A Paired Flow Cytometry–Pathology Assessment for Immune Cell Detection in Intestinal Biopsies: Proof of Principle
Abstract
1. Introduction
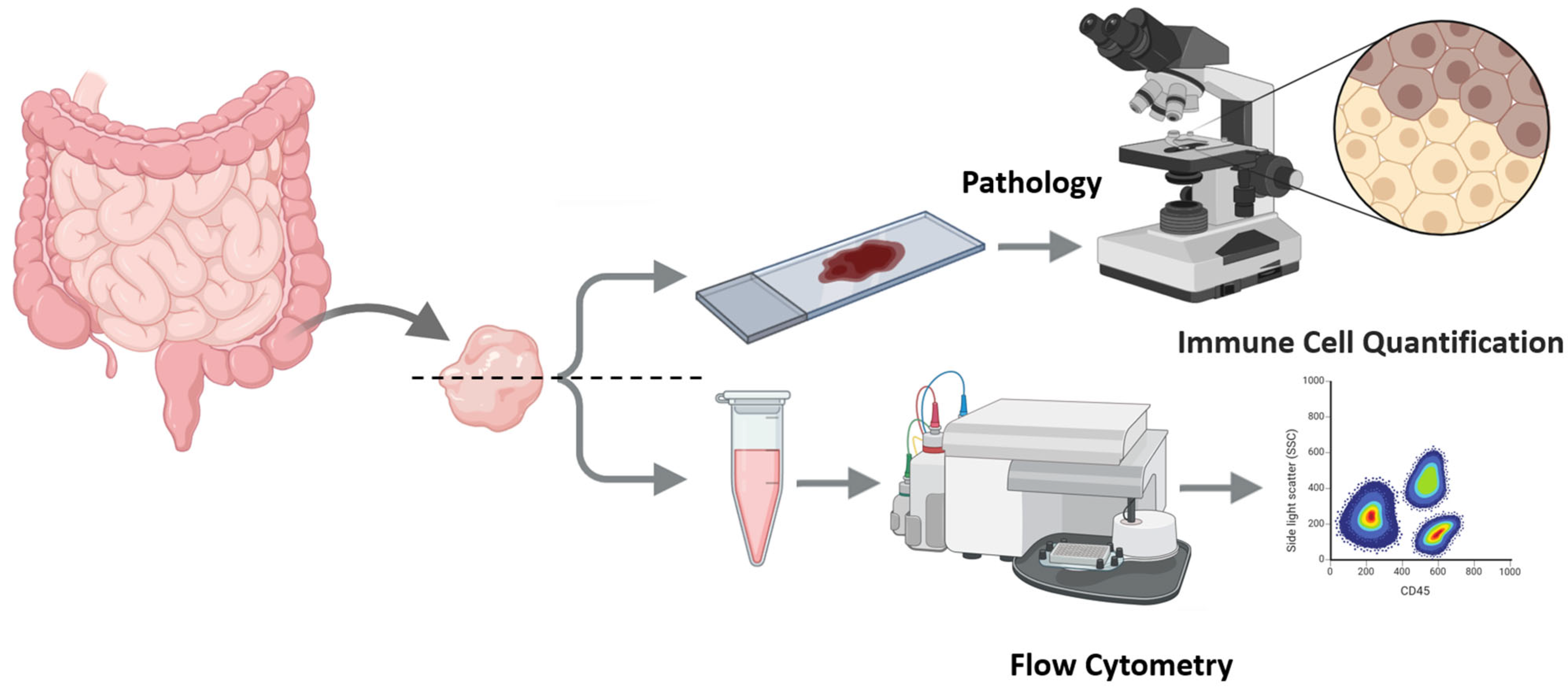
2. Experimental Design
2.1. Materials
- Sterile single-pack CellTrics® filters (Sysmex, Hyogo, Japan, Cat.no. 04-004-2323).
- Antibodies for immunohistochemistry:
- ○
- CD3 (Clone SP7) (Zytomed Systems, Berlin, Germany);
- ○
- CD4 (Clone SP35) (Cell Marque, Merck KGaA, Darmstadt, Germany);
- ○
- CD8 (Clone C8/144B) (Dako, Agilent, Santa Clara, CA, USA).
