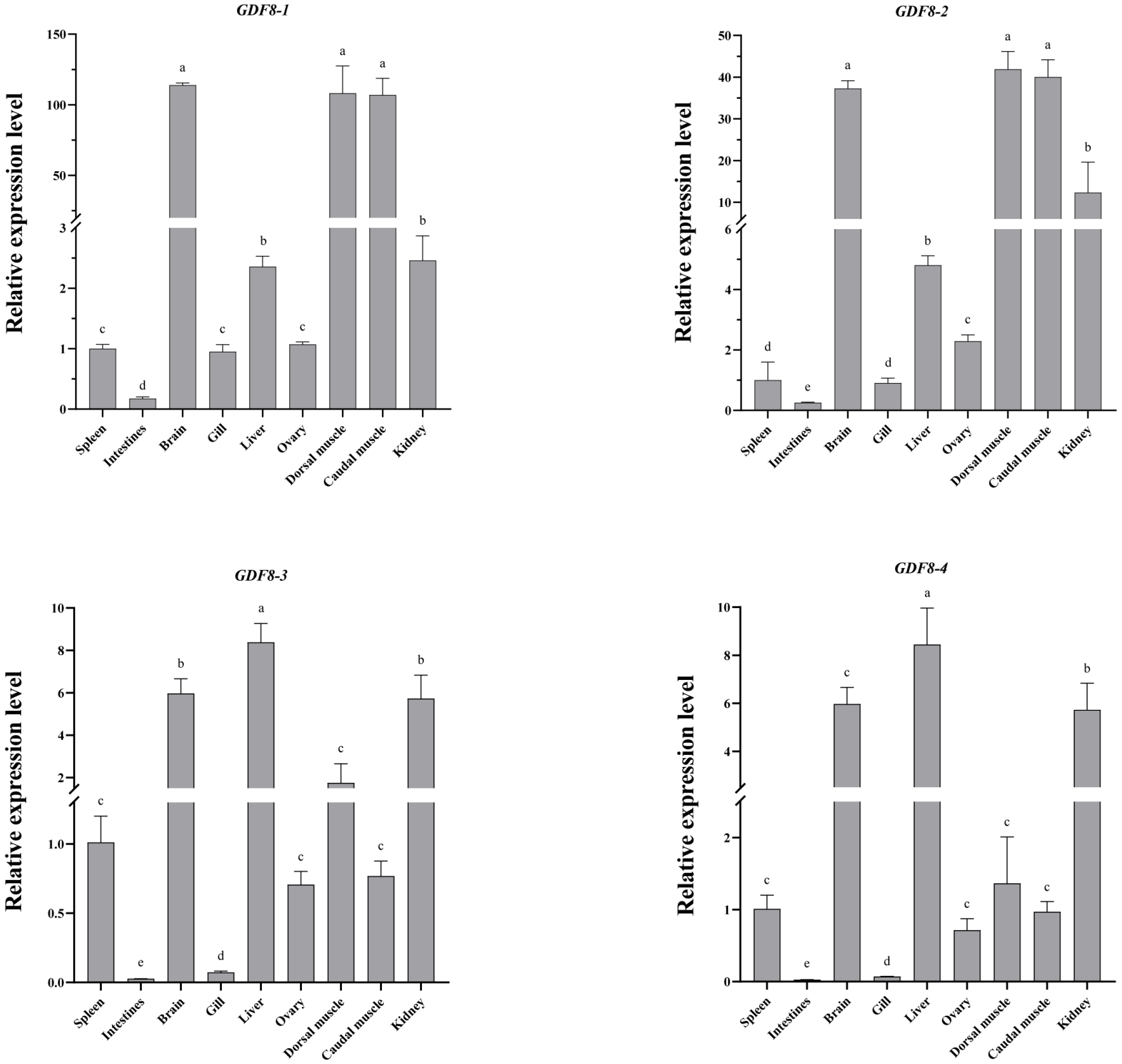
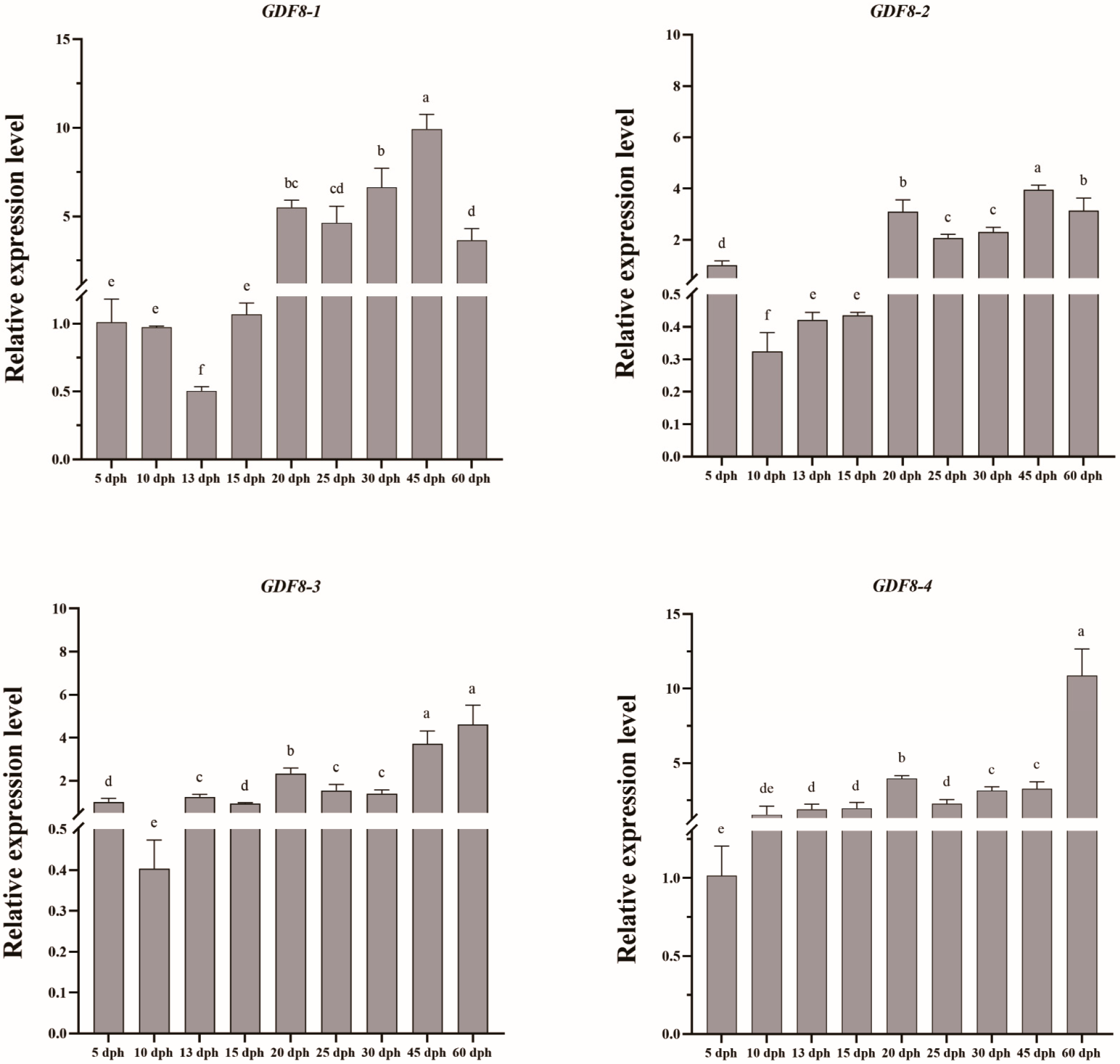
3.6. Spatio-Temporal Specific Expression of GDF8 Gene
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, Y.; Li, X.Y.; Xu, W.J.; Wang, K.; Wu, B.; Xu, M.; Chen, Y.; Miao, L.J.; Wang, Z.W.; Li, Z.; et al. Comparative genome anatomy reveals evolutionary insights into a unique amphitriploid fish. Nat. Ecol. Evol. 2022, 6, 1354–1366. [Google Scholar] [CrossRef]
- Zhang, J.; Zhu, Y.; Chen, Z.; Li, C.; Zhao, X.; Kong, X. Molecular cloning and expression analysis of MyD88 and TRAF6 in Qihe crucian carp Carassius auratus. Fish. Shellfish. Immunol. 2019, 87, 829–838. [Google Scholar] [CrossRef]
- Frangogiannis, N.G. Transforming growth factor-β in myocardial disease. Nat. Rev. Cardiol. 2022, 19, 435–455. [Google Scholar] [CrossRef]
- Kokabu, S.; Kodama, N.; Miyawaki, A.; Tsuji, K.; Hino, J.; Ono, Y.; Matsubara, T. Excessive BMP3b suppresses skeletal muscle differentiation. Biochem. Biophys. Res. Commun. 2025, 746, 151261. [Google Scholar] [CrossRef]
- Guignabert, C.; Humbert, M. Targeting transforming growth factor-β receptors in pulmonary hypertension. Eur. Respir. J. 2021, 57, 2002341. [Google Scholar] [CrossRef]
- Veitia, R.A.; Caburet, S. Extensive sequence turnover of the signal peptides of members of the GDF/BMP family: Exploring their evolutionary landscape. Biol. Direct 2009, 4, 22. [Google Scholar] [CrossRef] [PubMed]
- Hogan, B.L. Bone morphogenetic proteins: Multifunctional regulators of vertebrate development. Genes. Dev. 1996, 10, 1580–1594. [Google Scholar] [CrossRef] [PubMed]
- Plikus, M.V.; Guerrero-Juarez, C.F.; Ito, M.; Li, Y.R.; Dedhia, P.H.; Zheng, Y.; Shao, M.; Gay, D.L.; Ramos, R.; Hsi, T.C.; et al. Regeneration of fat cells from myofibroblasts during wound healing. Science 2017, 355, 748–752. [Google Scholar] [CrossRef]
- Traoré, M.; Noviello, C.; Vergnol, A.; Gentil, C.; Halliez, M.; Saillard, L.; Gelin, M.; Forand, A.; Lemaitre, M.; Guesmia, Z.; et al. GDF5 as a rejuvenating treatment for age-related neuromuscular failure. Brain 2024, 147, 3834–3848. [Google Scholar] [CrossRef] [PubMed]
