Molecular Tools for Lynx spp. qPCR Identification and STR-Based Individual Identification of Eurasian Lynx (Lynx lynx) in Forensic Casework
Abstract
1. Introduction
2. Materials Studied, Methods, Techniques
2.1. Specimens Used for the Analyses
2.2. Quantification System and Species Identification
2.3. STR System for Individual Identification
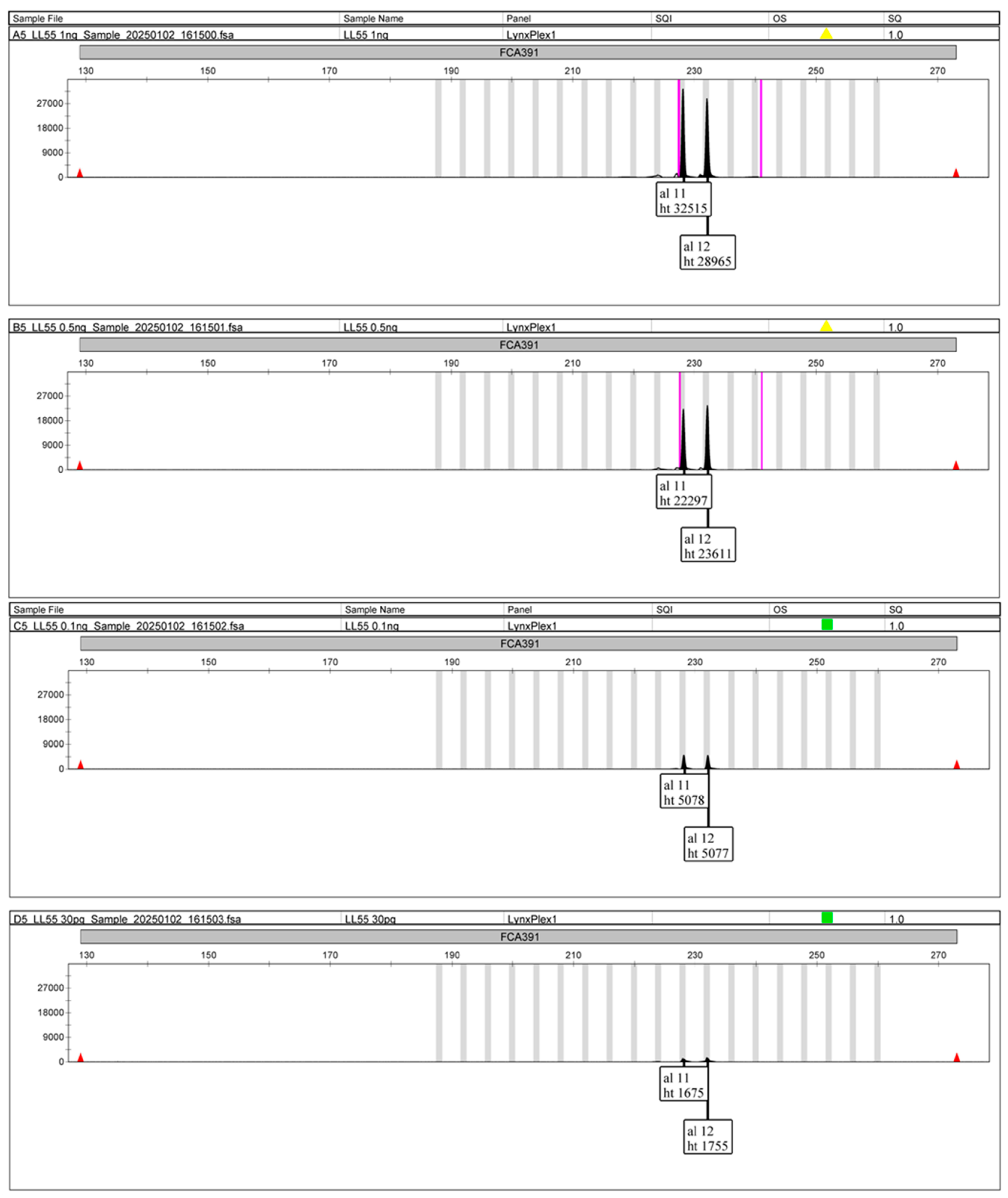
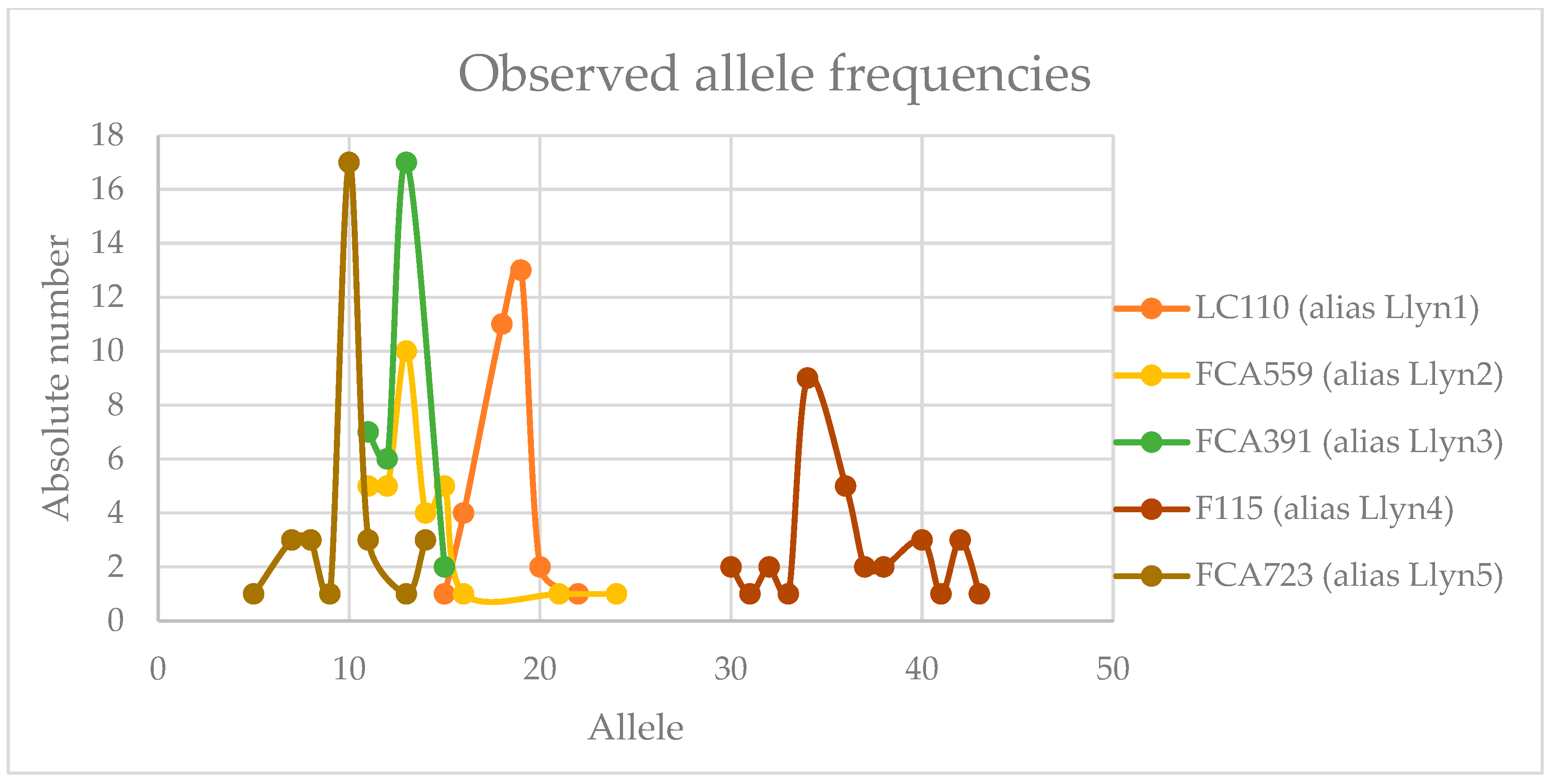
3. Results
4. Validation
4.1. Llynx Qplex
4.2. Llyn STRPlex
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wilson-Wilde, L. Wildlife Crime: A Global Problem. Forensic Sci. Med. Pathol. 2010, 6, 221–222. [Google Scholar] [CrossRef] [PubMed]
- Moreto, W.D.; Van Uhm, D.P. Nested Complex Crime: Assessing the Convergence of Wildlife Trafficking, Organized Crime and Loose Criminal Networks. Br. J. Criminol. 2021, 61, 1334–1353. [Google Scholar] [CrossRef]
- Zimmerman, M.E. The Black Market for Wildlife: Combating Transnational Organized Crime in the Illegal Wildlife Trade. Vanderbilt J. Transnatl. Law 2003, 36, 1656–1689. [Google Scholar]
- Bartlett, S.E.; Davidson, W.S. Identification of Thunnus Tuna Species by the Polymerase Chain Reaction and Direct Sequence Analysis of Their Mitochondrial Cytochrome b Genes. Can. J. Fish. Aquat. Sci. 1991, 48, 309–317. [Google Scholar] [CrossRef]
- Leeton, P.; Christidis, L.; Westerman, M. Feathers from Museum Bird Skins: A Good Source of DNA for Phylogenetic Studies. Condor 1993, 95, 465. [Google Scholar] [CrossRef]
- Savolainen, V.; Cuénoud, P.; Spichiger, R.; Martinez, M.D.P.; Crèvecoeur, M.; Manen, J.-F. The Use of Herbarium Specimens in DNA Phylogenetics: Evaluation and Improvement. Plant Syst. Evol. 1995, 197, 87–98. [Google Scholar] [CrossRef]
- Spooner, D.M.; Anderson, G.J.; Jansen, R.K. Chloroplast DNA Evidence for the Interrelationships of Tomtoes, Potatoes, and Pepinos (Solanaceae). Am. J. Bot. 1993, 80, 676–678. [Google Scholar] [CrossRef]
- Remsen, J.V. The Importance of Continued Collecting of Bird Specimens to Ornithology and Bird Conservation. Bird Conserv. Int. 1995, 5, 146–180. [Google Scholar] [CrossRef]
