A Practical Guide to Developing and Troubleshooting Patient-Derived “Mini-Gut” Colorectal Organoids for Clinical Research
Abstract
1. Introduction
2. Materials and Methods
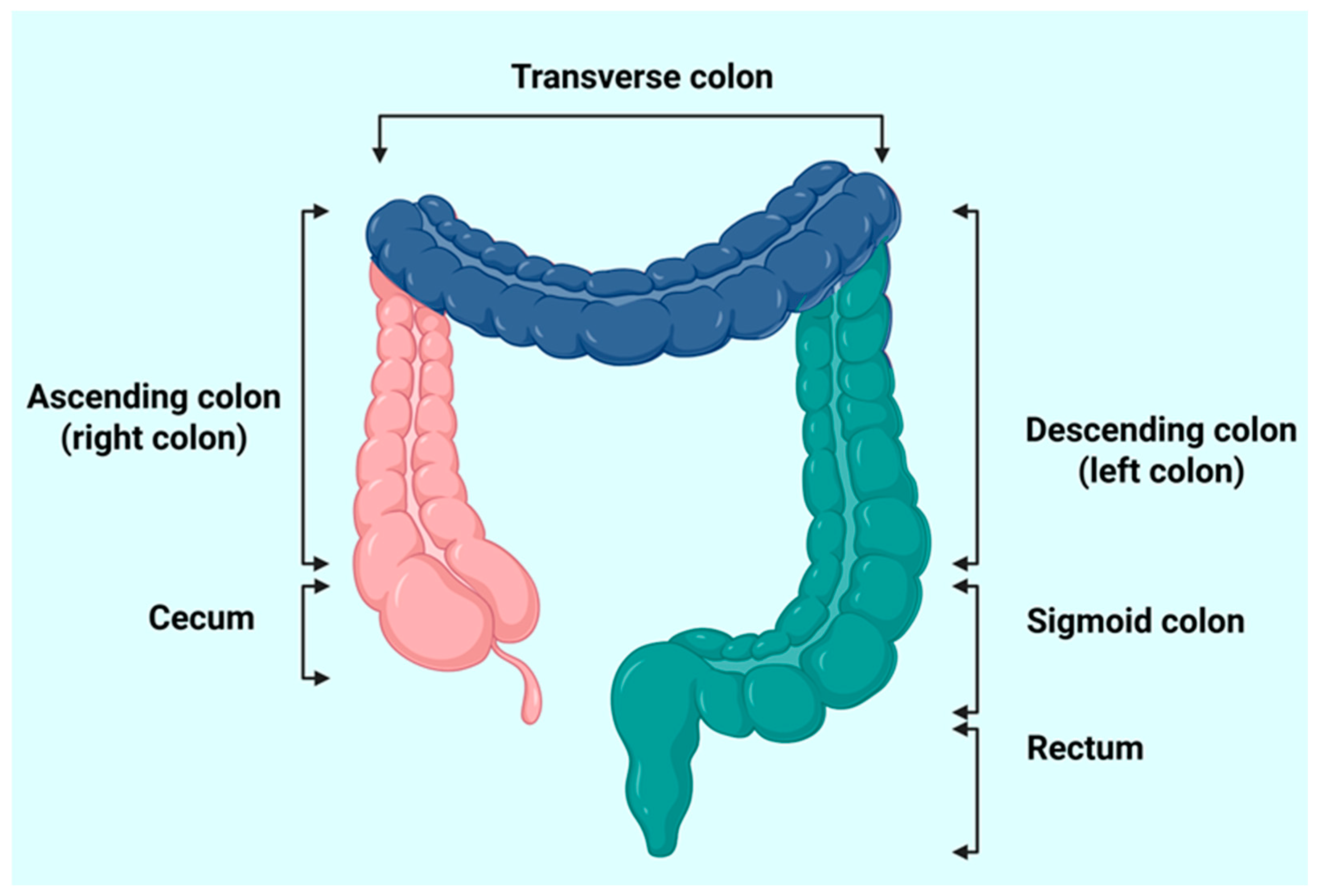
2.1. Identifying Target Colon Tissue Regions for Sample Collection
2.2. Tissue Procurement and Initial Processing (Approximately 2 h)
 CRITICAL STEP: Transfer samples in a 15 mL Falcon tube containing 5–10 mL of cold Advanced DMEM/F12 medium supplemented with antibiotics (e.g., penicillin-streptomycin) to avoid microbial contamination during transit. Delays in tissue processing reduce cell viability and impact organoid formation efficiency. Cryopreservation is an additional option to preserve the tissues for future organoid development.
CRITICAL STEP: Transfer samples in a 15 mL Falcon tube containing 5–10 mL of cold Advanced DMEM/F12 medium supplemented with antibiotics (e.g., penicillin-streptomycin) to avoid microbial contamination during transit. Delays in tissue processing reduce cell viability and impact organoid formation efficiency. Cryopreservation is an additional option to preserve the tissues for future organoid development. CRITICAL STEP: In some laboratories, same-day sample processing is not possible; this is often a challenge, particularly when there is a disconnect between the clinical site and the research lab. To minimize sample loss and increase reproducibility, we used two methods. (1) Interim cold storage (6–10 h) with antibiotics: if the delay was within 6–10 h, tissue collected at night was given an antibiotic wash, stored at 4 °C in RPMI or DMEM containing antibiotics, and processed the following morning. (2) Cryopreservation: after an antibiotic wash, tissue was cryopreserved in an appropriate medium for later processing. We observed a 20–30% variability in live-cell viability between these two preservation methods. Based on this experience, we recommend selecting the method according to the expected delay; when the delay exceeds 14 h, cryopreserving the tissue and processing it later is preferable. Two validated preservation methods are given below.
CRITICAL STEP: In some laboratories, same-day sample processing is not possible; this is often a challenge, particularly when there is a disconnect between the clinical site and the research lab. To minimize sample loss and increase reproducibility, we used two methods. (1) Interim cold storage (6–10 h) with antibiotics: if the delay was within 6–10 h, tissue collected at night was given an antibiotic wash, stored at 4 °C in RPMI or DMEM containing antibiotics, and processed the following morning. (2) Cryopreservation: after an antibiotic wash, tissue was cryopreserved in an appropriate medium for later processing. We observed a 20–30% variability in live-cell viability between these two preservation methods. Based on this experience, we recommend selecting the method according to the expected delay; when the delay exceeds 14 h, cryopreserving the tissue and processing it later is preferable. Two validated preservation methods are given below.3. Cryopreservation
3.1. Required Items and Media
- DMEM/F12 (Gibco, Thermo Fisher Scientific; cat. no. 11320033) + 20% fetal bovine serum (FBS) (Gibco, Thermo Fisher Scientific; cat. no. A5670801) + Penicillin/Streptomycin (50 mL conical aliquot) (Gibco, Thermo Fisher Scientific; cat. no. 15140122);
- 1× Phosphate buffered saline (PBS) (Sigma-Aldrich D8537);
- Human colon media: 50% L-WRN conditioned media;
- Sterile filtered FBS;
- Dimethyl sulfoxide (DMSO) (Sigma-Aldrich D2650);
- Normocin (InvivoGen ant-nr-1);
- Gentamicin (Amresco E737);
- Antibiotic antimycotic solution (AB/AM) (GenDEPOT, Cat. No. C980B07);
- Cryogenic vials (Corning Inc. Cat. No. 430488);
- Freezing container (Nalgene Cryo 1 °C Freezing Container, Cat. No. 5100-0001).
 CRITICAL STEPS
CRITICAL STEPS3.2. Sample Processing Procedure
- Transfer samples directly to a 15 mL conical tube containing 5 mL ice-cold PBS. Surgical specimens: Wash with PBS and transfer to a Petri dish on ice.
- Dissect and remove fat section to isolate the mucosa or cancer tissues, section into small fragments. Transfer processed tissue to a 15 mL conical tube and wash samples twice with chilled PBS and resuspend in 5 mL chilled PBS.
- Prepare antibiotic solution by adding the following to 5 mL PBS: 10 μL normocin (from 50 mg/mL stock) 5 μL gentamicin (from 50 mg/mL stock), and 50 μL AB/AM (from 100× stock).
 PAUSE STEP Incubate samples in antibiotic solution for 15 min at room temperature.
PAUSE STEP Incubate samples in antibiotic solution for 15 min at room temperature.- 4.
- Wash samples twice with chilled PBS, removing all of the supernatant after the final wash.
- 5.
- Distribute samples into appropriately labeled cryogenic vials (Lab ID, Tissue Type, Date). Colonoscopy samples: one cryogenic vial per tissue type. Surgical samples: multiple cryogenic vials per tissue type as needed.
- 6.
- Add 1000 μL freezing media (given in Step 3) to each sample vial and gently suspend tissue samples in freezing media to ensure uniform distribution. Place cryogenic vials in a freezing container designed to achieve a −1 °C/minute cooling rate. Example: Mr. Frosty™ Freezing Container (Thermo Fisher, Cat no. 5100-0001) or CoolCell™ Freezer Container (Corning®, Cat no. CLS432000-1EA). Initial freezing: store container in a −80 °C freezer for 24 h. For long-term storage, transfer vials from −80 °C to liquid nitrogen for long-term cryopreservation.
4. Conditioned Medium Preparation
4.1. Cell Line and Medium
- The conditioned medium is obtained from a genetically modified L-WRN (ATCC CRL-3276) cell line that simultaneously produces Wnt3a, R-spondin 3, and Noggin, which is extensively utilized in gastrointestinal stem cell cultures.
- L-WRN cells were maintained in high-glucose DMEM supplemented with 10% FBS, 0.5 mg/mL G418 (Geneticin; Cat. No. 10-131-035), and 0.5 mg/mL hygromycin (InvivoGen; Cat. No. ant-hg-1).