- Monoclonal antibodies for flow cytometry (BD Biosciences, Franklin Lakes, NJ, USA):
- ○
- CD45-APC (Clone 2D1);
- ○
- CD3-PE (Clone SK7);
- ○
- CD4-FITC (Clone SK3);
- ○
- CD8-PerCP (Clone SK1).
2.2. Equipment
- BD FACSCaliBur Flow Cytometer (BD Biosciences).
3. Detailed Procedure
- Tissue procurement, splitting, and parallel preparation (IHC and FC) from intestinal biopsies.
- 2.
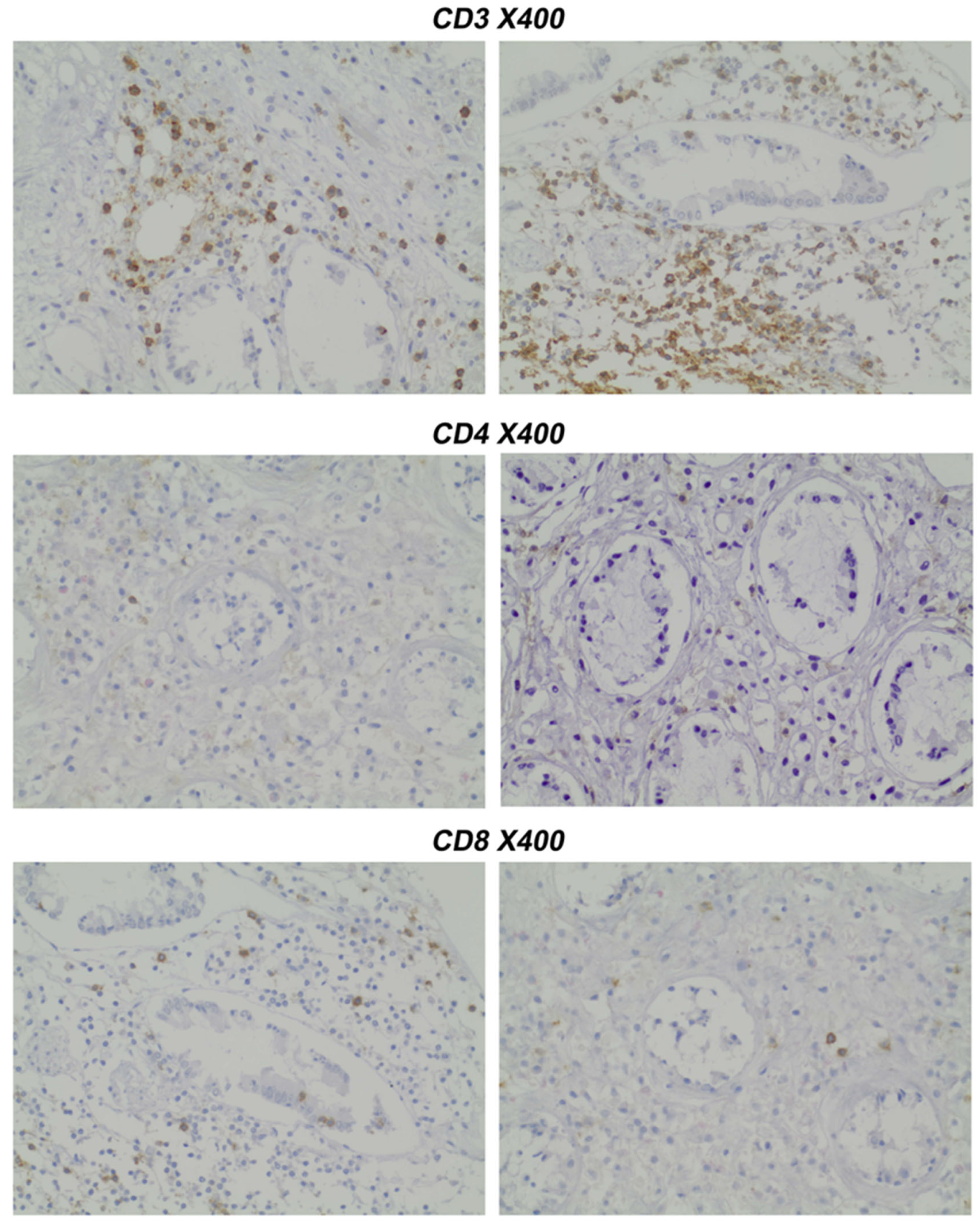
- Pathology Assessment: Paraffin-embedded slides (prepared under standard conditions) were stained immunohistochemically with CD4 and CD8 antibodies.