- Ellegren, H. Polymerase-Chain-Reaction (PCR) Analysis of Microsatellites: A New Approach to Studies of Genetic Relationships in Birds. Auk 1992, 109, 886–895. [Google Scholar] [CrossRef]
- Primmer, C.R.; Møller, A.P.; Ellegren, H. Resolving Genetic Relationships with Microsatellite Markers: A Parentage Testing System for the Swallow Hirundo rustica. Mol. Ecol. 1995, 4, 493–498. [Google Scholar] [CrossRef]
- Edwards, S.V.; Grahn, M.; Potts, W.K. Dynamics of Mhc Evolution in Birds and Crocodilians: Amplification of Class II Genes with Degenerate Primers. Mol. Ecol. 1995, 4, 719–730. [Google Scholar] [CrossRef] [PubMed]
- Blank, R.J.; Huss, V.A.R. DNA Divergency and Speciation In Symbiodinium (Dinophyceae). Plant Syst. Evol. 1989, 163, 153–163. [Google Scholar] [CrossRef]
- Avise, J.C.; Bowen, B.W.; Lamb, T.; Meylan, A.B.; Bermingham, E. Mitochondrial DNA Evolution at a Turtle’s Pace: Evidence for Low Genetic Variability and Reduced Microevolutionary Rate in the Testudines. Mol. Biol. Evol. 1992, 9, 457–473. [Google Scholar] [CrossRef]
- MacFadden, B.J. Fossil Horses: Systematics, Paleobiology, and Evolution of the Family Equidae; Cambridge University Press: Cambridge, UK, 1994. [Google Scholar]
- Higuchi, R.G.; Wrischnik, L.A.; Oakes, E.; George, M.; Tong, B.; Wilson, A.C. Mitochondrial DNA of the Extinct Quagga: Relatedness and Extent of Postmortem Change. J. Mol. Evol. 1987, 25, 283–287. [Google Scholar] [CrossRef] [PubMed]
- Guglich, E.; Wilson, P.; White, B. Forensic Application of Repetitive DNA Markers to the Species Identification of Animal Tissues. J. Forensic Sci. 1994, 39, 353–361. [Google Scholar] [CrossRef]
- Cronin, M.A.; Palmisciano, D.A.; Vyse, E.R.; Cameron, D.G. Mitochondrial DNA in Wildlife Forensic Science: Species Identification of Tissues. Wildl. Soc. Bull. 1991, 19, 94–105. [Google Scholar]
- Alford, R.L.; Caskey, C.T. DNA Analysis in Forensics, Disease and Animal/Plant Identification. Curr. Opin. Biotechnol. 1994, 5, 29–33. [Google Scholar] [CrossRef]