- Tajer, B.; Dutko, J.A.; Little, S.C.; Mullins, M.C. BMP heterodimers signal via distinct type I receptor class functions. Proc. Natl. Acad. Sci. USA 2021, 118, e2017952118. [Google Scholar] [CrossRef]
- Katoh, Y.; Katoh, M. Comparative integromics on BMP/GDF family. Int. J. Mol. Med. 2006, 17, 951–955. [Google Scholar] [CrossRef]
- Whitfield, C.W.; Band, M.R.; Bonaldo, M.F.; Kumar, C.G.; Liu, L.; Pardinas, J.R.; Robertson, H.M.; Soares, M.B.; Robinson, G.E. Annotated Expressed Sequence Tags and cDNA Microarrays for Studies of Brain and Behavior in the Honey Bee. Genome Res. 2002, 12, 555–566. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Xu, W.; Xiang, S.; Tao, L.; Fu, W.; Liu, J.; Liu, W.; Xiao, Y.; Peng, L. Defining the Pluripotent Marker Genes for Identification of Teleost Fish Cell Pluripotency During Reprogramming. Front. Genet. 2022, 13, 819682. [Google Scholar] [CrossRef]
- Xu, J.; Liu, S.; Ai, Y.; Zhang, Y.; Li, S.; Li, Y. Establishment and transcriptome analysis of single blastomere-derived cell lines from zebrafish. J. Genet. Genom. 2024, 51, 957–969. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Cui, X.; Chen, S.; Xu, J.; Li, Y.; Zhang, Q.; Sun, Y. Mechanistic Insights into Nonylphenol Stress on BMP2 and BMP4 Gene Expression in Red Crucian Carp (Carassius auratus Red var.). Fishes 2024, 9, 159. [Google Scholar] [CrossRef]
- Kuang, Y.; Zheng, X.; Cao, D.; Sun, Z.; Tong, G.; Xu, H.; Yan, T.; Tang, S.; Chen, Z.; Zhang, T. Generate a new crucian carp (Carassius auratus) strain without intermuscular bones by knocking out bmp6. Aquaculture 2023, 569, 14. [Google Scholar] [CrossRef]
- Ye, D.; Wang, X.; Wei, C.; He, M.; Sun, Y. Marcksb plays a key role in the secretory pathway of zebrafish Bmp2b. PLOS Genet. 2019, 15, e1008306. [Google Scholar] [CrossRef]
- Zhong, S.; Li, H.; Wang, Y.; Wang, Y.; Ji, G.; Li, H.; Zhang, S.; Liu, Z. Bmp8a is an essential positive regulator of antiviral immunity in zebrafish. Commun. Biol. 2021, 4, 318. [Google Scholar] [CrossRef]