- 3.
- Sample preparation for Flow Cytometry (FC)
- 3.1.
- Dissociate samples using a scalpel and 150 μL PBS buffer. Resuspend cells by gently pipetting 20–30 times to liberate infiltrating leukocytes; avoid harsh trituration/enzymes. Allow gravity to settle for 60 s; carefully transfer the supernatant to a fresh tube. Pass through a CellTrics® filter (or analog) to remove tissue remnants and obtain a single-cell suspension.
 CRITICAL STEP: The mix should be resuspended gently to avoid cell destruction. The main goal is to dissociate immune infiltrating cells (and to quantify them thereafter) from the tissue and not to prepare a single tissue cell suspension.
CRITICAL STEP: The mix should be resuspended gently to avoid cell destruction. The main goal is to dissociate immune infiltrating cells (and to quantify them thereafter) from the tissue and not to prepare a single tissue cell suspension.- 3.2.
- Aliquot 100 µL (or 1–5 × 105 events where available). Incubate for 30 min at 4 °C in the dark with fluorochrome-conjugated antibodies: CD45-APC (leukocytes), CD3-PE (T cells), CD4-FITC, and CD8-PerCP.
- 3.3.
- Lyse red blood cells by adding 1 × RBC lysis buffer, incubate for 2–3 min at RT, and analyze immediately white blood cell subpopulations. Acquire on FACSCalibur (or equivalent) with standard filters for FITC/PE/PerCP/APC. Targets: ≥50,000 total events and ≥5000 CD45+ events per sample whenever yield permits (record actuals).
- 4.
- Flow Cytometry Acquisition and Analysis
- 4.1.
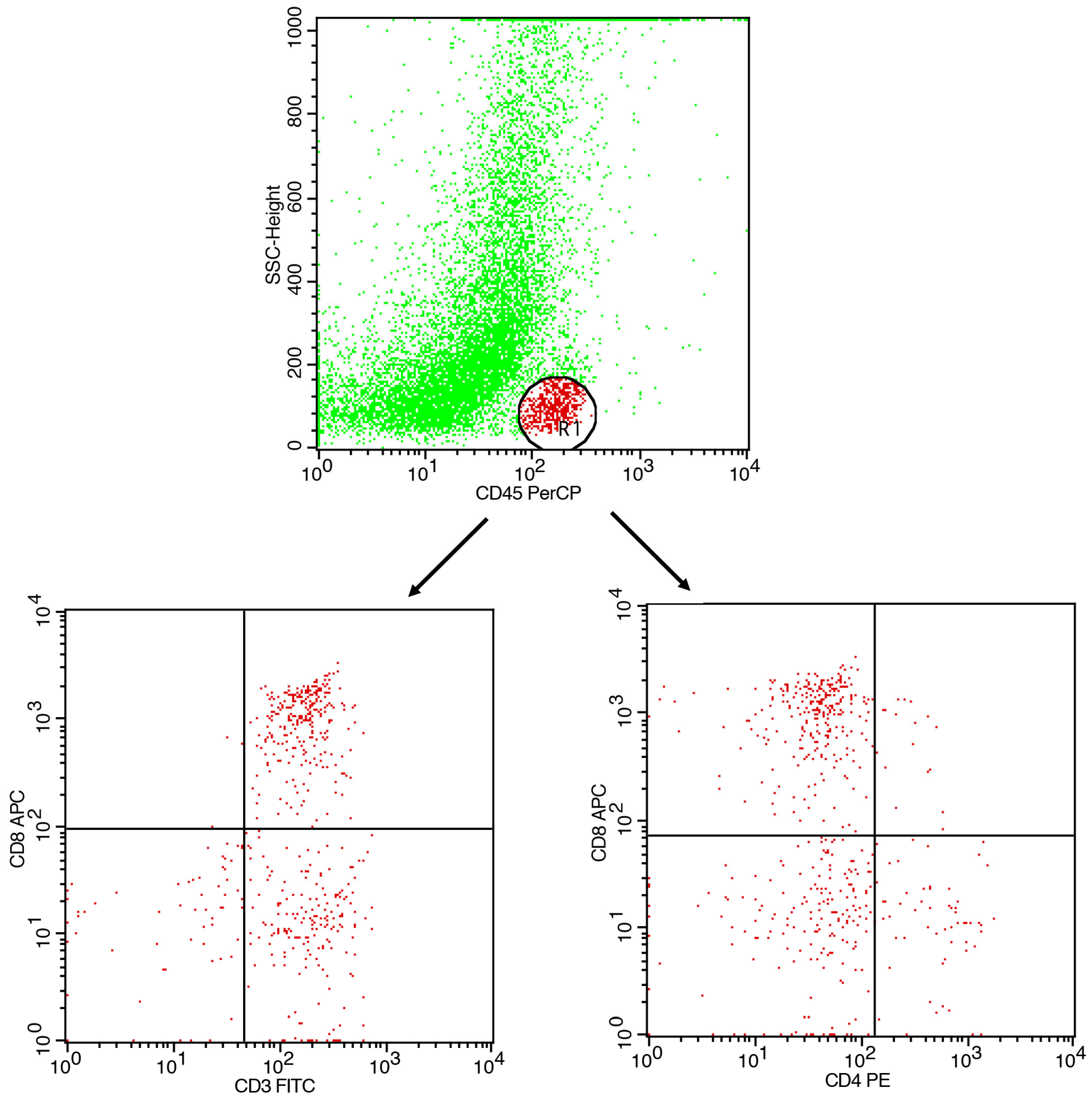
- Gating strategy: CD45+ events were gated to identify leukocytes. CD3+ cells were gated as total T cells. CD4+ and CD8+ subsets were analyzed within the CD3+ population.
- CD45 vs. SSC-A→leukocyte gate.
- CD3 within CD45+→T-cell gate.
