Nanopore-Based Metagenomic Approaches for Detection of Bacterial Pathogens in Recirculating Aquaculture Systems
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Sampling
2.3. DNA Extraction and Full 16S rRNA Amplification
2.4. Nanopore Library Synthesis and Sequencing
2.5. Bioinformatic Analysis
2.6. Functional Prediction
3. Results
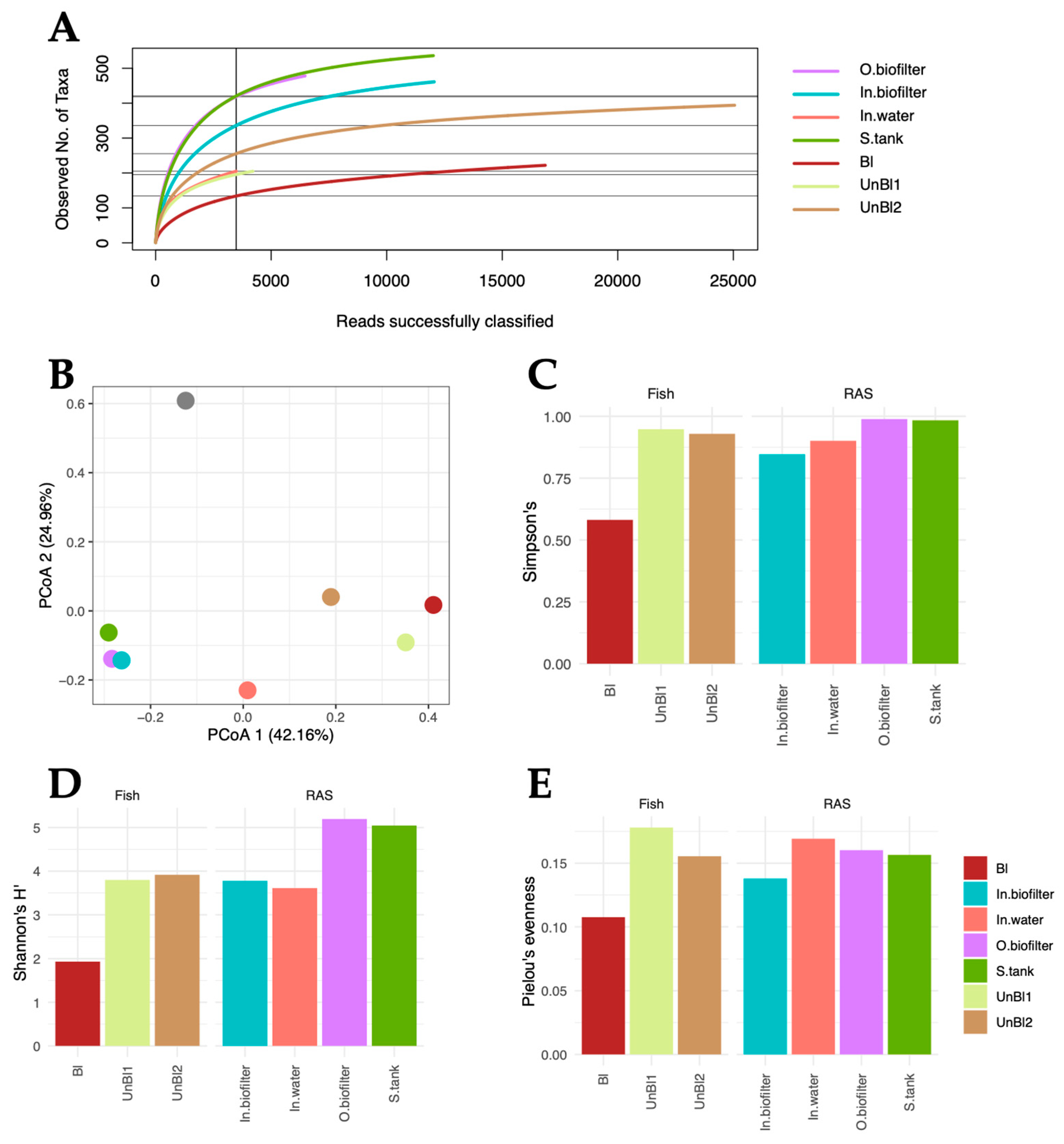
3.1. Microbiota Diversity Indexes
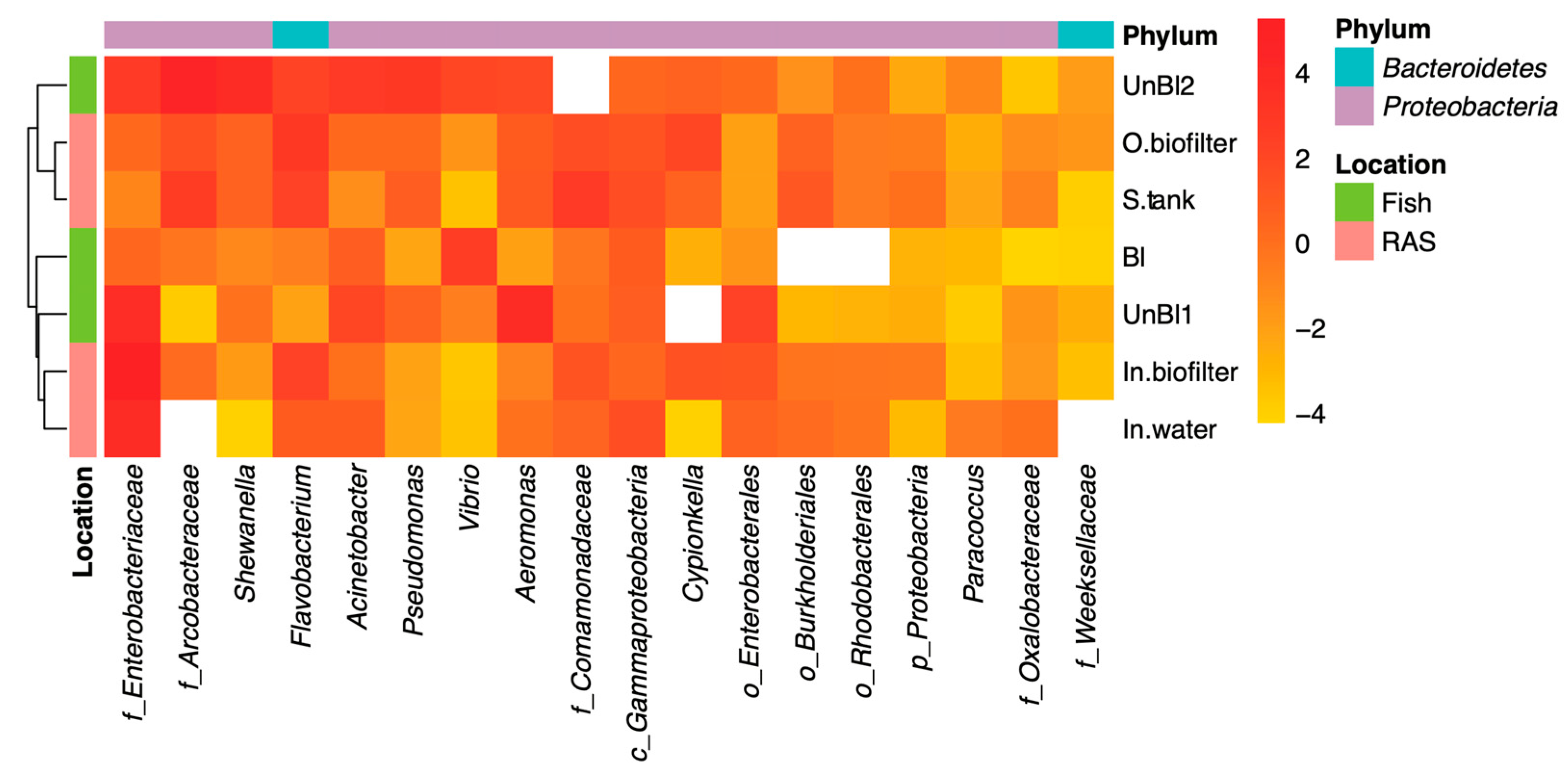
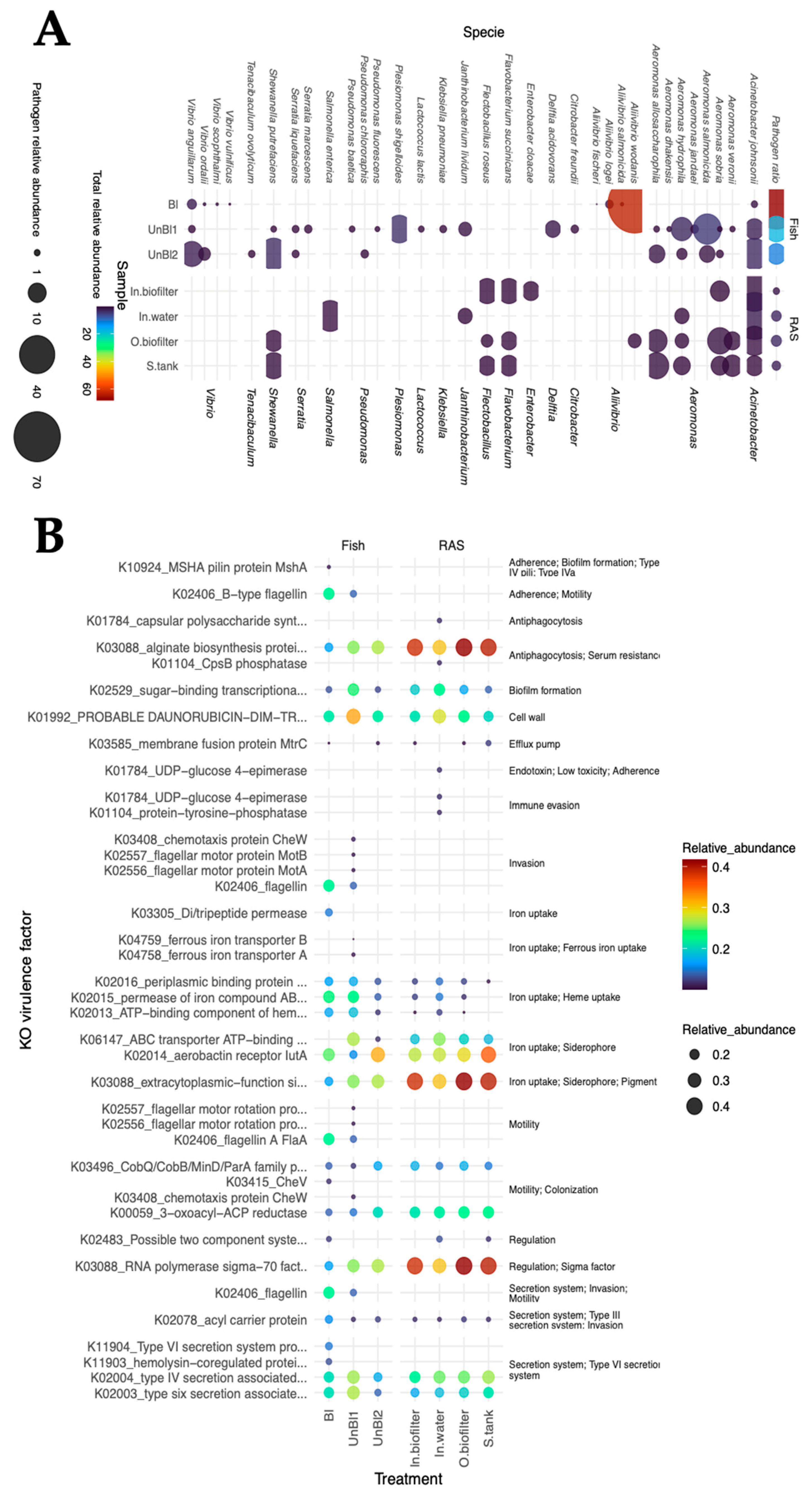
3.2. Potential Pathogens in RAS Microbiota
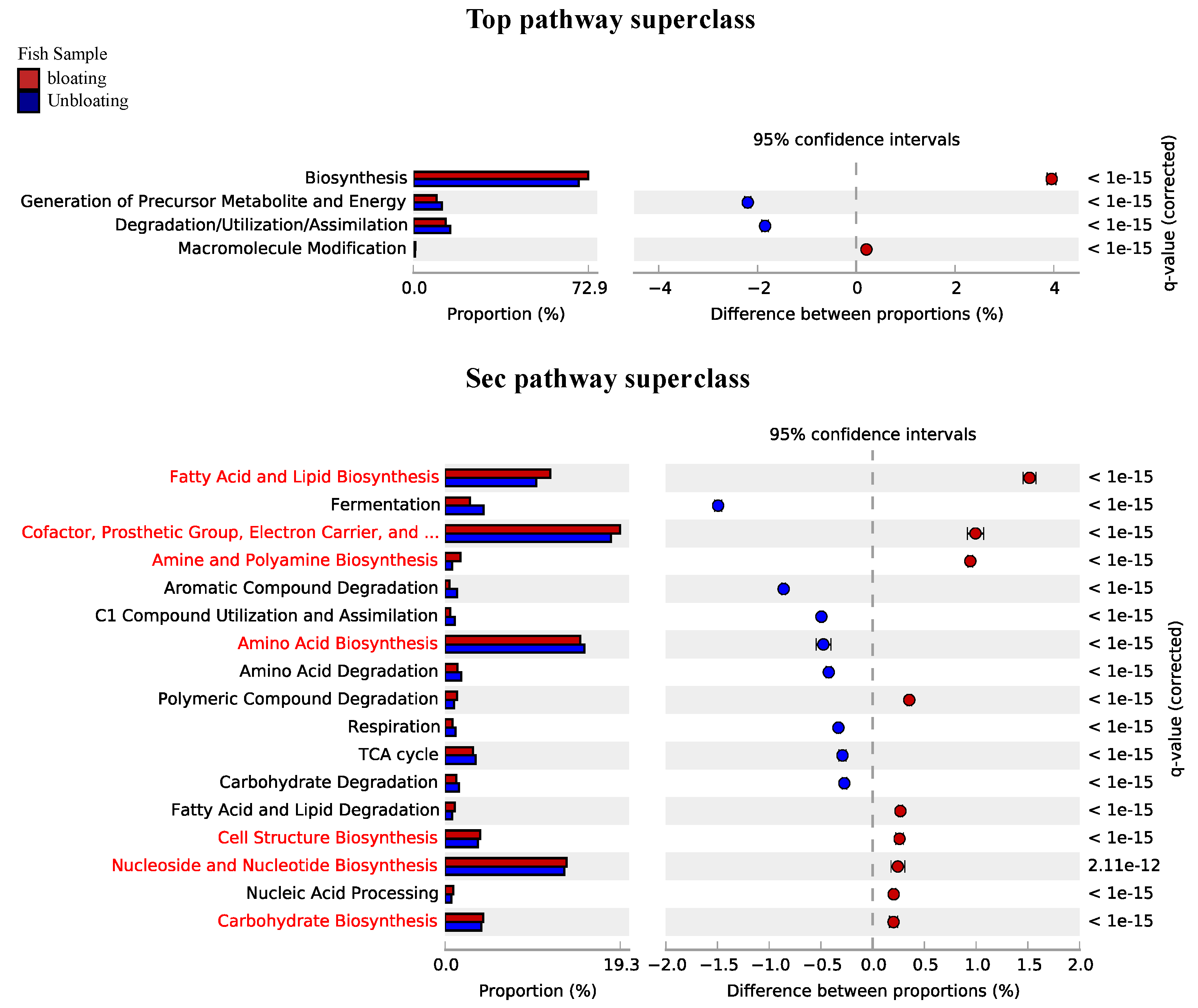
3.3. Functional Role of RAS Microbiota
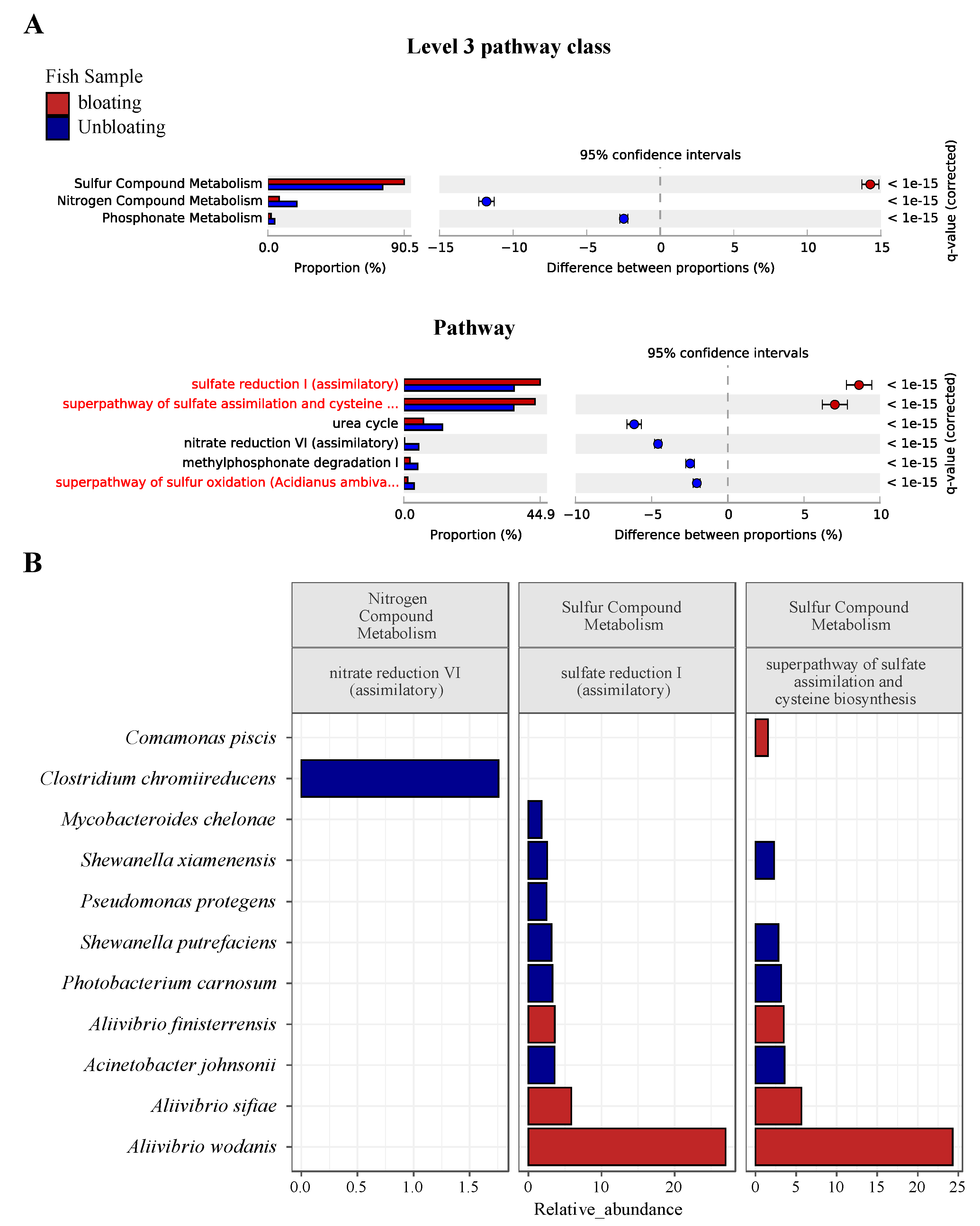
3.4. Sulfur and Nitrogen Metabolism
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Schreier, H.J.; Mirzoyan, N.; Saito, K. Microbial diversity of biological filters in recirculating aquaculture systems. Curr. Opin. Biotechnol. 2010, 21, 318–325. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, N.; Turchini, G.M. Recirculating aquaculture systems (RAS): Environmental solution and climate change adaptation. J. Clean. Prod. 2021, 297, 126604. [Google Scholar] [CrossRef]
- Yogev, U.; Gross, A. Reducing environmental impact of recirculating aquaculture systems by introducing a novel microaerophilic assimilation reactor: Modeling and proof of concept. J. Clean. Prod. 2019, 226, 1042–1050. [Google Scholar] [CrossRef]