- Wang, W.; Yang, N.; Wang, L.; Zhu, Y.; Chu, X.; Xu, W.; Li, Y.; Xu, Y.; Gao, L.; Zhang, B.; et al. The TET-Sall4-BMP regulatory axis controls craniofacial cartilage development. Br. J. Med. Med. Res. 2024, 43, 113873. [Google Scholar] [CrossRef]
- Ravenscroft, T.A.; Phillips, J.B.; Fieg, E.; Bajikar, S.S.; Bellen, H.J. Heterozygous loss-of-function variants significantly expand the phenotypes associated with loss of GDF11. Genet. Med. 2021, 23, 1889–1900. [Google Scholar] [CrossRef]
- Bandyopadhyay, A.; Tsuji, K.; Cox, K.; Harfe, B.D.; Tabin, C.J. Genetic Analysis of the Roles of BMP2, BMP4, and BMP7 in Limb Patterning and Skeletogenesis. PLoS Genet. 2007, 2, e216. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-J. Myostatin: A skeletal muscle chalone. Annu. Rev. Physiol. 2023, 85, 269–291. [Google Scholar] [CrossRef]
- Shi, R.; Li, X.; Xu, X.; Chen, Z.; Zhu, Y.; Wang, N. Genome-wide analysis of BMP/GDF family and DAP-seq of YY1 suggest their roles in Cynoglossus semilaevis sexual size dimorphism. Int. J. Biol. Macromol. 2023, 253 Pt 5, 127201. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39 (Suppl. S2), W29–W37. [Google Scholar] [CrossRef]
- Letunic, I.; Doerks, T.; Bork, P. SMART: Recent updates, new developments and status in 2015. Nucleic Acids Res. 2015, 43, D257–D260. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Liu, Z.; Wei, Y.; Peng, R. Genome-wide identification of the MIOX gene family and their expression profile in cotton development and response to abiotic stress. PLoS ONE 2021, 16, e0254111. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one”bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; Wang, X.; Sun, Y.; Joseph, P.V.; Paterson, A.H. Detection of colinear blocks and synteny and evolutionary analyses based on utilization of MCScanX. Nat. Protoc. 2024, 19, 2206–2229. [Google Scholar] [CrossRef]