- CD4 vs. CD8 within CD3+→compute CD4:CD8 ratio; optionally report CD3+CD4−CD8− fraction.
- Report % within parent and the CD4:CD8 ratio.
- 4.2.
- Quality control: Include fluorescence-minus-one (FMO) controls and isotype controls.
 CRITICAL STEP: Following standard quality control, use peripheral blood for standardization of leukocyte subpopulation in the instrument.
CRITICAL STEP: Following standard quality control, use peripheral blood for standardization of leukocyte subpopulation in the instrument.- Low CD45+ events (<5000): increase gentle pipetting cycles; pool two rinse fractions.
- Excess RBCs: verify timely RBC lysis (2–3 min) and analyze immediately.
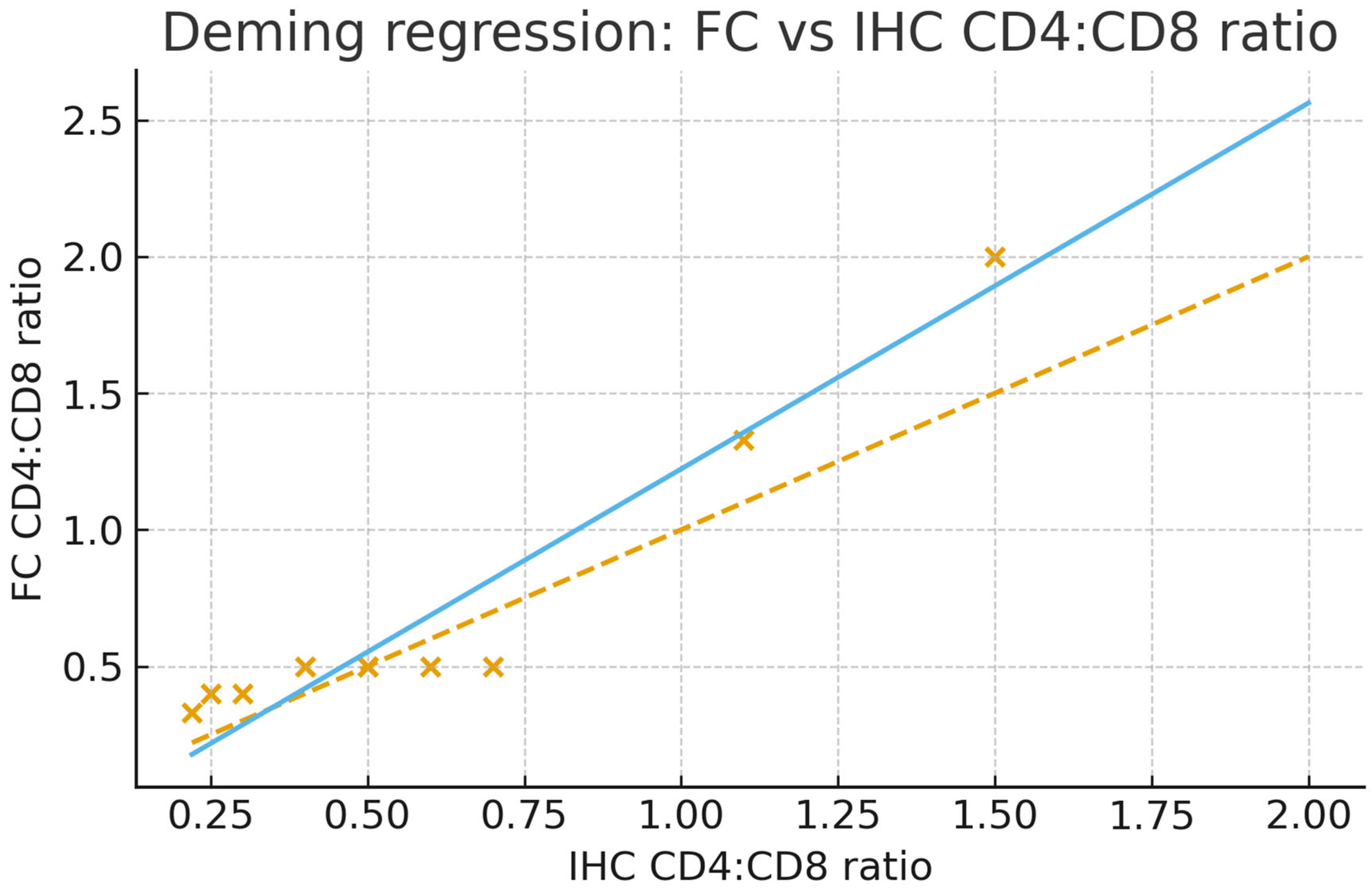
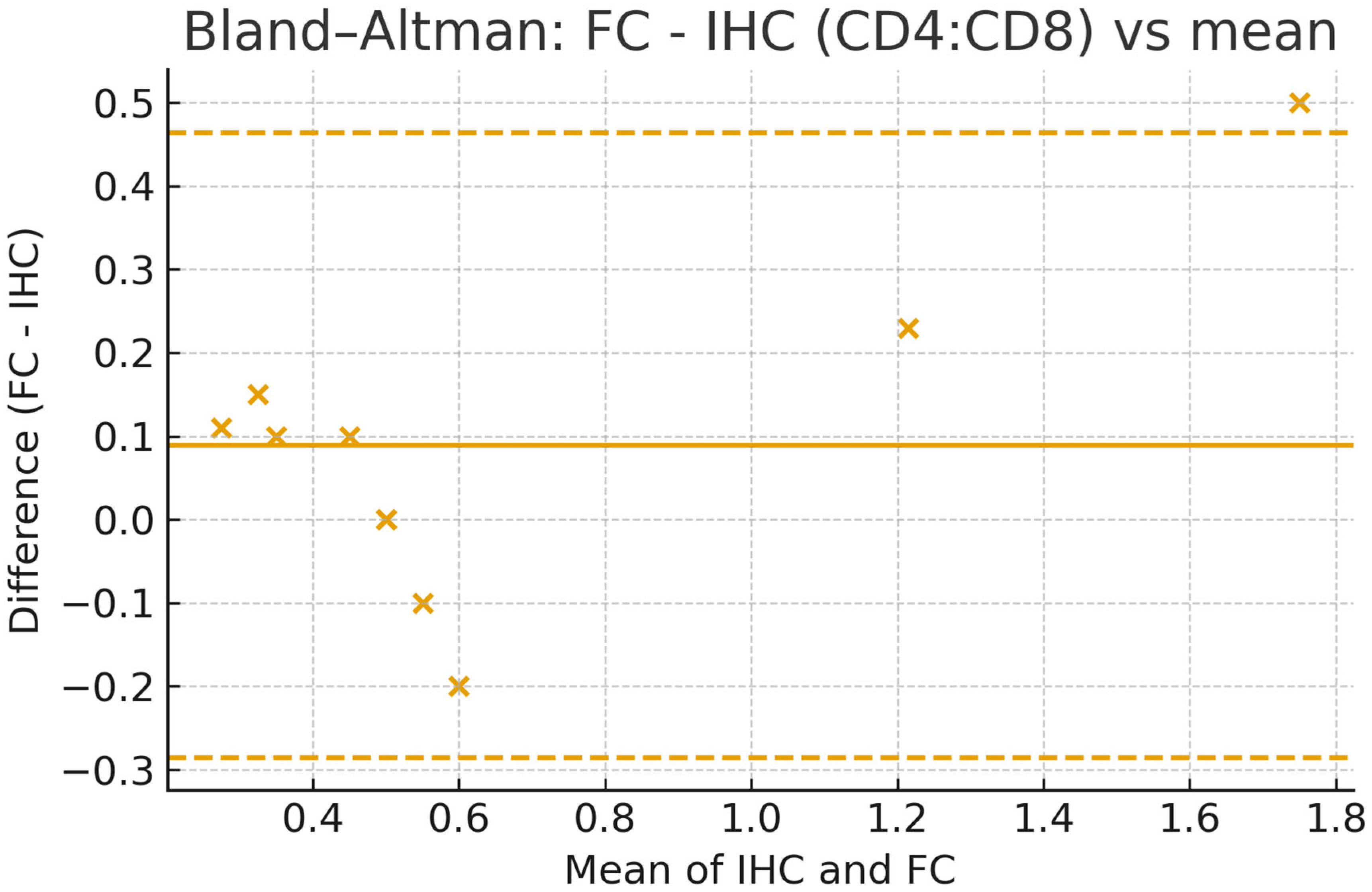
4. Expected Results
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Domínguez Conde, C.; Xu, C.; Jarvis, L.B.; Rainbow, D.B.; Wells, S.B.; Gomes, T.; Howlett, S.; Suchanek, O.; Polanski, K.; King, H. Cross-tissue immune cell analysis reveals tissue-specific features in humans. Science 2022, 376, eabl5197. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, H.M. Practical Flow Cytometry; John Wiley & Sons: New York, NY, USA, 2005. [Google Scholar]
- Cossarizza, A.; Chang, H.D.; Radbruch, A.; Acs, A.; Adam, D.; Adam-Klages, S.; Agace, W.W.; Aghaeepour, N.; Akdis, M.; Allez, M. Guidelines for the use of flow cytometry and cell sorting in immunological studies. Eur. J. Immunol. 2019, 49, 1457–1973. [Google Scholar] [CrossRef] [PubMed]
- Dimitriadis, S.; Dova, L.; Kotsianidis, I.; Hatzimichael, E.; Kapsali, E.; Markopoulos, G.S. Imaging Flow Cytometry: Development, Present Applications, and Future Challenges. Methods Protoc. 2024, 7, 28. [Google Scholar] [CrossRef]