 CRITICAL STEPS
CRITICAL STEPS4.2. Step by Step L-WRN Conditioned Medium Preparation Protocol
- Cell revival: Thaw cryopreserved L-WRN cells rapidly at 37 °C. Add directly into pre-warmed DMEM + 10% FBS without antibiotics, and plate immediately in T25 flask. After 24 h, replace with complete medium containing 0.5 mg/mL G418 and 0.5 mg/mL hygromycin B. Optional: Record cell viability at thawing (e.g., Trypan Blue count) to check cell viability.
- Passaging: Subculture when cells are subconfluent (~70–80%) using trypsin-EDTA. Recommended seeding density: 1.5–3 × 104 cells/cm2. Change medium every 2–3 days.
- Maintain L-WRN cells in high-glucose DMEM + 10% FBS, 0.5 mg/mL G418, and 0.5 mg/mL hygromycin B.
- Preparation for L-WRN CM collection: Grow L-WRN cells to confluence in T-150 flasks. Once cells are confluent, switch to collection medium (DMEM + 10% FBS, without G418/hygromycin).Note: Rinse the cells with PBS then antibiotic-free DMEM before changing the medium; this will reduce antibiotic carryover.
- L-WRN CM collectionAllow cells to reach over-confluence (typically it takes 3–4 days). Start collecting CM medium for 4 consecutive days.
- Remove and discard old medium;
- Add fresh collection medium;
- After 24 h, collect medium into a sterile 50 mL Falcon tube.
- Centrifuge collected medium at 2000× g for 5 min at room temperature to pellet cell debris.
CRITICAL STEP: Carefully transfer the supernatant to a new sterile tube (50 mL falcon tube) without disturbing the pellet. Filter the supernatant through a 0.22 µm filter into a sterile container to remove residual debris and contaminants. Label the tube with the date and passage number. Maintain passage records and avoid extended passaging (more than 20 passages can reduce CM activity).
- Store aliquots at −20 °C for short-term use (within 3 months). For long-term storage, −80 °C is recommended. Avoid repeated freeze-thaw cycles by aliquoting into 50 mL falcon tube.
5. Normal Human Colon Crypt Isolation for Organoid Establishment
 CRITICAL STEP
CRITICAL STEP- Thaw Matrigel on ice, plan on 45 µL of Matrigel per dome. Thaw time is two to three h for a 1.7 mL Eppendorf aliquot. Alternatively, thaw Matrigel overnight on ice if needed first thing in the morning the next day.
- Note: All of the organoid experiments were performed using Corning Matrigel® Growth Factor Reduced (GFR) Basement Membrane Matrix (Cat. No. 356255). Each lot was accompanied by a certificate of analysis indicating a protein concentration in the range of ~9–12 mg/mL To ensure reproducibility, a minimal lot-acceptance test was performed based on organoid revival efficiency, monitored daily until day 7; only lots that supported robust organoid formation and expansion were retained for downstream experiments.
- Thaw antibiotics on ice.
- Warm a 24-well non-treated plate in a 37 °C incubator.
- Warm human colon media (50% L-WRN CM + 50% Advanced DMEM/F12 with supplements) in a 37 °C water bath for 30 min. (see Table 1 for medium composition).
- Place PBS, DMEM/F12 media, and 10% FBS media on ice.
5.1. List of Required Reagents and Materials
- Human colon media: 50% L-WRNCM;
- 1 × PBS (Sigma-Aldrich D8537);
- Normocin (Invivogen ant-nr-1);
- Gentamicin (Amresco E737);
- Antibiotic-antimycotic solution (AB/AM) (Sigma C980B07);
- Collagenase type I (Invitrogen 17100-017);
- Corning® Matrigel® GFR Growth Factor Reduced (GFR) (Product #356231, 1.7 mL aliquots);
- DMEM/F12 without FBS (50 mL conical);
- DMEM/F12 + 10% FBS + Pen/Strep (50 mL conical);
- 24-well non-treated cell culture plate;
- 5 mM EDTA.
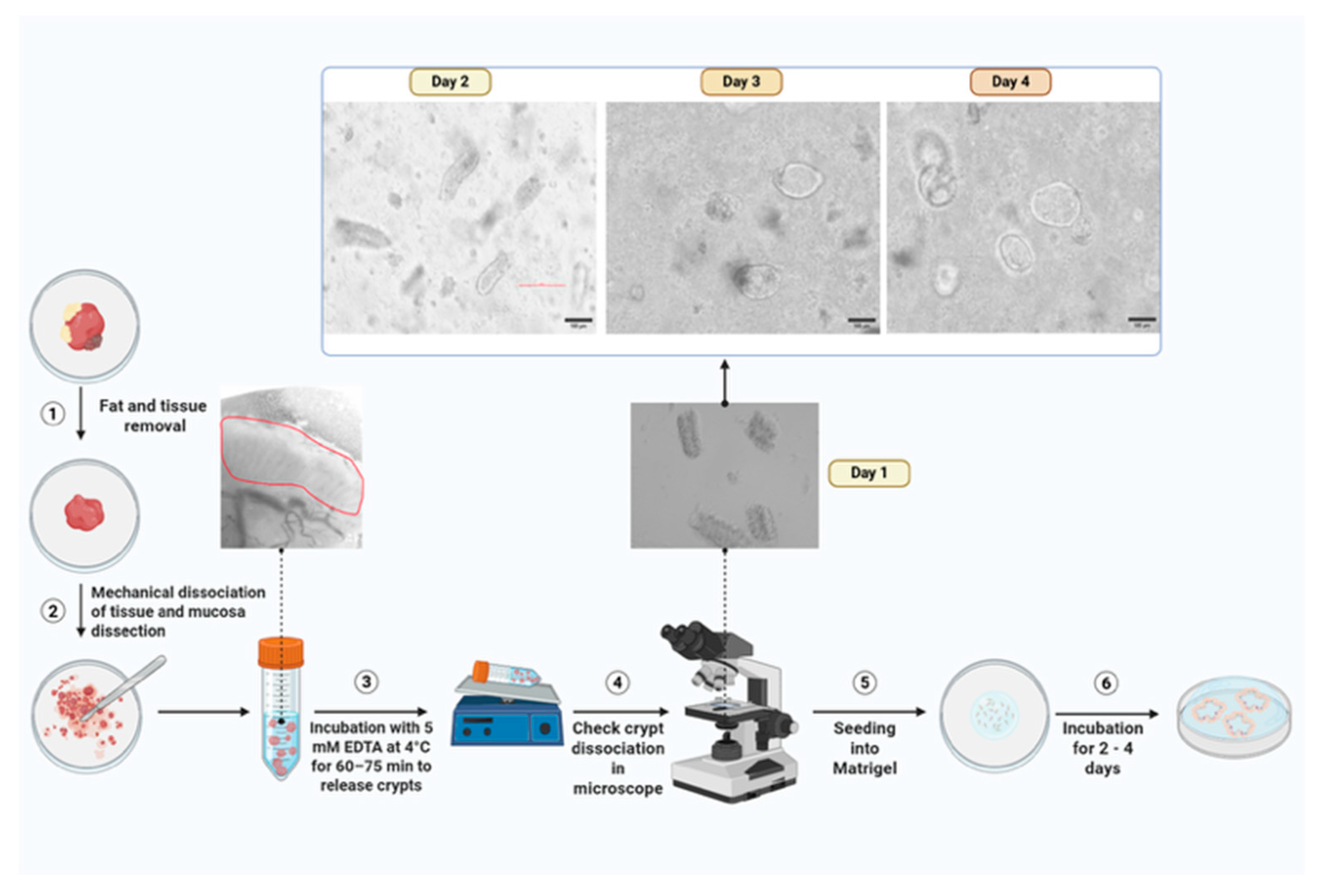
5.2. Sample Processing Procedure
- Transfer colonoscopy samples into a 15 mL conical tube containing 5 mL ice-cold PBS. For surgical specimens, rinse with PBS, dissect to isolate the mucosa containing crypts, mince into small fragments, and transfer to a 15 mL conical tube with 5 mL PBS. Add 10 µL normocin, 5 µL gentamicin, and 50 µL AB/AM to 5 mL PBS and incubate tissue in the antibiotic mix for 15 min. Wash samples twice with cold PBS, removing all liquid after the final wash.