- Maren, N.A.; Duduit, J.R.; Huang, D.; Zhao, F.; Ranney, T.G.; Liu, W. Stepwise Optimization of Real-Time RT-PCR Analysis. Methods Mol. Biol. 2023, 2653, 317–332. [Google Scholar] [CrossRef]
- Barnett, M.J.; Doroudgar, S.; Khosraviani, V.; Ip, E.J. Multiple comparisons: To compare or not to compare, that is the question. Res. Soc. Adm. Pharm. 2022, 18, 2331–2334. [Google Scholar] [CrossRef]
- Jiang, S.-Y.; Wang, Y.; Zhou, L.; Chen, F.; Li, Z.; Gui, J.-F. Molecular characteristics, genomic structure and expression patterns of diverse BMP 15 alleles in polyploid gibel carp clone F. ACTA Hydrobiol. Sin. 2020, 44, 518–527. [Google Scholar]
- Reichwald, K.; Petzold, A.; Koch, P.; Downie, B.R.; Hartmann, N.; Pietsch, S.; Baumgart, M.; Chalopin, D.; Felder, M.; Bens, M.; et al. Insights into Sex Chromosome Evolution and Aging from the Genome of a Short-Lived Fish. Cell 2015, 163, 1527–1538. [Google Scholar] [CrossRef]
- Yang, F.; Liu, S.; Qu, J.; Zhang, Q. Identification and functional characterization of Pomstna in Japanese flounder (Paralichthys olivaceus). Gene 2022, 837, 146675. [Google Scholar] [CrossRef]
- Kuhl, H.; Du, K.; Schartl, M.; Kalous, L.; Stöck, M.; Lamatsch, D.K. Equilibrated evolution of the mixed auto-/allopolyploid haplotype-resolved genome of the invasive hexaploid Prussian carp. Nat. Commun. 2022, 13, 4092. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Chai, J.; Wen, Y.; Tao, M.; Lin, G.; Liu, X.; Ren, L.; Chen, Z.; Wu, S.; Li, S.; et al. From asymmetrical to balanced genomic diversification during rediploidization: Subgenomic evolution in allotetraploid fish. Sci. Adv. 2020, 6, eaaz7677. [Google Scholar] [CrossRef]
- Shuqing, Z.; Juan, L.; Zhilong, L.; Wenjing, T.; Deshou, W. Identification and Evolution of TGF-β Signaling Pathway Members in Twenty-Four Animal Species and Expression in Tilapia. Int. J. Mol. Sci. 2018, 19, 1154. [Google Scholar]
- Gabillard, J.C.; Biga, P.R.; Rescan, P.Y.; Seiliez, I. Revisiting the paradigm of myostatin in vertebrates: Insights from fishes. Gen. Comp. Endocrinol. 2013, 194, 45–54. [Google Scholar] [CrossRef]