- Chen, R.; Li, C.; Zheng, J.; Fan, Z.; Li, L.; Chen, M.; Chen, B.; Zhang, S. Lymphocyte subsets for predicting inflammatory bowel disease progression and treatment response: A systematic review. Front. Immunol. 2024, 15, 1403420. [Google Scholar] [CrossRef]
- Jiménez, J.M.; Contreras-Riquelme, J.S.; Vidal, P.M.; Prado, C.; Bastías, M.; Meneses, C.; Martín, A.J.M.; Perez-Acle, T.; Pacheco, R. Identification of master regulator genes controlling pathogenic CD4+ T cell fate in inflammatory bowel disease through transcriptional network analysis. Sci. Rep. 2024, 14, 10553. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Gu, X.; Yang, S.; Tao, R.; Fan, M.; Bao, W.; Wang, X. Research progress on intestinal tissue-resident memory T cells in inflammatory bowel disease. Scand. J. Immunol. 2023, 98, e13332. [Google Scholar] [CrossRef]
- Uniken Venema, W.T.C.; Ramírez-Sánchez, A.D.; Bigaeva, E.; Withoff, S.; Jonkers, I.; McIntyre, R.E.; Ghouraba, M.; Raine, T.; Weersma, R.K.; Franke, L.; et al. Gut mucosa dissociation protocols influence cell type proportions and single-cell gene expression levels. Sci. Rep. 2022, 12, 9897. [Google Scholar] [CrossRef]
- DaMata, J.P.; Zelkoski, A.E.; Nhan, P.B.; Ennis, K.H.E.; Kim, J.S.; Lu, Z.; Malloy, A.M.W. Dissociation protocols influence the phenotypes of lymphocyte and myeloid cell populations isolated from the neonatal lymph node. Front. Immunol. 2024, 15, 1368118. [Google Scholar] [CrossRef]
- Lu, W.; Mehraj, V.; Vyboh, K.; Cao, W.; Li, T.; Routy, J.-P. CD4:CD8 ratio as a frontier marker for clinical outcome, immune dysfunction and viral reservoir size in virologically suppressed HIV-positive patients. J. Int. AIDS Soc. 2015, 18, 20052. [Google Scholar] [CrossRef]
- Mehandru, S.; Poles, M.A.; Tenner-Racz, K.; Horowitz, A.; Hurley, A.; Hogan, C.; Boden, D.; Racz, P.; Markowitz, M. Primary HIV-1 infection is associated with preferential depletion of CD4+ T lymphocytes from effector sites in the gastrointestinal tract. J. Exp. Med. 2004, 200, 761–770. [Google Scholar] [CrossRef]