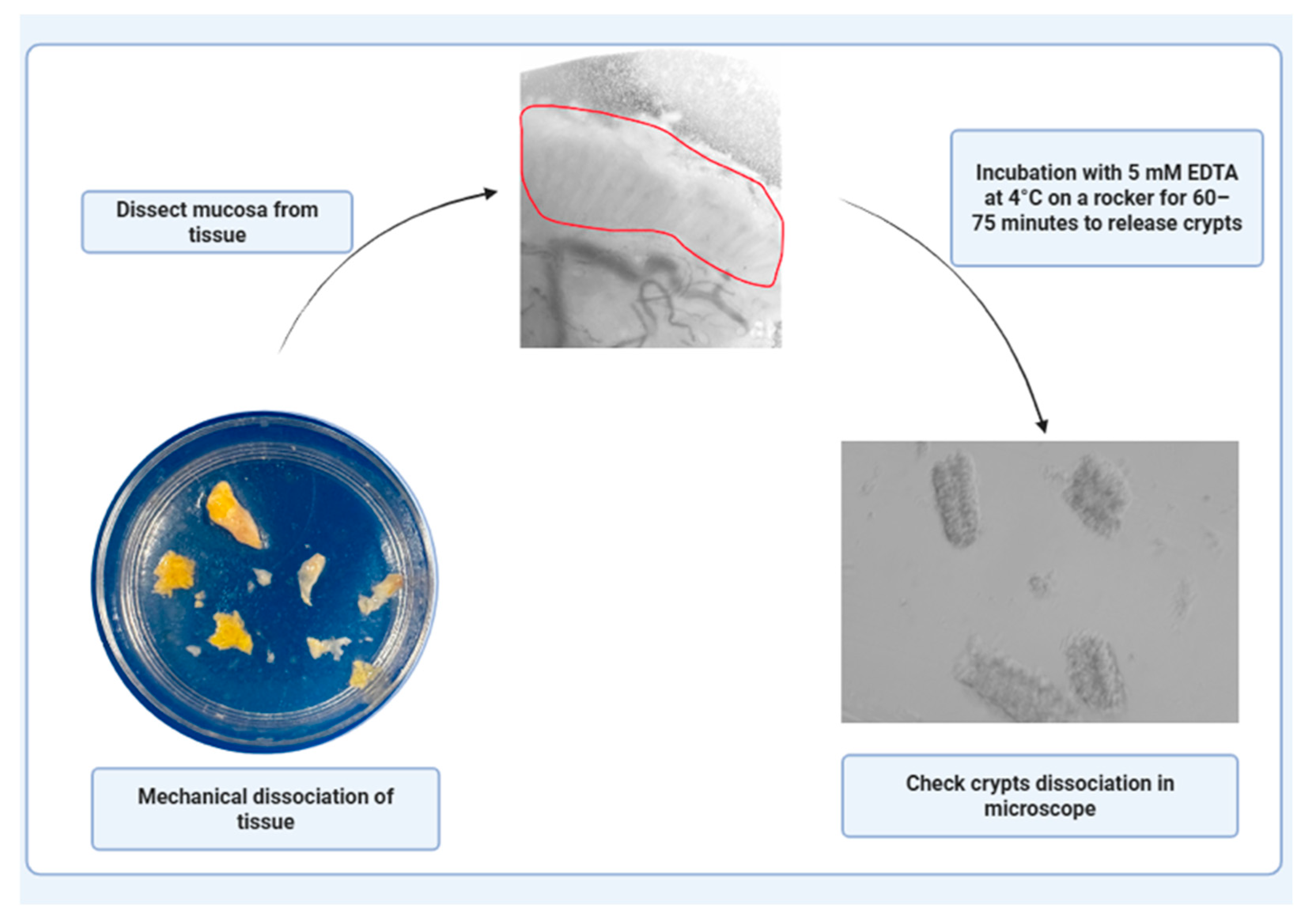
 PAUSE STEP: Incubate tissue in 10 mL of 5 mM EDTA at 4 °C on a rocker for 60–75 min. Carefully decant the EDTA and replace it with 5 mL fresh PBS, ensuring that the tissue remains undisturbed. Coat a 10 mL serological pipette with 10% FBS (or FBS/BSA-containing media) to reduce sticking to pipette wall.
PAUSE STEP: Incubate tissue in 10 mL of 5 mM EDTA at 4 °C on a rocker for 60–75 min. Carefully decant the EDTA and replace it with 5 mL fresh PBS, ensuring that the tissue remains undisturbed. Coat a 10 mL serological pipette with 10% FBS (or FBS/BSA-containing media) to reduce sticking to pipette wall.- 2.
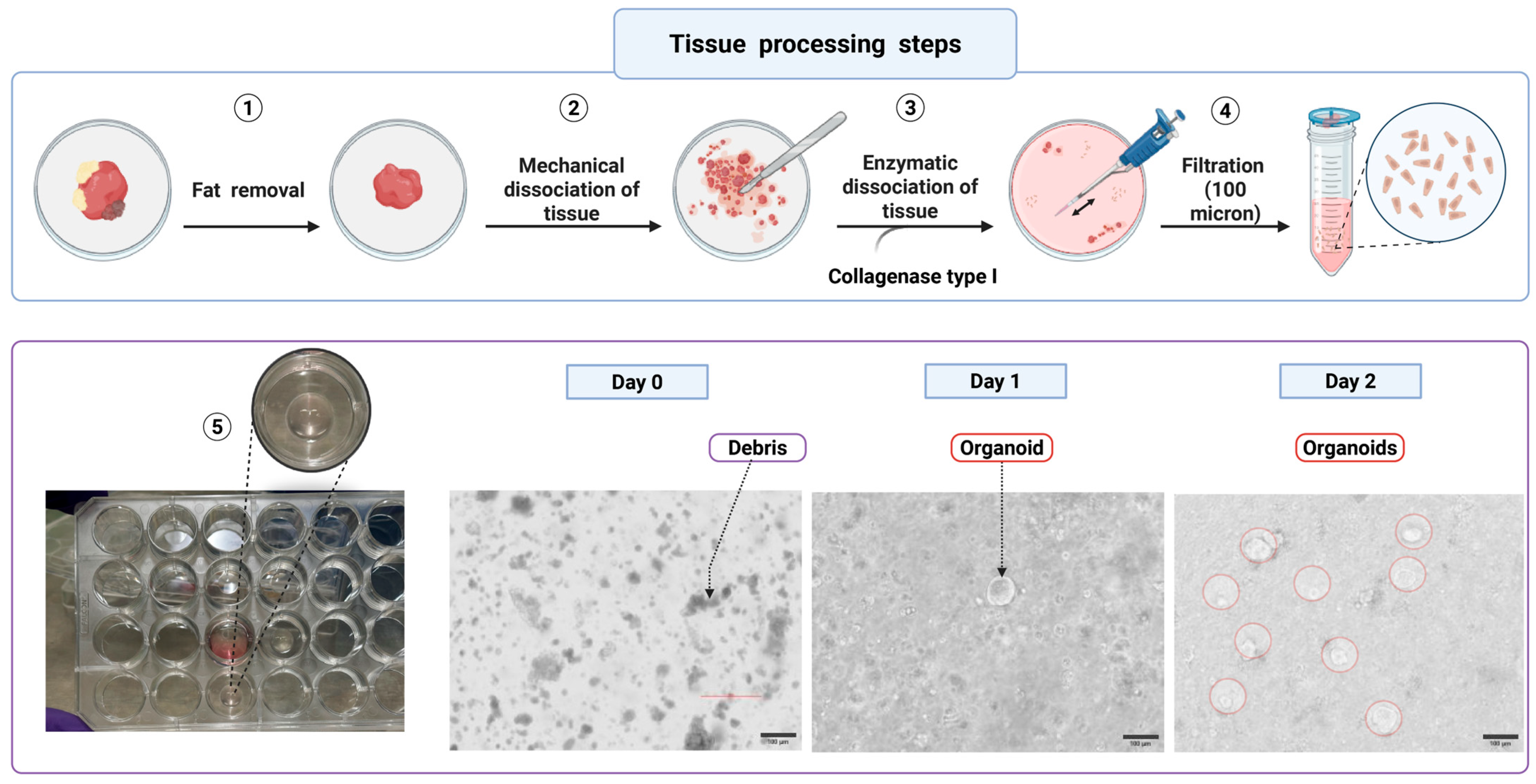
- Mechanical disruption by pipetting tissue up and down with the coated pipette to release crypts. Check the dissociated crypts under a microscope to confirm successful crypt isolation (Figure 2).
- 3.
- Centrifuge at 500× g for 5 min at 4 °C.
- 4.
- Discard the supernatant carefully, avoiding disturbance of the crypt pellet. Resuspend the crypts in DMEM/F12 (without FBS) using ~10 µL per dome. Pre-wet the pipette tip with DMEM/F12 + 10% FBS before resuspending to reduce cell loss.
- 5.
- Mix the resuspended crypts with cold Matrigel (40–50 µL per dome). Plate the cells immediately and avoid introducing bubbles.
- 6.
- Pipette 40–50 µL of the Matrigel–crypt mixture into the center of each well in the pre-warmed 24-well plate to form domes. Allow domes to polymerize: 5 min at room temperature and then 20–30 min at 37 °C. Add 750 µL of pre-warmed organoid complete media to each well and incubate.
5.3. Key Steps to Remember Before Plating the Crypts
- Mix pellet with ice-cold Matrigel (approx. 40 µL per dome) carefully to avoid air bubbles.
- Pipette Matrigel onto the pellet slowly. To avoid introducing air bubbles, do not remove the pipette tip from liquid when pipetting up and down.
- Pipette 50 µL of the media, Matrigel, and crypts into a well of the pre-warmed plate.
- Pipette slowly to avoid introducing air bubbles and let the dome polymerize for 5 min at room temperature, then transfer to a 37 °C incubator for 15 min before adding the medium.
- After polymerization, add 750 µL of 50% L-WRN complete media to each well and place back in the incubator.