- Elashry, M.I.; Schneider, V.C.; Heimann, M.; Wenisch, S.; Arnhold, S. CRISPR/Cas9-Targeted Myostatin Deletion Improves the Myogenic Differentiation Parameters for Muscle-Derived Stem Cells in Mice. J. Dev. Biol. 2025, 13, 5. [Google Scholar] [CrossRef]
- Grobet, L.; Martin, L.J.; Poncelet, D.; Pirottin, D.; Brouwers, B.; Riquet, J.; Schoeberlein, A.; Dunner, S.; Ménissier, F.; Massabanda, J.; et al. A deletion in the bovine myostatin gene causes the double-muscled phenotype in cattle. Nat. Genet. 1997, 17, 71–74. [Google Scholar] [CrossRef] [PubMed]
- Esposito, P.; Picciotto, D.; Battaglia, Y.; Costigliolo, F.; Viazzi, F.; Verzola, D. Myostatin: Basic biology to clinical application. Adv. Clin. Chem. 2022, 106, 181–234. [Google Scholar] [CrossRef] [PubMed]
- Berrios, L.; Rentsch, J.D. Linking Reactive Oxygen Species (ROS) to Abiotic and Biotic Feedbacks in Plant Microbiomes: The Dose Makes the Poison. Int. J. Mol. Sci. 2022, 23, 4402. [Google Scholar] [CrossRef]
- Nadarajah, K.K. ROS Homeostasis in Abiotic Stress Tolerance in Plants. Int. J. Mol. Sci. 2020, 21, 5208. [Google Scholar] [CrossRef] [PubMed]
- Feiner, N.; Begemann, G.; Renz, A.J.; Meyer, A.; Kuraku, S. The origin of bmp16, a novel Bmp2/4 relative, retained in teleost fish genomes. BMC Evol. Biol. 2009, 9, 277. [Google Scholar] [CrossRef] [PubMed]
- Funkenstein, B.; Olekh, E. Growth/differentiation factor-11: An evolutionary conserved growth factor in vertebrates. Dev. Genes. Evol. 2010, 220, 129–137. [Google Scholar] [CrossRef]
- Rescan, P.Y.; Jutel, I.; Rallière, C. Two myostatin genes are differentially expressed in myotomal muscles of the trout (Oncorhynchus mykiss). J. Exp. Biol. 2001, 204 Pt 20, 3523–3529. [Google Scholar] [CrossRef]
- Xue, T.; Liangyan, W.; Lin, C.; Lei, W.; Xiao, M.A.; Cancan, H.U.; Xianghui, K.; Guoxing, N.; Xuejun, L.I. The mRNA and protein expression of gene SOST during ossification process of intermuscular bone in crucian carp (Carassius auratus) in Qihe River. J. Fish. China 2016, 40, 673–680. [Google Scholar]