- Hermans, I.F.; Atkinson, J.; Hamilton, J.F.; Chambers, G.K. Three Cases of Disputed Paternity in Dogs Resolved by the Use of DNA Fingerprinting. N. Z. Vet. J. 1991, 39, 61–64. [Google Scholar] [CrossRef]
- Yoon, C.K. Botanical Witness for the Prosecution. Science 1993, 260, 894–895. [Google Scholar] [CrossRef]
- Marklund, S.; Sandberg, K.; Andersson, L. Forensic Tracing of Horse Identities Using Urine Samples and DNA Markers. Anim. Biotechnol. 1996, 7, 145–153. [Google Scholar] [CrossRef]
- Dayton, M.; Koskinen, M.T.; Tom, B.K.; Mattila, A.-M.; Johnston, E.; Halverson, J.; Fantin, D.; DeNise, S.; Budowle, B.; Smith, D.G.; et al. Developmental Validation of Short Tandem Repeat Reagent Kit for Forensic DNA Profiling of Canine Biological Materials. Croat. Med. J. 2009, 50, 268–285. [Google Scholar] [CrossRef] [PubMed]
- Butler, J.M.; David, V.A.; O’Brien, S.J.; Menotti-Raymond, M. The MeowPlex: A New DNA Test Using Tetranucleotide STR Markers for the Domestic Cat. Profiles DNA 2002, 5, 7. [Google Scholar]
- Imaizumi, K.; Akutsu, T.; Miyasaka, S.; Yoshino, M. Development of Species Identification Tests Targeting the 16S Ribosomal RNA Coding Region in Mitochondrial DNA. Int. J. Leg. Med. 2007, 121, 184–191. [Google Scholar] [CrossRef]
- Budowle, B.; Garofano, P.; Hellman, A.; Ketchum, M.; Kanthaswamy, S.; Parson, W.; van Haeringen, W.; Fain, S.; Broad, T. Recommendations for Animal DNA Forensic and Identity Testing. Int. J. Leg. Med. 2005, 119, 295–302. [Google Scholar] [CrossRef]
- Linacre, A.; Gusmão, L.; Hecht, W.; Hellmann, A.P.; Mayr, W.R.; Parson, W.; Prinz, M.; Schneider, P.M.; Morling, N. ISFG: Recommendations Regarding the Use of Non-Human (Animal) DNA in Forensic Genetic Investigations. Forensic Sci. Int. Genet. 2011, 5, 501–505. [Google Scholar] [CrossRef] [PubMed]
- Harper, C.K. RhODIS® (The Rhinoceros DNA Index System): The Application of Simple Forensic and Genetic Tools Help Conserve African Rhinoceros. In Wildlife Biodiversity Conservation; Springer International Publishing: Cham, Switzerland, 2021; pp. 463–485. [Google Scholar]