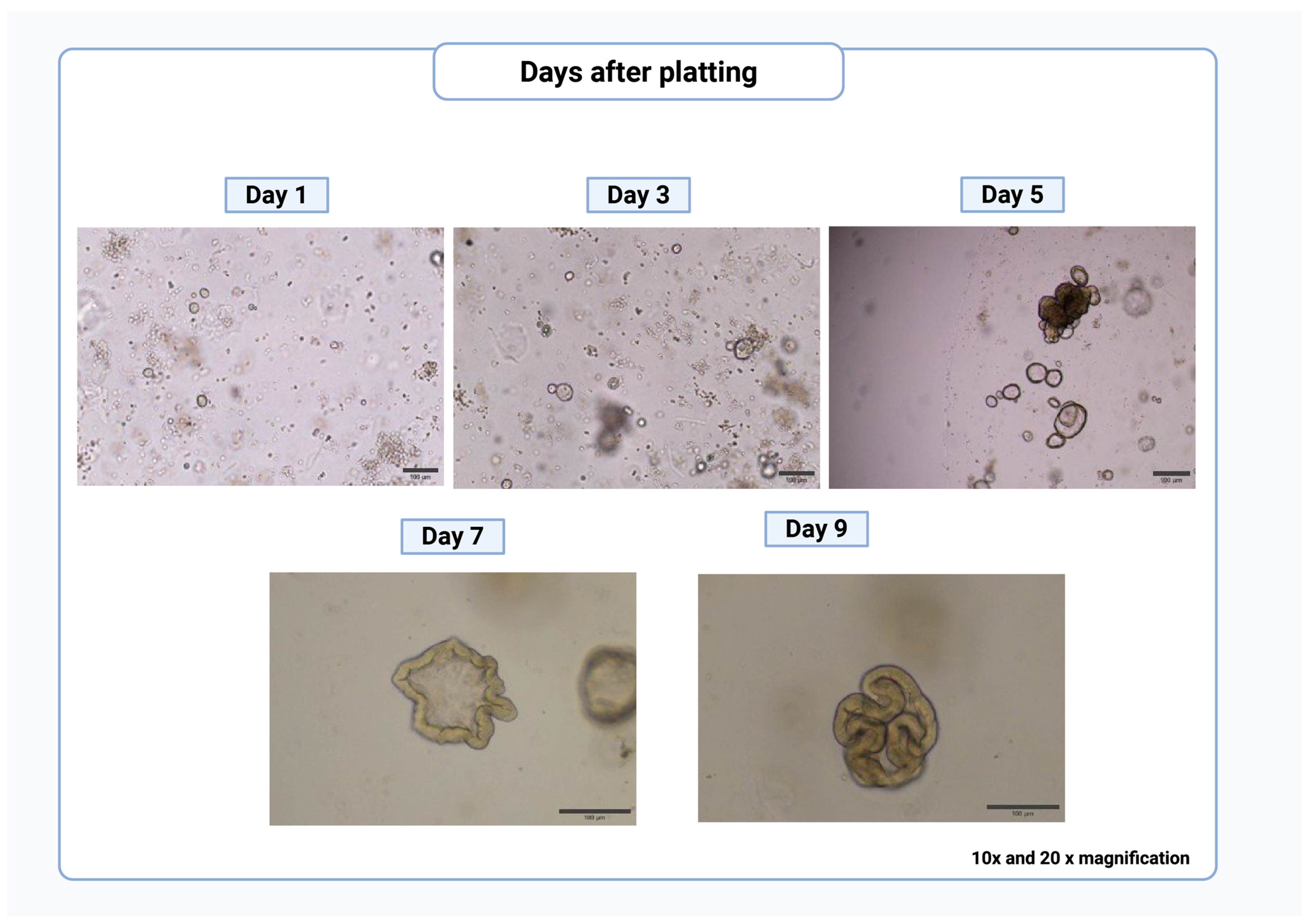
- Check the crypt plate after ~24 h to determine whether the organoids have started to form. Clean and combine wells of crypts to remove tissue debris and make sure the cultures are not too sparse by following the passage protocol to Step 5 and replate.
- Passage every ~7 days or as needed based on the growth of organoids (Figure 3).
6. Polyp/Cancer Cell Isolation
6.1. Pre-Experiment Preparation
- Transfer biopsy samples to 15 mL conical tube with 5 mL ice-cold PBS. For surgical specimens, wash in PBS, dissect mucosa on ice, mince into small pieces, and transfer to a 15 mL tube with 5 mL ice-cold PBS.
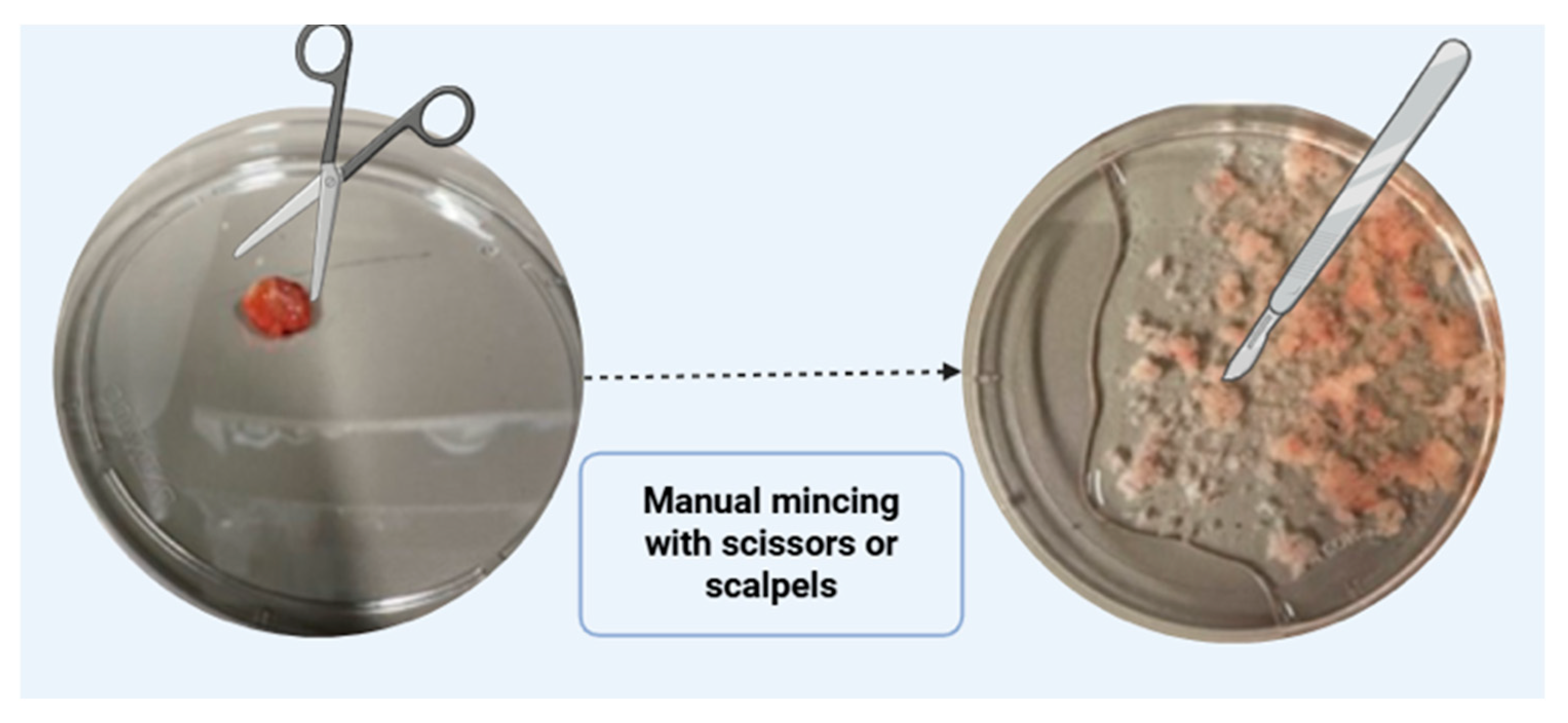
- Incubate sample in antibiotics for 15 min. Add to 5 mL PBS containing 10 µL of normocin (from 50 mg/mL stock), 5 µL of gentamicin (from 50 mg/mL stock), and 50 µL of AB/AM (from 100 × stock). Rinse tissue 2–3 times with cold PBS containing antibiotics to remove blood and debris. Trim away excess adipose tissue using sterile forceps and scissors. Mince cleaned tissue into small fragments using scalpels or fine scissors.
 CRITICAL STEP: Avoid over-mincing the tissue. Fragments should be small enough for efficient digestion but not lysed completely (Figure 4).
CRITICAL STEP: Avoid over-mincing the tissue. Fragments should be small enough for efficient digestion but not lysed completely (Figure 4).- 3.
- Add 250 μL of collagenase I (6 KU/mL stock; Invitrogen, 17100-017) to 4.75 mL of DMEM/F12 without FBS, yielding a final concentration of ~300 U/mL. For fibrotic or mucus-rich samples, supplement with DNase I (10–20 U/mL) to reduce DNA-mediated clumping. Place the sample in a 37 °C water bath for 20–30 min, agitating gently and monitoring digestion progress under a microscope every 5–10 min. Block the 5 mL collagenase digestion sample by adding 10 mL DMEM/F12 + 10% FBS (1:2 ratio of supernatant to blocking medium).
- 4.
- Centrifuge sample at 500× g for 5 min at 4 °C. Carefully discard the supernatant without disturbing the cells. Add the desired volume of DMEM/F12 without FBS to the cells.
 CRITICAL STEP: Pre-wet the pipette tip with DMEM/F12 + 10% FBS and resuspend polyp/cancer cells. Mix pellet with ice-cold Matrigel (approximately 40 μL per dome) carefully to avoid air bubbles because bubbles may interfere with imaging studies.
CRITICAL STEP: Pre-wet the pipette tip with DMEM/F12 + 10% FBS and resuspend polyp/cancer cells. Mix pellet with ice-cold Matrigel (approximately 40 μL per dome) carefully to avoid air bubbles because bubbles may interfere with imaging studies.- 5.
- Pipette 50 μL of the media + Matrigel + polyp/cancer cells into a well of the pre-warmed plate.
- 6.
- Pipette 40–50 µL of the Matrigel–cells combination into the center of each well in the pre-warmed 24-well plate to form domes. Optional: Invert the well plate to polymerize the dome. Allow domes to polymerize: 5 min at room temperature and then 20–30 min at 37 °C. Add 750 µL of pre-warmed organoid complete media to each well and incubate (Figure 5).
6.2. Key Steps to Remember Before Plating the Cells
- Mix pellet with ice-cold Matrigel (approx. 40 µL per dome) carefully to avoid air bubbles.
- Pipette Matrigel onto the pellet slowly and do not remove the tip from liquid when pipetting up and down; this will introduce air bubbles.
- Pipette 50 µL of the media + Matrigel + cells into a well of the pre-warmed plate.
- Pipette slowly to avoid introducing air bubbles, let the dome polymerize for 5 minutes at room temperature, then transfer to a 37 °C incubator for 15 min before adding medium.
- After polymerization, add 750 µL of human colon media (50% L-WRN complete media) to each well and place back in the incubator.
7. Passage Organoids with TrypLE Express
Sample Processing Procedure (Typical Passage Once Every 7 Days)
- Remove medium from wells (around the solid Matrigel).
 CRITICAL STEP: Direct pipetting will disturb the Matrigel and will lead to organoid loss.
CRITICAL STEP: Direct pipetting will disturb the Matrigel and will lead to organoid loss.- 2.
- Add 500 µL cold Advanced DMEM/F12 (or other base media with no organoid factors) to well and mechanically break up the Matrigel with pipetting P1000 up and down (3–4 times). Repeat this process if needed. The purpose of this step is to wash most of the Matrigel off the wells. Repeat with other wells and combine the domes in a 15 mL Falcon tube.