- Bauer, J.; Teitge, F.; Neffe, L.; Adamek, M.; Jung, A.; Peppler, C.; Steinhagen, D.; Jung-Schroers, V. Impact of a reduced water salinity on the composition of Vibrio spp. in recirculating aquaculture systems for Pacific white shrimp (Litopenaeus vannamei) and its possible risks for shrimp health and food safety. J. Fish Dis. 2021, 44, 89–105. [Google Scholar] [CrossRef]
- Blancheton, J.P.; Attramadal, K.J.K.; Michaud, L.; d’Orbcastel, E.R.; Vadstein, O. Insight into bacterial population in aquaculture systems and its implication. Aquacult. Eng. 2013, 53, 30–39. [Google Scholar] [CrossRef]
- Quiñones, R.A.; Fuentes, M.; Montes, R.M.; Soto, D.; León-Muñoz, J. Environmental issues in Chilean salmon farming: A review. Rev. Aquacult. 2019, 11, 375–402. [Google Scholar] [CrossRef]
- Bozzi, D.; Rasmussen, J.A.; Caroe, C.; Sveier, H.; Nordoy, K.; Gilbert, M.T.P.; Limborg, M.T. Salmon gut microbiota correlates with disease infection status: Potential for monitoring health in farmed animals. Anim. Microbiome 2021, 3, 30. [Google Scholar] [CrossRef]
- Dahle, S.W.; Gaarden, S.I.; Buhaug, J.F.; Netzer, R.; Attramadal, K.J.K.; Busche, T.; Aas, M.; Ribicic, D.; Bakke, I. Long-term microbial community structures and dynamics in a commercial RAS during seven production batches of Atlantic salmon fry. Aquaculture 2023, 565, 739155. [Google Scholar] [CrossRef]
- Zhao, R.; Symonds, J.E.; Walker, S.P.; Steiner, K.; Carter, C.G.; Bowman, J.P.; Nowak, B.F. Relationship between gut microbiota and Chinook salmon (Oncorhynchus tshawytscha) health and growth performance in freshwater recirculating aquaculture systems. Front. Microbiol. 2023, 14, 1065823. [Google Scholar] [CrossRef] [PubMed]