- Shacklett, B.L.; Cox, C.A.; Sandberg, J.K.; Stollman, N.H.; Jacobson, M.A.; Nixon, D.F. Trafficking of human immunodeficiency virus type 1-specific CD8+ T cells to gut-associated lymphoid tissue during chronic infection. J. Virol. 2003, 77, 5621–5631. [Google Scholar] [CrossRef] [PubMed]
- Geng, B.; Ding, X.; Li, X.; Liu, H.; Zhao, W.; Gong, H.; Tian, Z.; Guo, J. Peripheral blood T-lymphocyte subsets are potential biomarkers of disease severity and clinical outcomes in patients with ulcerative colitis: A retrospective study. BMC Gastroenterol. 2023, 23, 136. [Google Scholar] [CrossRef]
- Williams, C.J.M.; Gray, R.; Hills, R.K.; Shires, M.; Zhang, L.; Zhao, Z.; Gardner, T.; Sapanara, N.; Xu, X.-M.; Bai, I.; et al. Evaluation of CD3 and CD8 T-Cell Immunohistochemistry for Prognostication and Prediction of Benefit From Adjuvant Chemotherapy in Early-Stage Colorectal Cancer Within the QUASAR Trial. J. Clin. Oncol. 2024, 42, 3430–3442. [Google Scholar] [CrossRef] [PubMed]
- Hone Lopez, S.; Kats-Ugurlu, G.; Renken, R.J.; Buikema, H.J.; de Groot, M.R.; Visschedijk, M.C.; Dijkstra, G.; Jalving, M.; de Haan, J.J. Immune checkpoint inhibitor treatment induces colitis with heavy infiltration of CD8+ T cells and an infiltration pattern that resembles ulcerative colitis. Virchows Arch. 2021, 479, 1119–1129. [Google Scholar] [CrossRef]
- Reina-Campos, M.; Monell, A.; Ferry, A.; Luna, V.; Cheung, K.P.; Galletti, G.; Scharping, N.E.; Takehara, K.K.; Quon, S.; Challita, P.P.; et al. Tissue-resident memory CD8 T cell diversity is spatiotemporally imprinted. Nature 2025, 639, 483–492. [Google Scholar] [CrossRef]