- 3.
- Transfer organoids/media to a 15 mL Falcon tube.
- 4.
- Spin down in a refrigerated swing rotor centrifuge at 500× g for 5 min at 4 °C.
- 5.
- Remove supernatant (note: carefully not to disturb pellet) and resuspend pellet in 500 µL TrypLE Express and put tube in a 37 °C water/bead bath for 10 min. At the 5-minute mark, you can flick the tube (or pipette up and down to break up the organoids if they are particularly mature—this can be checked under a microscope).Note: If the sample appears viscous, add DNase I (e.g., 10 U/mL) to aid dissociation.
- 6.
- Add 4–5 mL Advanced DMEM/F12 on ice to dilute TrypLE and stop the dissociation of cells and mix the tube.
- 7.
- Spin down in a refrigerated swing rotor centrifuge at 500× g for 5 min at 4 °C.
- 8.
- Remove all supernatants (use 10-mL pipette to remove the supernatant, then the P1000 or P200 pipette, careful not to disturb pellet). Keep the tube on ice.
- 9.
- Resuspend the pellet in Matrigel (calculate the amount of Matrigel required, 40–50 µL/well) and plate. After the Matrigel solidifies, add complete 50% L-WRN organoid medium.
 CRITICAL STEPS
CRITICAL STEPS8. Colon Organoid Culture Medium
| Reagent | Stock Concentration | Volume from Stock (for 50 mL) | Final Concentration | Source | Catalog Number | Role in Colon Organoid Culture |
|---|---|---|---|---|---|---|
| L-WRN conditioned medium (100%) | - | 25 mL | 50% | In-house | - | Provides Wnt3A, R-spondin, and Noggin signals to maintain stemness and self-renewal and stimulate high levels of Wnt signaling. |
| Advanced DMEM/F12 | - | 25 mL | 50% | GIBCO (Thermo Fisher Scientific, Waltham, MA, USA) | 12634010 | Reduced-serum basal medium (1:1 mixture of DMEM and Ham’s F-12) providing essential nutrients for organoid culture. |
| N2 Supplement | 100× | 500 μL | 1× | GIBCO (Thermo Fisher Scientific, Waltham, MA, USA) | 17502048 | Basal micronutrients in N2 that support epithelial cell viability and stem/progenitor maintenance in colon PDOs. |
| B27 Supplement | 50× | 1000 μL | 1× | GIBCO (Thermo Fisher Scientific, Waltham, MA, USA) | 17504044 | Vitamins and antioxidants enhance cell growth. |
| EGF | 50 μg/mL (1:2) | 40 μL | 40 ng/mL | R&D Systems (Minneapolis, MN, USA) | 236EG200 | Activates the EGFR-MAPK signaling pathway to sustain cell proliferation and crypt-like growth of colon PDO epithelium. |
| SB202190 | 3 mM | 50 μL | 3 μM | Sigma-Aldrich (Darmstadt, Germany) | S7067-5MG | p38 MAPK inhibitor, which reduces stress-induced differentiation/apoptosis to maintain colon stem cell compartments. |
| A83-01 | 500 μM | 50 μL | 500 nM | Tocris Bioscience (Minneapolis, MN, USA) | 293910 | ALK4/5/7 (TGF-β) inhibitor; inhibits the premature differentiation of colon epithelium and supports expansion. |
| Y-27632 | 5 mM (5000 μM) | 100 μL | 10 μM | APExBio (APExBIO Technology LLC, Houston, TX, USA) | 501146540 | ROCK inhibitor: improves survival of dissociated colon organoid cells after revival or passaging (Note: use for the first 24–48 h post-thaw/passaging). |
| NAC (N-acetyl-L-cysteine) | 0.612 mM (1:1000) | 81.5 μL | 1 mM | Sigma-Aldrich (Darmstadt, Germany) | A9165-5G | Antioxidant that limits oxidative stress during colon PDO expansion. |
| Nicotinamide | 500 mM | 1000 μL | 10 mM | Sigma-Aldrich (Darmstadt, Germany) | N3376 | Promotes the expansion of colon epithelial progenitors; excessive or prolonged use may dampen differentiation. |
| Gastrin I | 6 μM (1:10) | 83.5 μL | 10 nM | Sigma-Aldrich (Darmstadt, Germany) | G9020-250UG | Gastrointestinal peptide that supports colon epithelial growth and enhances secretory lineage balance. |
| Primocin | 50 mg/mL | 100 μL | 100 μg/mL | Invivogen (San Diego, CA, USA) | ant-pm-1 | Antimicrobial to prevent bacterial, fungal, and mycoplasma contamination during colon PDO establishment. |
| Antibiotic/Antimycotic | 100× | 500 μL | 1× | Fisher Scientific (Waltham, MA, USA) | CA002-010 | Reduce contamination risk in colon PDO cultures. |
 CRITICAL STEP: All media should be sterile-filtered and stored at 4 °C for short-term use (up to 1 week); certain factors such as EGF and A83-01 may degrade and should be added fresh or from aliquots stored at −20 °C. For example, EGF is a small, soluble growth factor that quickly loses biological activity under standard culture conditions. It is susceptible to degradation at 37 °C, and the presence of serum and proteins as well as repeated freezing and thawing cycles further reduce its effectiveness. Adding new EGF to each media change guarantees consistent activation of the EGFR–MAPK pathway, reproducible proliferation, and reduced variability in the colon PDO experiments.
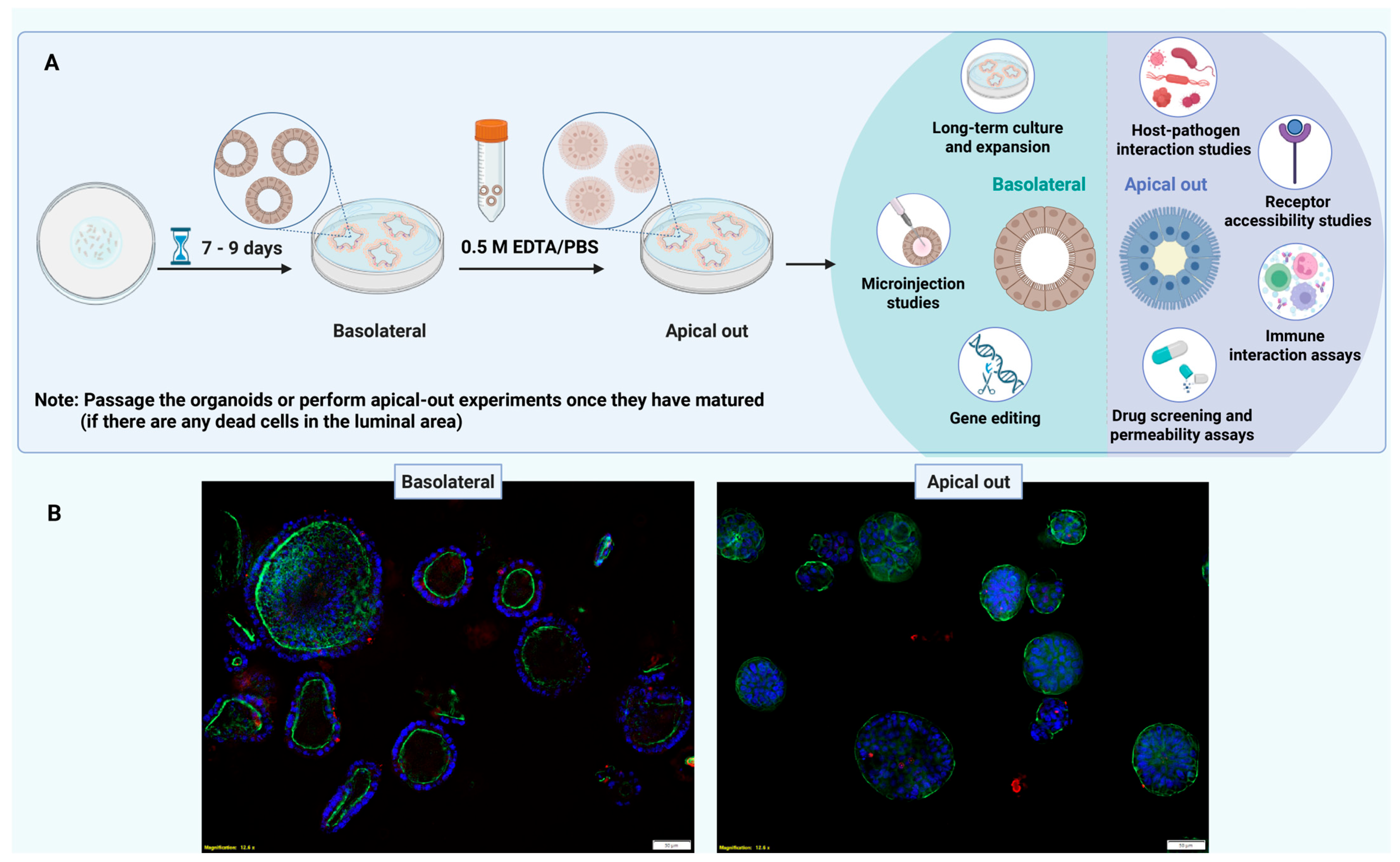
CRITICAL STEP: All media should be sterile-filtered and stored at 4 °C for short-term use (up to 1 week); certain factors such as EGF and A83-01 may degrade and should be added fresh or from aliquots stored at −20 °C. For example, EGF is a small, soluble growth factor that quickly loses biological activity under standard culture conditions. It is susceptible to degradation at 37 °C, and the presence of serum and proteins as well as repeated freezing and thawing cycles further reduce its effectiveness. Adding new EGF to each media change guarantees consistent activation of the EGFR–MAPK pathway, reproducible proliferation, and reduced variability in the colon PDO experiments.9. Apical-Out CRC Organoids
Timeline for Organoid Culture Establishment and Polarity Reversal
10. Immunostaining of Organoids
- Fixation solution:4% paraformaldehyde (PFA)1% Triton X-100 in PBSWorking solution: 1 mL 16% PFA + 2 mL 2% Triton X-100 + 1 mL PBS
- Washing solution:0.1% Triton X-100 in PBSWorking solution: 1 mL 2% Triton X-100 + 19 mL PBS
- Blocking solution:3% BSA + 0.1% Triton X-100 in PBS
- Working solution: 0.3 g BSA + 10 mL PBS + 10 μL Triton X-100
- Dilution buffer (for antibody incubation):1× PBS + 1% BSA + 0.1% Triton X-100Working solution: 0.1 g BSA + 10 mL PBS + 10 μL Triton X-100
Step by Step Organoids Processing Procedure
- Wash organoids (basolateral or apical out) with ice cold PBS.