- Minich, J.J.; Poore, G.D.; Jantawongsri, K.; Johnston, C.; Bowie, K.; Bowman, J.; Knight, R.; Nowak, B.; Allen, E.E. Microbial Ecology of Atlantic Salmon (Salmo salar) Hatcheries: Impacts of the Built Environment on Fish Mucosal Microbiota. Appl. Environ. Microbiol. 2020, 86, e00411–e00420. [Google Scholar] [CrossRef]
- Lu, J.; Zhang, Y.; Wu, J.; Luo, Y. Effects of microplastics on distribution of antibiotic resistance genes in recirculating aquaculture system. Ecotoxicol. Environ. Saf. 2019, 184, 109631. [Google Scholar] [CrossRef]
- Fossmark, R.O.; Attramadal, K.J.K.; Nordoy, K.; Osterhus, S.W.; Vadstein, O. A comparison of two seawater adaptation strategies for Atlantic salmon post-smolt (Salmo salar) grown in recirculating aquaculture systems (RAS): Nitrification, water and gut microbiota, and performance of fish. Aquaculture 2021, 532, 735973. [Google Scholar] [CrossRef]
- Sergaliev, N.K.; Kakishev, M.G.; Ginayatov, N.S.; Nurzhanova, F.K.; Andronov, E.E. Microbiome structure in a recirculating aquaculture system and its connection to infections in sturgeon fish. Vet. World 2021, 14, 661–668. [Google Scholar] [CrossRef]
- Vadstein, O.; Attramadal, K.J.K.; Bakke, I.; Olsen, Y. K-Selection as Microbial Community Management Strategy: A Method for Improved Viability of Larvae in Aquaculture. Front. Microbiol. 2018, 9, 2730. [Google Scholar] [CrossRef]