- Harper, C.K.; Vermeulen, G.J.; Clarke, A.B.; de Wet, J.I.; Guthrie, A.J. Extraction of Nuclear DNA from Rhinoceros Horn and Characterization of DNA Profiling Systems for White (Ceratotherium simum) and Black (Diceros bicornis) Rhinoceros. Forensic Sci. Int. Genet. 2013, 7, 428–433. [Google Scholar] [CrossRef]
- Wasser, S.K.; Mailand, C.; Booth, R.; Mutayoba, B.; Kisamo, E.; Clark, B.; Stephens, M. Using DNA to Track the Origin of the Largest Ivory Seizure since the 1989 Trade Ban. Proc. Natl. Acad. Sci. USA 2007, 104, 4228–4233. [Google Scholar] [CrossRef]
- Wasser, S.K.; Brown, L.; Mailand, C.; Mondol, S.; Clark, W.; Laurie, C.; Weir, B.S. Genetic Assignment of Large Seizures of Elephant Ivory Reveals Africa’s Major Poaching Hotspots. Science 2015, 349, 84–87. [Google Scholar] [CrossRef]
- Wasser, S.K.; Clark, W.J.; Drori, O.; Kisamo, S.E.; Mailand, C.; Mutayoba, B.; Stephens, M. Combating the Illegal Trade in African Elephant Ivory with DNA Forensics. Conserv. Biol. 2008, 22, 1065–1071. [Google Scholar] [CrossRef]
- Wasser, S.K.; Wolock, C.J.; Kuhner, M.K.; Brown, J.E.; Morris, C.; Horwitz, R.J.; Wong, A.; Fernandez, C.J.; Otiende, M.Y.; Hoareau, Y.; et al. Elephant Genotypes Reveal the Size and Connectivity of Transnational Ivory Traffickers. Nat. Hum. Behav. 2022, 6, 371–382. [Google Scholar] [CrossRef]
- Vankova, L.; Vanek, D. DNA-Based Identification of Big Cats and Traditional Chinese Medicine Artifacts in the Czech Republic. Forensic Sci. Int. Genet. Suppl. Ser. 2022, 8, 122–124. [Google Scholar] [CrossRef]
- Vankova, L.; Vanek, D. Capillary-Electrophoresis-Based Species Barcoding of Big Cats: CR-MtDNA-Length Polymorphism. Life 2024, 14, 497. [Google Scholar] [CrossRef]
- Vaněk, D.; Ehler, E.; Vaňková, L. Technical Note: Development of DNA Quantitation and STR Typing Systems for Panthera Tigris Species Determination and Individual Identification in Forensic Casework. Eur. J. Environ. Sci. 2021, 11, 113–118. [Google Scholar] [CrossRef]
- Hebenstreitova, K.; Salaba, O.; Trubac, J.; Kufnerova, J.; Vanek, D. The Influence of Tanning Chemical Agents on DNA Degradation: A Robust Procedure for the Analysis of Tanned Animal Hide—A Pilot Study. Life 2024, 14, 147. [Google Scholar] [CrossRef] [PubMed]