- Fix and permeabilize simultaneously at 4 °C through exposure with 4% paraformaldehyde and 1% Triton X-100 in PBS for 3 h.
- OPTIONAL STEP: Remove the supernatant and add 1 × PBS and store at 4 °C if you plan to process the sample the next day. If there is concern regarding epitope masking, perform a 30 min fixation with 4% PFA at room temperature followed by permeabilization with 0.1%Triton to minimize this risk [45,46].
- Wash solution (0.1% Triton in PBS): Wash the organoids with 0.1% Triton, flush the organoids three times, and then block overnight with 3% BSA and 0.1% Triton in PBS solution at 4 °C.
- Incubate organoid samples for 24 h at 4 °C with primary antibodies diluted in incubation solution (1 × PBS with 1% BSA and 0.1% Triton X-100).
- Primary antibody incubation (preparation)
- Prepare antibodies in dilution:
600 μL total: 20 μL Phalloidin-iFluor 488 (Abcam, ab176753, Abcam plc, Cambridge, UK) for F-actin, and top up with dilution buffer (PBS + 1% BSA + 0.1% Triton X-100). Incubate organoids for 24 h at 4 °C. Optional: You can also use ZO-1 monoclonal antibody (ZO1-1A12, mouse) instead of Phalloidin-iFluor 488. - Wash the samples 5 times and then expose them to secondary antibodies diluted in incubation solution for 24 h at 4 °C.
- Secondary antibody incubation (preparation)
- Prepare antibodies in dilution buffer:
- 80 μL total: 1 μL DAPI solution (1.0 mg/mL, BD Biosciences, 564907, Becton, Dickinson and Company, Franklin Lakes, NJ, USA), and top up with dilution buffer (1 × PBS + 1% BSA + 0.1% Triton X-100). Note: For unlabeled antibodies, use a fluorescent secondary antibody. For example, for ZO-1 (mouse monoclonal), use donkey anti-Mouse IgG (H+L), Alexa Fluor 546 (Thermo Fisher, A10036, Thermo Fisher Scientific, Waltham, MA, USA).Secondary antibody solution was then removed by flushing with washing solutions 5 times.
- Phalloidin can be added at 1:50 dilution where needed;
- Keep all steps at 4 °C unless otherwise stated;
- Use gentle handling to avoid damaging the organoids;
- Use wide-bore tips to prevent organoid damage;
- Inverted fluorescence microscope settings: Organoids were imaged with an inverted fluorescence microscope (IX-83, Olympus) using 20× long distance objective (LUCPLFLN PH 20 × /0.45). Z stacks were taken in steps of 2 µm and deconvoluted using the constrained iterative process in CellSens Dimensions 3.2 software (Olympus) to remove out-of-focus blur. Images are presented as maximum intensity projection over Z.
11. Organoid Cryopreservation Protocol
- Beginning at passage 2, reserve at least two wells from each passage specifically for cryopreservation.
- Gently scrape the Matrigel domes from the bottom of each selected well using a pipette tip and transfer the contents into a 15 mL conical tube.
- Rinse each well with 500 µL of complete organoid culture medium to collect any remaining organoids and add the rinse to the same tube.
- Add additional organoid culture medium to the tube to bring the total volume to 10 mL.
- Centrifuge the tube at 300× g for 5 min at 4 °C.
- Carefully aspirate the supernatant, leaving a small volume above the Matrigel pellet to avoid disturbing the organoids.
- Prepare a cryopreservation medium consisting of:
- 800 µL of complete organoid culture medium;
- 100 µL of FBS;
- 100 µL of DMSO (note: use 1 mL of this freezing mixture per two wells).
- Gently resuspend the organoid pellet in the cryopreservation medium and transfer 1 mL of the suspension into each cryovial.
- Place the cryovials at −80 °C for a minimum of 24 h. For long-term storage, transfer the vials to liquid nitrogen within a few days.
12. Thawing Human Colon Thawing Organoids
12.1. Materials Required
- Advanced DMEM/F12;
- 15 mL conical centrifuge tubes;
- Human colon organoid medium: 50% L-WRN-conditioned media;
- Corning® Matrigel® GFR;
- 24-well non-treated cell culture plate.
12.2. Sample Processing Procedure
- Remove the cryovial containing frozen colon organoids from liquid nitrogen storage. Immediately place it in a 37 °C water bath and gently swirl until completely thawed (avoid prolonged exposure).
- Transfer the thawed contents into a 15 mL conical tube containing 10 mL of room temperature Advanced DMEM/F12 to dilute the cryoprotectant.
- Centrifuge the tube at 300× g for 5 min at room temperature.
- Carefully aspirate the supernatant without disturbing the pellet.
- Gently resuspend the organoid pellet in chilled Matrigel. Avoid creating bubbles. Plate the Matrigel drop (dome) into the center of a well in a 24-well plate.
 CRITICAL STEP: Typically, the entire thawed vial is plated into one dome, unless it originally contained multiple domes.
CRITICAL STEP: Typically, the entire thawed vial is plated into one dome, unless it originally contained multiple domes.- 6.
- Leave the plate at room temperature (on the bench) for 5 min to allow the Matrigel to settle. Then, transfer the plate to a 37 °C incubator for 30 min to allow for full polymerization.
- 7.
- Once the Matrigel dome has solidified, gently add 750 µL of pre-warmed complete human colon organoid medium to each well.
- 8.
- Monitor organoid recovery over the next few days. Change medium if needed. Passage or clean the dome once organoids are healthy and have expanded; timing depends on the specific organoid line. The inclusion of a ROCK inhibitor during the first 24–48 h after thawing to support organoid survival and we advise refreshing the domes once morphological evidence of recovery is observed.
13. Recovery and Maintenance During Tissue Processing, Collection Medium Collection, and Co-Culture: Use of Antibiotics
- Tissue processing: Antibiotic/antimycotic, normocin, and gentamicin were used to help control bacterial contamination during tissue sample collection and processing.
- Cells recovery and maintenance (first 24–72 h post-thaw or post-passage): G418 and hygromycin antibiotics were used only to maintain the L-WRN cells.
- Washout prior to experiments: The antibiotics should be removed from culture at least 72 h before initiating conditioned medium (CM) collection or performing microbiome and immune co-culture assays. Cells should be washed twice with PBS, transferred to antibiotic-free medium, and the medium renewed daily for 2–3 days before the experiment.
- Conditioned medium collection and co-culture assays: Antibiotics were completely removed during CM production and throughout all of the co-culture experiments. This approach reduces selection pressure, prevents antibiotic carryover into CM, and preserves microbial and immune cell viability and signaling.