- Minich, J.J.; Petrus, S.; Michael, J.D.; Michael, T.P.; Knight, R.; Allen, E.E. Temporal, Environmental, and Biological Drivers of the Mucosal Microbiome in a Wild Marine Fish. mSphere 2020, 5, e00401-20. [Google Scholar] [CrossRef]
- Shephard, K.L. Functions for Fish Mucus. Rev. Fish Biol. Fish. 1994, 4, 401–429. [Google Scholar] [CrossRef]
- Curry, K.D.; Wang, Q.; Nute, M.G.; Tyshaieva, A.; Reeves, E.; Soriano, S.; Wu, Q.; Graeber, E.; Finzer, P.; Mendling, W.; et al. Emu: Species-level microbial community profiling of full-length 16S rRNA Oxford Nanopore sequencing data. Nat. Methods 2022, 19, 845–853. [Google Scholar] [CrossRef]
- Langille, M.G.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Wan, Q.; Li, L.; Liu, B.; Zhang, Z.; Liu, Y.; Xie, M. Different and unified responses of soil bacterial and fungal community composition and predicted functional potential to 3 years’ drought stress in a semiarid alpine grassland. Front. Microbiol. 2023, 14, 1104944. [Google Scholar] [CrossRef]
- Laroche, O.; Pochon, X.; Wood, S.A.; Keeley, N. Beyond taxonomy: Validating functional inference approaches in the context of fish-farm impact assessments. Mol. Ecol. Resour. 2021, 21, 2264–2277. [Google Scholar] [CrossRef] [PubMed]
- Letelier-Gordo, C.O.; Aalto, S.L.; Suurnäkki, S.; Pedersen, P.B. Increased sulfate availability in saline water promotes hydrogen sulfide production in fish organic waste. Aquacult. Eng. 2020, 89, 102062. [Google Scholar] [CrossRef]