- Xia, X.; Huang, Z.; Xu, C.; Fu, H.; Wang, S.; Tian, J.; Rui, K. Regulation of intestinal tissue-resident memory T cells: A potential target for inflammatory bowel disease. Cell Commun. Signal. 2024, 22, 610. [Google Scholar] [CrossRef]
- Soteriou, D.; Kubánková, M.; Schweitzer, C.; López-Posadas, R.; Pradhan, R.; Thoma, O.-M.; Györfi, A.-H.; Matei, A.-E.; Waldner, M.; Distler, J.H.W.; et al. Rapid single-cell physical phenotyping of mechanically dissociated tissue biopsies. Nat. Biomed. Eng. 2023, 7, 1392–1403. [Google Scholar] [CrossRef]
- Georvasili, V.K.; Markopoulos, G.S.; Batistatou, A.; Mitsis, M.; Messinis, T.; Lianos, G.D.; Alexiou, G.; Vartholomatos, G.; Bali, C.D. Detection of cancer cells and tumor margins during colorectal cancer surgery by intraoperative flow cytometry. Int. J. Surg. 2022, 104, 106717. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.P. Flow cytometry: Past and future. Biotechniques 2022, 72, 159–169. [Google Scholar] [CrossRef]
- Ferrer-Font, L.; Small, S.J.; Lewer, B.; Pilkington, K.R.; Johnston, L.K.; Park, L.M.; Lannigan, J.; Jaimes, M.C.; Price, K.M. Panel optimization for high-dimensional immunophenotyping assays using full-spectrum flow cytometry. Curr. Protoc. 2021, 1, e222. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Zheng, Y.; Sheng, J.; Han, Y.; Yang, Y.; Pan, H.; Yao, J. CD3+CD4-CD8- (Double-negative) T cells in inflammation, immune disorders and cancer. Front. Immunol. 2022, 13, 816005. [Google Scholar] [CrossRef] [PubMed]
- Ferrer-Font, L.; Small, S.J.; Hyde, E.; Pilkington, K.R.; Price, K.M. Panel Design and Optimization for Full Spectrum Flow Cytometry. Methods Mol. Biol. 2024, 2779, 99–124. [Google Scholar] [CrossRef] [PubMed]
- Park, L.M.; Lannigan, J.; Low, Q.; Jaimes, M.C.; Bonilla, D.L. OMIP-109: 45-color full spectrum flow cytometry panel for deep immunophenotyping of the major lineages present in human peripheral blood mononuclear cells with emphasis on the T cell memory compartment. Cytom. Part A 2024, 105, 807–815. [Google Scholar] [CrossRef] [PubMed]