14. Bridging 3D Organoids to 2D and Organ-on-Chip Platforms
15. Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| PDOs | Patient-derived organoids |
| iPSC | Induced pluripotent stem cells |
| DMSO | Dimethyl sulfoxide |
| PBS | Phosphate buffered saline |
| AB/AM | Antibiotic antimycotic solution |
| DMEM | Dulbecco’s modified Eagle medium |
| 3D | Three-dimensional |
| MSI-H | Microsatellite instability-high |
| CIMP-H | CpG island methylator phenotype-high |
| PFA | Paraformaldehyde |
References
- Khaled, W.T.; Liu, P. Cancer mouse models: Past, present and future. Semin. Cell Dev. Biol. 2014, 27, 54–60. [Google Scholar] [CrossRef]
- Sajjad, H.; Imtiaz, S.; Noor, T.; Siddiqui, Y.H.; Sajjad, A.; Zia, M. Cancer models in preclinical research: A chronicle review of advancement in effective cancer research. Anim. Model. Exp. Med. 2021, 4, 87–103. [Google Scholar] [CrossRef] [PubMed]
- Stribbling, S.M.; Ryan, A.J. The cell-line-derived subcutaneous tumor model in preclinical cancer research. Nat. Protoc. 2022, 17, 2108–2128. [Google Scholar] [CrossRef]
- Heydari, Z.; Devasahayam Arokia Balaya, R.; Sarkar, G.; Boardman, L. The Role of Organoids in Advancing Colorectal Cancer Research: Insights and Future Directions. Cancers 2025, 17, 2129. [Google Scholar] [CrossRef]
- Roper, J.; Martin, E.S.; Hung, K.E. Overview of genetically engineered mouse models of colorectal carcinoma to enable translational biology and drug development. Curr. Protoc. Pharmacol. 2014, 65, 1–10. [Google Scholar] [CrossRef]
- Shah, S.; D’Souza, G.G.M. Modeling Tumor Microenvironment Complexity In Vitro: Spheroids as Physiologically Relevant Tumor Models and Strategies for Their Analysis. Cells 2025, 14, 732. [Google Scholar] [CrossRef]
- Liu, X.; Zhou, Z.; Zhang, Y.; Zhong, H.; Cai, X.; Guan, R. Recent progress on the organoids: Techniques, advantages and applications. Biomed. Pharmacother. 2025, 185, 117942. [Google Scholar] [CrossRef]
- Papaccio, F.; Cabeza-Segura, M.; Garcia-Mico, B.; Gimeno-Valiente, F.; Zuniga-Trejos, S.; Gambardella, V.; Gutierrez-Bravo, M.F.; Martinez-Ciarpaglini, C.; Rentero-Garrido, P.; Fleitas, T.; et al. Decoding chromosomal instability insights in CRC by integrating omics and patient-derived organoids. J. Exp. Clin. Cancer Res. 2025, 44, 77. [Google Scholar] [CrossRef]
- Papaccio, F.; Cabeza-Segura, M.; Garcia-Mico, B.; Tarazona, N.; Roda, D.; Castillo, J.; Cervantes, A. Will Organoids Fill the Gap towards Functional Precision Medicine? J. Pers. Med. 2022, 12, 1939. [Google Scholar] [CrossRef] [PubMed]
- Vlachogiannis, G.; Hedayat, S.; Vatsiou, A.; Jamin, Y.; Fernandez-Mateos, J.; Khan, K.; Lampis, A.; Eason, K.; Huntingford, I.; Burke, R.; et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 2018, 359, 920–926. [Google Scholar] [CrossRef] [PubMed]
- Clevers, H. The intestinal crypt, a prototype stem cell compartment. Cell 2013, 154, 274–284. [Google Scholar] [CrossRef] [PubMed]
- Corro, C.; Novellasdemunt, L.; Li, V.S.W. A brief history of organoids. Am. J. Physiol. Cell Physiol. 2020, 319, C151–C165. [Google Scholar] [CrossRef] [PubMed]
- Pleguezuelos-Manzano, C.; Puschhof, J.; van den Brink, S.; Geurts, V.; Beumer, J.; Clevers, H. Establishment and Culture of Human Intestinal Organoids Derived from Adult Stem Cells. Curr. Protoc. Immunol. 2020, 130, e106. [Google Scholar] [CrossRef]
- Sato, T.; Clevers, H. Primary mouse small intestinal epithelial cell cultures. Methods Mol. Biol. 2013, 945, 319–328. [Google Scholar]
- van de Wetering, M.; Francies, H.E.; Francis, J.M.; Bounova, G.; Iorio, F.; Pronk, A.; van Houdt, W.; van Gorp, J.; Taylor-Weiner, A.; Kester, L.; et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 2015, 161, 933–945. [Google Scholar] [CrossRef]
- Drost, J.; van Boxtel, R.; Blokzijl, F.; Mizutani, T.; Sasaki, N.; Sasselli, V.; de Ligt, J.; Behjati, S.; Grolleman, J.E.; van Wezel, T.; et al. Use of CRISPR-modified human stem cell organoids to study the origin of mutational signatures in cancer. Science 2017, 358, 234–238. [Google Scholar] [CrossRef] [PubMed]
- Fujii, M.; Matano, M.; Nanki, K.; Sato, T. Author Correction: Efficient genetic engineering of human intestinal organoids using electroporation. Nat. Protoc. 2019, 14, 2595. [Google Scholar] [CrossRef]
- Fujii, M.; Matano, M.; Toshimitsu, K.; Takano, A.; Mikami, Y.; Nishikori, S.; Sugimoto, S.; Sato, T. Human Intestinal Organoids Maintain Self-Renewal Capacity and Cellular Diversity in Niche-Inspired Culture Condition. Cell Stem Cell 2018, 23, 787–793.e6. [Google Scholar] [CrossRef]
- Daoud, A.; Munera, J.O. Generation of human colonic organoids from human pluripotent stem cells. Methods Cell Biol. 2020, 159, 201–227. [Google Scholar]
- Lee, I.T.; Takahashi, Y.; Sasaki, T.; Yamauchi, Y.; Sato, R. Human colon organoid differentiation from induced pluripotent stem cells using an improved method. FEBS Lett. 2025, 599, 912–924. [Google Scholar] [CrossRef]
- Dotti, I.; Mayorgas, A.; Salas, A. Generation of human colon organoids from healthy and inflammatory bowel disease mucosa. PLoS ONE 2022, 17, e0276195. [Google Scholar] [CrossRef]
- Wijesekara, P.; Patel, K.Z.; Otto, E.L.; Campbell, P.G.; Ren, X. Protocol to engineer apical-out airway organoids using suspension culture of human airway basal stem cell aggregates. STAR Protoc. 2023, 4, 102154. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.G.; Lee, B.R.; Lee, P.; Choi, S.; Kim, J.H.; Oh, M.H.; Yoo, J.G. Apical-out intestinal organoids as an alternative model for evaluating deoxynivalenol toxicity and Lactobacillus detoxification in bovine. Sci. Rep. 2024, 14, 31373. [Google Scholar] [CrossRef]
- Co, J.Y.; Margalef-Català, M.; Monack, D.M.; Amieva, M.R. Controlling the polarity of human gastrointestinal organoids to investigate epithelial biology and infectious diseases. Nat. Protoc. 2021, 16, 5171–5192. [Google Scholar] [CrossRef]
- Saorin, G.; Caligiuri, I.; Rizzolio, F. Microfluidic organoids-on-a-chip: The future of human models. Semin. Cell Dev. Biol. 2023, 144, 41–54. [Google Scholar] [CrossRef]
- Hetzel, L.A.; Ali, A.; Corbo, V.; Hankemeier, T. Microfluidics and Organoids, the Power Couple of Developmental Biology and Oncology Studies. Int. J. Mol. Sci. 2023, 24, 10882. [Google Scholar] [CrossRef]
- Yin, Y.; Liu, P.Y.; Shi, Y.; Li, P. Single-Cell Sequencing and Organoids: A Powerful Combination for Modelling Organ Development and Diseases. Rev. Physiol. Biochem. Pharmacol. 2021, 179, 189–210. [Google Scholar]
- Spiller, E.R.; Ung, N.; Kim, S.; Patsch, K.; Lau, R.; Strelez, C.; Doshi, C.; Choung, S.; Choi, B.; Juarez Rosales, E.F.; et al. Imaging-Based Machine Learning Analysis of Patient-Derived Tumor Organoid Drug Response. Front. Oncol. 2021, 11, 771173. [Google Scholar] [CrossRef]
- Kahveci, B.; Polatli, E.; Bastanlar, Y.; Guven, S. OrganoLabeler: A Quick and Accurate Annotation Tool for Organoid Images. ACS Omega 2024, 9, 46117–46128. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Flinders, C.; Mumenthaler, S.M.; Hummon, A.B. MALDI Mass Spectrometry Imaging for Evaluation of Therapeutics in Colorectal Tumor Organoids. J. Am. Soc. Mass. Spectrom. 2018, 29, 516–526. [Google Scholar] [CrossRef] [PubMed]
- Jukes, Z.; Freier, A.; Glymenaki, M.; Brown, R.; Parry, L.; Want, E.; Vorkas, P.A.; Li, J.V. Lipid profiling of mouse intestinal organoids for studying APC mutations. Biosci. Rep. 2021, 41, BSR20202915. [Google Scholar] [CrossRef]
- Zuo, J.; Fang, Y.; Wang, R.; Liang, S. High-throughput solutions in tumor organoids: From culture to drug screening. Stem Cells 2025, 43, sxae070. [Google Scholar] [CrossRef]
- Walocha, R.; Kim, M.; Wong-Ng, J.; Gobaa, S.; Sauvonnet, N. Organoids and organ-on-chip technology for investigating host-microorganism interactions. Microbes Infect. 2024, 26, 105319. [Google Scholar] [CrossRef]
- Sun, S.; Xue, X.; Fu, J. Modeling development using microfluidics: Bridging gaps to foster fundamental and translational research. Curr. Opin. Genet. Dev. 2023, 82, 102097. [Google Scholar] [CrossRef]
- Olawade, D.B.; Oisakede, E.O.; Egbon, E.; Ovsepian, S.V.; Boussios, S. Immune Organoids: A Review of Their Applications in Cancer and Autoimmune Disease Immunotherapy. Curr. Issues Mol. Biol. 2025, 47, 653. [Google Scholar] [CrossRef]
- Gomez, D.; Dalal, Z.; Raw, E.; Roberts, C.; Lyndon, P.J. Anatomical distribution of colorectal cancer over a 10 year period in a district general hospital: Is there a true “rightward shift”? Postgrad. Med. J. 2004, 80, 667–669. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Duan, B.; Zhao, Y.; Bai, J.; Wang, J.; Duan, X.; Luo, X.; Zhang, R.; Pu, Y.; Kou, M.; Lei, J.; et al. Colorectal Cancer: An Overview. In Gastrointestinal Cancers; Morgado-Diaz, J.A., Ed.; JExon Publications: Brisbane, Australia, 2022. [Google Scholar][Green Version]