- Zhang, W.; Liu, B.; Sun, Z.; Wang, T.; Tan, S.; Fan, X.; Zou, D.; Zhuang, Y.; Liu, X.; Wang, Y.; et al. Comparision of nitrogen removal characteristic and microbial community in freshwater and marine recirculating aquaculture systems. Sci. Total Environ. 2023, 878, 162870. [Google Scholar] [CrossRef]
- Attramadal, K.J.K.; Oien, J.V.; Kristensen, E.; Evjemo, J.O.; Kjorsvik, E.; Vadstein, O.; Bakke, I. UV treatment in RAS influences the rearing water microbiota and reduces the survival of European lobster larvae (Homarus gammarus). Aquacult. Eng. 2021, 94, 102176. [Google Scholar] [CrossRef]
- Chen, Z.; Chang, Z.; Zhang, L.; Jiang, Y.; Ge, H.; Song, X.; Chen, S.; Zhao, F.; Li, J. Effects of water recirculation rate on the microbial community and water quality in relation to the growth and survival of white shrimp (Litopenaeus vannamei). BMC Microbiol. 2019, 19, 192. [Google Scholar] [CrossRef]
- Schmidt, V.; Amaral-Zettler, L.; Davidson, J.; Summerfelt, S.; Good, C. Influence of Fishmeal-Free Diets on Microbial Communities in Atlantic Salmon (Salmo salar) Recirculation Aquaculture Systems. Appl. Environ. Microbiol. 2016, 82, 4470–4481. [Google Scholar] [CrossRef] [PubMed]
- Lane, D.J. 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Stackebrandt, E., Goodfellow, M., Eds.; Wiley: Hoboken, NJ, USA, 1991; Volume 4, pp. 115–175. [Google Scholar]
- Wick, R. Porechop: An Adapter Trimmer for Oxford Nanopore Reads. 2018. Available online: https://github.com/rrwick/Porechop (accessed on 1 June 2024).