| cd4:cd8 (FC) | ||
| cd4:cd8 (FC) | Pearson Correlation | 1 |
| Sig. (2-tailed) | ||
| Sum of Squares and Cross-products | 1.486 | |
| Covariance | 0.165 | |
| N | 10 | |
| cd4:cd8 (IHC) | Pearson Correlation | 0.956 ** |
| Sig. (2-tailed) | 0.000 | |
| Sum of Squares and Cross-products | 1.881 | |
| Covariance | 0.209 | |
| N | 10 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Skamnelos, A.; Markopoulos, G.S.; Dova, L.; Tragani, I.; Katsipaneli, M.; Christodoulou, D.; Katsanos, K.; Lampri, E. A Paired Flow Cytometry–Pathology Assessment for Immune Cell Detection in Intestinal Biopsies: Proof of Principle. Methods Protoc. 2025, 8, 122. https://doi.org/10.3390/mps8050122
Skamnelos A, Markopoulos GS, Dova L, Tragani I, Katsipaneli M, Christodoulou D, Katsanos K, Lampri E. A Paired Flow Cytometry–Pathology Assessment for Immune Cell Detection in Intestinal Biopsies: Proof of Principle. Methods and Protocols. 2025; 8(5):122. https://doi.org/10.3390/mps8050122
Chicago/Turabian StyleSkamnelos, Alexandros, Georgios S. Markopoulos, Lefkothea Dova, Ioulia Tragani, Meropi Katsipaneli, Dimitrios Christodoulou, Konstantinos Katsanos, and Evangeli Lampri. 2025. "A Paired Flow Cytometry–Pathology Assessment for Immune Cell Detection in Intestinal Biopsies: Proof of Principle" Methods and Protocols 8, no. 5: 122. https://doi.org/10.3390/mps8050122
APA StyleSkamnelos, A., Markopoulos, G. S., Dova, L., Tragani, I., Katsipaneli, M., Christodoulou, D., Katsanos, K., & Lampri, E. (2025). A Paired Flow Cytometry–Pathology Assessment for Immune Cell Detection in Intestinal Biopsies: Proof of Principle. Methods and Protocols, 8(5), 122. https://doi.org/10.3390/mps8050122