- Zhao, R.; Ren, S.; Li, C.; Guo, K.; Lu, Z.; Tian, L.; He, J.; Zhang, K.; Cao, Y.; Liu, S.; et al. Biomarkers for pancreatic cancer based on tissue and serum metabolomics analysis in a multicenter study. Cancer Med. 2023, 12, 5158–5171. [Google Scholar] [CrossRef] [PubMed]
- Mauri, G.; Sartore-Bianchi, A.; Russo, A.G.; Marsoni, S.; Bardelli, A.; Siena, S. Early-onset colorectal cancer in young individuals. Mol. Oncol. 2019, 13, 109–131. [Google Scholar] [CrossRef]
- Yang, Q.; Qu, R.; Lu, S.; Zhang, Y.; Zhang, Z.; Fu, W. Biological and Clinical Characteristics of Proximal Colon Cancer: Far from Its Anatomical Subsite. Int. J. Med. Sci. 2024, 21, 1824–1839. [Google Scholar] [CrossRef] [PubMed]
- Murphy, N.; Ward, H.A.; Jenab, M.; Rothwell, J.A.; Boutron-Ruault, M.C.; Carbonnel, F.; Kvaskoff, M.; Kaaks, R.; Kuhn, T.; Boeing, H.; et al. Heterogeneity of Colorectal Cancer Risk Factors by Anatomical Subsite in 10 European Countries: A Multinational Cohort Study. Clin. Gastroenterol. Hepatol. 2019, 17, 1323–1331.e6. [Google Scholar] [CrossRef]
- Wang, L.; Lo, C.H.; He, X.; Hang, D.; Wang, M.; Wu, K.; Chan, A.T.; Ogino, S.; Giovannucci, E.L.; Song, M. Risk Factor Profiles Differ for Cancers of Different Regions of the Colorectum. Gastroenterology 2020, 159, 241–256.e13. [Google Scholar] [CrossRef]
- Munro, M.J.; Tan, S.T.; Gray, C. Applications for colon organoid models in cancer research. Organoids 2023, 2, 37–49. [Google Scholar] [CrossRef]
- Puschhof, J.; Pleguezuelos-Manzano, C.; Martinez-Silgado, A.; Akkerman, N.; Saftien, A.; Boot, C.; de Waal, A.; Beumer, J.; Dutta, D.; Heo, I. Intestinal organoid cocultures with microbes. Nat. Protoc. 2021, 16, 4633–4649. [Google Scholar] [CrossRef]
- Gwon, K.; Choi, D.; de Hoyos-Vega, J.M.; Baskaran, H.; Gonzalez-Suarez, A.M.; Lee, S.; Hong, H.J.; Nguyen, K.M.; Dharmesh, E.; Sugahara, G.; et al. Function of hepatocyte spheroids in bioactive microcapsules is enhanced by endogenous and exogenous hepatocyte growth factor. Bioact. Mater. 2023, 28, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Fattahi, P.; de Hoyos-Vega, J.M.; Choi, J.H.; Duffy, C.D.; Gonzalez-Suarez, A.M.; Ishida, Y.; Nguyen, K.M.; Gwon, K.; Peterson, Q.P.; Saito, T.; et al. Guiding Hepatic Differentiation of Pluripotent Stem Cells Using 3D Microfluidic Co-Cultures with Human Hepatocytes. Cells 2023, 12, 1982. [Google Scholar] [CrossRef] [PubMed]
- Fatehullah, A.; Appleton, P.L.; Nathke, I.S. Cell and tissue polarity in the intestinal tract during tumourigenesis: Cells still know the right way up, but tissue organization is lost. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013, 368, 20130014. [Google Scholar] [CrossRef]
- Co, J.Y.; Margalef-Catala, M.; Li, X.; Mah, A.T.; Kuo, C.J.; Monack, D.M.; Amieva, M.R. Controlling Epithelial Polarity: A Human Enteroid Model for Host-Pathogen Interactions. Cell Rep. 2019, 26, 2509–2520.e4. [Google Scholar] [CrossRef]
- Yamashita, T.; Inui, T.; Yokota, J.; Kawakami, K.; Morinaga, G.; Takatani, M.; Hirayama, D.; Nomoto, R.; Ito, K.; Cui, Y.; et al. Monolayer platform using human biopsy-derived duodenal organoids for pharmaceutical research. Mol. Ther. Methods Clin. Dev. 2021, 22, 263–278. [Google Scholar] [CrossRef]
- de Hoyos-Vega, J.M.; Yu, X.; Gonzalez-Suarez, A.M.; Chen, S.; Mercado-Perez, A.; Krueger, E.; Hernandez, J.; Fedyshyn, Y.; Druliner, B.R.; Linden, D.R.; et al. Modeling gut neuro-epithelial connections in a novel microfluidic device. Microsyst. Nanoeng. 2023, 9, 144. [Google Scholar] [CrossRef]
- Wang, L.; Han, J.; Su, W.; Li, A.; Zhang, W.; Li, H.; Hu, H.; Song, W.; Xu, C.; Chen, J. Gut-on-a-chip for exploring the transport mechanism of Hg(II). Microsyst. Nanoeng. 2023, 9, 2. [Google Scholar] [CrossRef] [PubMed]
- Piacentini, L.; Wainwright, C.L.; Parratt, J.R. Effects of Bordetella pertussis toxin pretreatment on the antiarrhythmic action of ischaemic preconditioning in anaesthetized rats. Br. J. Pharmacol. 1995, 114, 755–760. [Google Scholar] [CrossRef]
- Jelinsky, S.A.; Derksen, M.; Bauman, E.; Verissimo, C.S.; van Dooremalen, W.T.M.; Roos, J.L.; Higuera Baron, C.; Caballero-Franco, C.; Johnson, B.G.; Rooks, M.G.; et al. Molecular and Functional Characterization of Human Intestinal Organoids and Monolayers for Modeling Epithelial Barrier. Inflamm. Bowel Dis. 2023, 29, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Hamdan, F.H.; Perez, I.; Kossick, K.; Smith, H.; Edwinson, A.; de Hoyos-Vega, J.M.; Gonzalez, M.; Chiang, D.; Klatt, E.; Rumer, K.; et al. Intestinal Stem Cells from Patients with Inflammatory Bowel Disease Retain an Epigenetic Memory of Inflammation. bioRxiv 2025. [Google Scholar] [CrossRef]
- Teo, A.J.T.; Ng, S.K.; Khoo, K.; Wong, S.H.; Li, K.H.H. Microfluidic Gastrointestinal Cell Culture Technologies-Improvements in the Past Decade. Biosensors 2024, 14, 449. [Google Scholar] [CrossRef]
- Shin, W.; Ambrosini, Y.M.; Shin, Y.C.; Wu, A.; Min, S.; Koh, D.; Park, S.; Kim, S.; Koh, H.; Kim, H.J. Robust Formation of an Epithelial Layer of Human Intestinal Organoids in a Polydimethylsiloxane-Based Gut-on-a-Chip Microdevice. Front. Med. Technol. 2020, 2, 2. [Google Scholar] [CrossRef]
- Thomas, D.P.; Zhang, J.; Nguyen, N.T.; Ta, H.T. Microfluidic Gut-on-a-Chip: Fundamentals and Challenges. Biosensors 2023, 13, 136. [Google Scholar] [CrossRef]
- Zhang, J.; Hernandez-Gordillo, V.; Trapecar, M.; Wright, C.; Taketani, M.; Schneider, K.; Chen, W.L.K.; Stas, E.; Breault, D.T.; Carrier, R.L.; et al. Coculture of primary human colon monolayer with human gut bacteria. Nat. Protoc. 2021, 16, 3874–3900. [Google Scholar] [CrossRef]
- Angus, H.C.; Urbano, P.C.; Laws, G.A.; Fan, S.; Gadeock, S.; Schultz, M.; Butt, G.; Highton, A.J.; Kemp, R.A. An autologous colonic organoid-derived monolayer model to study immune: Bacterial interactions in Crohn’s disease patients. Clin. Transl. Immunol. 2022, 11, e1407. [Google Scholar] [CrossRef]
- Wang, C.M.; Oberoi, H.S.; Law, D.; Li, Y.; Kassis, T.; Griffith, L.G.; Breault, D.T.; Carrier, R.L. Human Mesofluidic Intestinal Model for Studying Transport of Drug Carriers and Bacteria Through a Live Mucosal Barrier. bioRxiv 2024. [Google Scholar] [CrossRef] [PubMed]
- Shin, Y.C.; Shin, W.; Koh, D.; Wu, A.; Ambrosini, Y.M.; Min, S.; Eckhardt, S.G.; Fleming, R.Y.D.; Kim, S.; Park, S.; et al. Three-Dimensional Regeneration of Patient-Derived Intestinal Organoid Epithelium in a Physiodynamic Mucosal Interface-on-a-Chip. Micromachines 2020, 11, 663. [Google Scholar] [CrossRef] [PubMed]
- Cenhrang, K.; Robart, L.; Castiaux, A.D.; Martin, R.S. 3D printed devices with integrated collagen scaffolds for cell culture studies including transepithelial/transendothelial electrical resistance (TEER) measurements. Anal. Chim. Acta 2022, 1221, 340166. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.B.; Hu, L.S.; Huang, W.J.; Zhou, Z.Z.; Luo, H.Y.; Tian, X.P. Comparative investigation of neoadjuvant immunotherapy versus adjuvant immunotherapy in perioperative patients with cancer: A global-scale, cross-sectional, and large-sample informatics study. Int. J. Surg. 2024, 110, 4660–4671. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Devasahayam Arokia Balaya, R.; Heydari, Z.; Sarkar, G.; Cruz Garcia, E.M.; de Hoyos-Vega, J.M.; Krueger, E.; Helgeson, L.; Revzin, A.; Ros, A.; Pandey, A.; et al. A Practical Guide to Developing and Troubleshooting Patient-Derived “Mini-Gut” Colorectal Organoids for Clinical Research. Methods Protoc. 2025, 8, 121. https://doi.org/10.3390/mps8050121
Devasahayam Arokia Balaya R, Heydari Z, Sarkar G, Cruz Garcia EM, de Hoyos-Vega JM, Krueger E, Helgeson L, Revzin A, Ros A, Pandey A, et al. A Practical Guide to Developing and Troubleshooting Patient-Derived “Mini-Gut” Colorectal Organoids for Clinical Research. Methods and Protocols. 2025; 8(5):121. https://doi.org/10.3390/mps8050121
Chicago/Turabian StyleDevasahayam Arokia Balaya, Rex, Zahra Heydari, Gobinda Sarkar, Estela Mariel Cruz Garcia, Jose M. de Hoyos-Vega, Eugene Krueger, Lauren Helgeson, Alexander Revzin, Alexandra Ros, Akhilesh Pandey, and et al. 2025. "A Practical Guide to Developing and Troubleshooting Patient-Derived “Mini-Gut” Colorectal Organoids for Clinical Research" Methods and Protocols 8, no. 5: 121. https://doi.org/10.3390/mps8050121
APA StyleDevasahayam Arokia Balaya, R., Heydari, Z., Sarkar, G., Cruz Garcia, E. M., de Hoyos-Vega, J. M., Krueger, E., Helgeson, L., Revzin, A., Ros, A., Pandey, A., & Boardman, L. (2025). A Practical Guide to Developing and Troubleshooting Patient-Derived “Mini-Gut” Colorectal Organoids for Clinical Research. Methods and Protocols, 8(5), 121. https://doi.org/10.3390/mps8050121