- De Coster, W.; D’Hert, S.; Schultz, D.T.; Cruts, M.; Van Broeckhoven, C. NanoPack: Visualizing and processing long-read sequencing data. Bioinformatics 2018, 34, 2666–2669. [Google Scholar] [CrossRef]
- Dixon, P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2003, 14, 927–930. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Shen, W.; Ren, H. TaxonKit: A practical and efficient NCBI taxonomy toolkit. J. Genet. Genom. 2021, 48, 844–850. [Google Scholar] [CrossRef]
- Austin, B.; Austin, D.A. Bacterial Fish Pathogens, Disease of Farmed and Wild Fish. In Bacterial Fish Pathogens, Disease of Farmed and Wild Fish; Springer: Cham, Switzerland, 2016. [Google Scholar]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef] [PubMed]
- Caspi, R.; Billington, R.; Fulcher, C.A.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Latendresse, M.; Midford, P.E.; Ong, Q.; Ong, W.K.; et al. The MetaCyc database of metabolic pathways and enzymes. Nucleic Acids Res. 2018, 46, D633–D639. [Google Scholar] [CrossRef]
- Parks, D.H.; Tyson, G.W.; Hugenholtz, P.; Beiko, R.G. STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics 2014, 30, 3123–3124. [Google Scholar] [CrossRef]
- Yadav, S.; Kapley, A. Exploration of activated sludge resistome using metagenomics. Sci. Total Environ. 2019, 692, 1155–1164. [Google Scholar] [CrossRef]
- Preena, P.G.; Kumar, V.J.R.; Singh, I.S.B. Nitrification and denitrification in recirculating aquaculture systems: The processes and players. Rev. Aquacult. 2021, 13, 2053–2075. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y. KEGG Mapper for inferring cellular functions from protein sequences. Protein Sci. 2020, 29, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Kawashima, M. KEGG mapping tools for uncovering hidden features in biological data. Protein Sci. 2022, 31, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Pattaroni, C.; Kieser, S.; Marsland, B. Virulence Factor Database for 16S metagenomics. Mendeley Data 2018, 1. [Google Scholar] [CrossRef]
- D’Orbcastel, E.R.; Blancheton, J.-P.; Aubin, J. Towards environmentally sustainable aquaculture: Comparison between two trout farming systems using Life Cycle Assessment. Aquacult. Eng. 2009, 40, 113–119. [Google Scholar] [CrossRef]
- Aja-Macaya, P.; Conde-Pérez, K.; Trigo-Tasende, N.; Buetas, E.; Nasser-Ali, M.; Nión, P.; Rumbo-Feal, S.; Ladra, S.; Bou, G.; Mira, Á.; et al. Nanopore full length 16S rRNA gene sequencing increases species resolution in bacterial biomarker discovery. Sci. Rep. 2025, 15, 26486. [Google Scholar] [CrossRef]
- Winand, R.; Bogaerts, B.; Hoffman, S.; Lefevre, L.; Delvoye, M.; Van Braekel, J.; Fu, Q.; Roosens, N.H.; De Keersmaecker, S.C.; Vanneste, K. Targeting the 16S rRNA Gene for Bacterial Identification in Complex Mixed Samples: Comparative Evaluation of Second (Illumina) and Third (Oxford Nanopore Technologies) Generation Sequencing Technologies. Int. J. Mol. Sci. 2020, 21, 298. [Google Scholar] [CrossRef]
- Liao, J.-J.; Chen, Y.-S.; Lin, H.-C.; Chen, Y.-J.; Lai, K.-L.; Mao, Y.-C.; Liu, P.-Y.; Chuang, H.-N. Nanopore 16S-Full Length and ITS Sequencing for Microbiota Identification in Intra-Abdominal Infections. Diagnostics 2025, 15, 2257. [Google Scholar] [CrossRef]
- Ofek, T.; Lalzar, M.; Izhaki, I.; Halpern, M. Intestine and spleen microbiota composition in healthy and diseased tilapia. Anim. Microbiome 2022, 4, 50. [Google Scholar] [CrossRef] [PubMed]
- Valdes, A.M.; Walter, J.; Segal, E.; Spector, T.D. Role of the gut microbiota in nutrition and health. BMJ 2018, 361, k2179. [Google Scholar] [CrossRef] [PubMed]
- Almeida, D.B.; Magalhaes, C.; Sousa, Z.; Borges, M.T.; Silva, E.; Blanquet, I.; Mucha, A.P. Microbial community dynamics in a hatchery recirculating aquaculture system (RAS) of sole. Aquaculture 2021, 539, 736592. [Google Scholar] [CrossRef]
- Dehler, C.E.; Secombes, C.J.; Martin, S.A.M. Seawater transfer alters the intestinal microbiota profiles of Atlantic salmon (Salmo salar L.). Sci. Rep. 2017, 7, 13877. [Google Scholar] [CrossRef]