- Gan, R.-H.; Li, Z.; Wang, Z.-W.; Li, X.-Y.; Wang, Y.; Zhang, X.-J.; Tong, J.-F.; Wu, Y.; Xia, L.-Y.; Gao, Z.-X. Creation of intermuscular bone-free mutants in amphitriploid gibel carp by editing two duplicated runx2b homeologs. Aquaculture 2023, 567, 739300. [Google Scholar] [CrossRef]
| Genes | Primer Sequences (5′→3′) | Annealing Temperature/°C | Product Length/bp | Applications |
|---|---|---|---|---|
| GDF8-1 | F1: GGATTGGACTGCGACGAGAA | 60 | 294 | qRT-PCR |
| R1: TGAGGGGATCTTGCCGTAGA | ||||
| GDF8-2 | F: GACCAACTGGGGCATTGAGA | 60 | 274 | qRT-PCR |
| R: TCCCGAACAGTAATTCGCCT | ||||
| GDF8-3 | F: ACTGAGGGAATGCCGAAGTG | 60 | 264 | qRT-PCR |
| R: AGGCTGTTTGAGCCACAACT | ||||
| GDF8-4 | F: GACACAGGCCTCGATTGTGA | 60 | 181 | qRT-PCR |
| R: TGGCCTTGTTGACGATGTGA | ||||
| β-actin | F: ACCATCTACCCCGGTATTGC | 60 | 149 | qRT-PCR |
| R: TGGAAGGTGGACAGGGAAGC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shan, J.; Liu, Y.; Lian, K.; Xiao, X.; Ma, J.; Ren, R.; Li, X.; Wei, G.; Kuang, Y.; Peng, R. Genome-Wide Identification of the DVR Gene Family and Expression Analysis of GDF8 Genes in Qihe Gibel Carp. Fishes 2025, 10, 529. https://doi.org/10.3390/fishes10100529
Shan J, Liu Y, Lian K, Xiao X, Ma J, Ren R, Li X, Wei G, Kuang Y, Peng R. Genome-Wide Identification of the DVR Gene Family and Expression Analysis of GDF8 Genes in Qihe Gibel Carp. Fishes. 2025; 10(10):529. https://doi.org/10.3390/fishes10100529
Chicago/Turabian StyleShan, Jinyan, Yuling Liu, Kaiqi Lian, Xianghui Xiao, Jun Ma, Ren Ren, Xiaolong Li, Guoqiang Wei, Youyi Kuang, and Renhai Peng. 2025. "Genome-Wide Identification of the DVR Gene Family and Expression Analysis of GDF8 Genes in Qihe Gibel Carp" Fishes 10, no. 10: 529. https://doi.org/10.3390/fishes10100529
APA StyleShan, J., Liu, Y., Lian, K., Xiao, X., Ma, J., Ren, R., Li, X., Wei, G., Kuang, Y., & Peng, R. (2025). Genome-Wide Identification of the DVR Gene Family and Expression Analysis of GDF8 Genes in Qihe Gibel Carp. Fishes, 10(10), 529. https://doi.org/10.3390/fishes10100529