- Fossmark, R.O.; Vadstein, O.; Rosten, T.W.; Bakke, I.; Koseto, D.; Bugten, A.V.; Helberg, G.A.; Nesje, J.; Jorgensen, N.O.G.; Raspati, G.; et al. Effects of reduced organic matter loading through membrane filtration on the microbial community dynamics in recirculating aquaculture systems (RAS) with Atlantic salmon parr (Salmo salar). Aquaculture 2020, 524, 735268. [Google Scholar] [CrossRef]
- Gajardo, K.; Rodiles, A.; Kortner, T.M.; Krogdahl, A.; Bakke, A.M.; Merrifield, D.L.; Sorum, H. A high-resolution map of the gut microbiota in Atlantic salmon (Salmo salar): A basis for comparative gut microbial research. Sci. Rep. 2016, 6, 30893. [Google Scholar] [CrossRef]
- Kim, P.S.; Shin, N.R.; Lee, J.B.; Kim, M.S.; Whon, T.W.; Hyun, D.W.; Yun, J.H.; Jung, M.J.; Kim, J.Y.; Bae, J.W. Host habitat is the major determinant of the gut microbiome of fish. Microbiome 2021, 9, 166. [Google Scholar] [CrossRef]
- Lorgen-Ritchie, M.; Clarkson, M.; Chalmers, L.; Taylor, J.F.; Migaud, H.; Martin, S.A.M. A Temporally Dynamic Gut Microbiome in Atlantic Salmon During Freshwater Recirculating Aquaculture System (RAS) Production and Post-seawater Transfer. Front. Mar. Sci. 2021, 8, 711797. [Google Scholar] [CrossRef]
- Wang, J.; Jaramillo-Torres, A.; Li, Y.; Kortner, T.M.; Gajardo, K.; Brevik, O.J.; Jakobsen, J.V.; Krogdahl, A. Microbiota in intestinal digesta of Atlantic salmon (Salmo salar), observed from late freshwater stage until one year in seawater, and effects of functional ingredients: A case study from a commercial sized research site in the Arctic region. Anim. Microbiome 2021, 3, 14. [Google Scholar] [CrossRef]
- Karlsen, C.; Vanberg, C.; Mikkelsen, H.; Sorum, H. Co-infection of Atlantic salmon (Salmo salar), by Moritella viscosa and Aliivibrio wodanis, development of disease and host colonization. Vet. Microbiol. 2014, 171, 112–121. [Google Scholar] [CrossRef]
- Lunder, T.; Evensen, Ã.; Holstad, G.; HÃ¥stein, T. “Winter ulcer” in the Atlantic salmon Salmo salar. Pathological and bacteriological investigations and transmission experiments. Dis. Aquat. Organ. 1995, 23, 39–49. [Google Scholar] [CrossRef]
- Padra, J.T.; Murugan, A.V.M.; Sundell, K.; Sundh, H.; Benktander, J.; Linden, S.K. Fish pathogen binding to mucins from Atlantic salmon and Arctic char differs in avidity and specificity and is modulated by fluid velocity. PLoS ONE 2019, 14, e0215583. [Google Scholar] [CrossRef] [PubMed]
- Rojas-Tirado, P.; Aalto, S.L.; Åtland, Å.; Letelier-Gordo, C. Biofilters are potential hotspots for H2S production in brackish and marine water RAS. Aquaculture 2021, 536, 736490. [Google Scholar] [CrossRef]
- Bakke, I.; Åm, A.L.; Kolarevic, J.; Ytrestoyl, T.; Vadstein, O.; Attramadal, K.J.K.; Terjesen, B.F. Microbial community dynamics in semi-commercial RAS for production of Atlantic salmon post-smolts at different salinities. Aquacult. Eng. 2017, 78, 42–49. [Google Scholar] [CrossRef]
- King, R.K.; Flick, G.J., Jr.; Pierson, D.; Smith, S.A.; Boardman, G.D.; Coale, C.W., Jr. Identification of Bacterial Pathogens in Biofilms of Recirculating Aquaculture Systems. J. Aquat. Food Prod. Technol. 2004, 13, 125–133. [Google Scholar] [CrossRef]
- Huang, Q.; Sham, R.C.; Deng, Y.; Mao, Y.; Wang, C.; Zhang, T.; Leung, K.M.Y. Diversity of gut microbiomes in marine fishes is shaped by host-related factors. Mol. Ecol. 2020, 29, 5019–5034. [Google Scholar] [CrossRef]
- Roques, S.; Deborde, C.; Richard, N.; Skiba-Cassy, S.; Moing, A.; Fauconneau, B. Metabolomics and fish nutrition: A review in the context of sustainable feed development. Rev. Aquacult. 2020, 12, 261–282. [Google Scholar] [CrossRef]
- Huyben, D.; Roehe, B.K.; Bekaert, M.; Ruyter, B.; Glencross, B. Dietary Lipid:Protein Ratio and n-3 Long-Chain Polyunsaturated Fatty Acids Alters the Gut Microbiome of Atlantic Salmon Under Hypoxic and Normoxic Conditions. Front. Microbiol. 2020, 11, 589898. [Google Scholar] [CrossRef]
- Kihara, M.; Sakata, T. Fermentation of dietary carbohydrates to short-chain fatty acids by gut microbes and its influence on intestinal morphology of a detritivorous teleost tilapia (Oreochromis niloticus). Comp. Biochem. Physiol. Part A Physiol. 1997, 118, 1201–1207. [Google Scholar] [CrossRef]
- Lorgen-Ritchie, M.; Clarkson, M.; Chalmers, L.; Taylor, J.F.; Migaud, H.; Martin, S.A.M. Temporal changes in skin and gill microbiomes of Atlantic salmon in a recirculating aquaculture system—Why do they matter? Aquaculture 2022, 558, 738352. [Google Scholar] [CrossRef]
- Barton, L.L.; Ritz, N.L.; Fauque, G.D.; Lin, H.C. Sulfur Cycling and the Intestinal Microbiome. Dig. Dis. Sci. 2017, 62, 2241–2257. [Google Scholar] [CrossRef]
- Walsh, B.J.C.; Giedroc, D.P. H2S and reactive sulfur signaling at the host-bacterial pathogen interface. J. Biol. Chem. 2020, 295, 13150–13168. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Bayliss, S.C.; Feil, E.J.; Liu, Y.; Wang, C.; Zhang, G.; Zhou, D.; Wei, D.; Tang, N.; Leclercq, S.O.; et al. Real time monitoring of Aeromonas salmonicida evolution in response to successive antibiotic therapies in a commercial fish farm. Environ. Microbiol. 2019, 21, 1113–1123. [Google Scholar] [CrossRef]
- Shatalin, K.; Shatalina, E.; Mironov, A.; Nudler, E. H2S: A universal defense against antibiotics in bacteria. Science 2011, 334, 986–990. [Google Scholar] [CrossRef] [PubMed]
- Abdollahi, M.; Hosseini, A. Hydrogen Sulfide. In Encyclopedia of Toxicology; Elsevier: Amsterdam, The Netherlands, 2014; pp. 971–974. [Google Scholar]
- Kazmierczak, M.J.; Wiedmann, M.; Boor, K.J. Alternative sigma factors and their roles in bacterial virulence. Microbiol. Mol. Biol. Rev. 2005, 69, 527–543. [Google Scholar] [CrossRef] [PubMed]
- Grove, A. Extracytoplasmic Function Sigma Factors Governing Production of the Primary Siderophores in Pathogenic Burkholderia Species. Front. Microbiol. 2022, 13, 851011. [Google Scholar] [CrossRef]
- Todor, H.; Osadnik, H.; Campbell, E.A.; Myers, K.S.; Li, H.; Donohue, T.J.; Gross, C.A. Rewiring the specificity of extracytoplasmic function sigma factors. Proc. Natl. Acad. Sci. USA 2020, 117, 33496–33506. [Google Scholar] [CrossRef]
- Lemos, M.L.; Balado, M. Iron uptake mechanisms as key virulence factors in bacterial fish pathogens. J. Appl. Microbiol. 2020, 129, 104–115. [Google Scholar] [CrossRef]
- Sheng, L.; Wang, L. The microbial safety of fish and fish products: Recent advances in understanding its significance, contamination sources, and control strategies. Compr. Rev. Food Sci. Food Saf. 2021, 20, 738–786. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valenzuela-Miranda, D.; Morales-Rivera, M.; Mancilla-Schutz, J.; Sandoval, A.; Valenzuela-Muñoz, V.; Gallardo-Escárate, C. Nanopore-Based Metagenomic Approaches for Detection of Bacterial Pathogens in Recirculating Aquaculture Systems. Fishes 2025, 10, 496. https://doi.org/10.3390/fishes10100496
Valenzuela-Miranda D, Morales-Rivera M, Mancilla-Schutz J, Sandoval A, Valenzuela-Muñoz V, Gallardo-Escárate C. Nanopore-Based Metagenomic Approaches for Detection of Bacterial Pathogens in Recirculating Aquaculture Systems. Fishes. 2025; 10(10):496. https://doi.org/10.3390/fishes10100496
Chicago/Turabian StyleValenzuela-Miranda, Diego, María Morales-Rivera, Jorge Mancilla-Schutz, Alberto Sandoval, Valentina Valenzuela-Muñoz, and Cristian Gallardo-Escárate. 2025. "Nanopore-Based Metagenomic Approaches for Detection of Bacterial Pathogens in Recirculating Aquaculture Systems" Fishes 10, no. 10: 496. https://doi.org/10.3390/fishes10100496
APA StyleValenzuela-Miranda, D., Morales-Rivera, M., Mancilla-Schutz, J., Sandoval, A., Valenzuela-Muñoz, V., & Gallardo-Escárate, C. (2025). Nanopore-Based Metagenomic Approaches for Detection of Bacterial Pathogens in Recirculating Aquaculture Systems. Fishes, 10(10), 496. https://doi.org/10.3390/fishes10100496