- Morf, N.V.; Kopps, A.M.; Nater, A.; Lendvay, B.; Vasiljevic, N.; Webster, L.M.I.; Fautley, R.G.; Ogden, R.; Kratzer, A. STRoe Deer: A Validated Forensic STR Profiling System for the European Roe Deer (Capreolus capreolus). Forensic Sci. Int. Anim. Environ. 2021, 1, 100023. [Google Scholar] [CrossRef]
- Meredith, E.P.; Adkins, J.K.; Rodzen, J.A. UrsaPlex: An STR Multiplex for Forensic Identification of North American Black Bear (Ursus americanus). Forensic Sci. Int. Genet. 2020, 44, 102161. [Google Scholar] [CrossRef]
- Friedenberger, A.; Doyle, C.; Couillard, L.; Kyle, C.J. The Bear Necessities: A Sensitive QPCR Assay for Bear DNA Detection from Bile and Derived Products to Complement Wildlife Forensic Enforcement. Forensic Sci. Int. Genet. 2023, 67, 102935. [Google Scholar] [CrossRef]
- Hrebianchuk, A.E.; Parfionava, N.S.; Zabauskaya, T.V.; Tsybovsky, I.S. A Panel of Tetranucleotide STR Markers as an Alternative Approach to Forensic DNA Identification of Wolf and Dog. Anim. Genet. 2024, 55, 440–451. [Google Scholar] [CrossRef]
- Berger, B.; Berger, C.; Hecht, W.; Hellmann, A.; Rohleder, U.; Schleenbecker, U.; Parson, W. Validation of Two Canine STR Multiplex-Assays Following the ISFG Recommendations for Non-Human DNA Analysis. Forensic Sci. Int. Genet. 2014, 8, 90–100. [Google Scholar] [CrossRef]
- Selkoe, K.A.; Toonen, R.J. Microsatellites for Ecologists: A Practical Guide to Using and Evaluating Microsatellite Markers. Ecol. Lett. 2006, 9, 615–629. [Google Scholar] [CrossRef]
- Rueness, E.K.; Jorde, P.E.; Hellborg, L.; Stenseth, N.C.; Ellegren, H.; Jakobsen, K.S. Cryptic Population Structure in a Large, Mobile Mammalian Predator: The Scandinavian Lynx. Mol. Ecol. 2003, 12, 2623–2633. [Google Scholar] [CrossRef]
- Herrero, A.; Klütsch, C.F.C.; Holmala, K.; Maduna, S.N.; Kopatz, A.; Eiken, H.G.; Hagen, S.B. Genetic Analysis Indicates Spatial-Dependent Patterns of Sex-Biased Dispersal in Eurasian Lynx in Finland. PLoS ONE 2021, 16, e0246833. [Google Scholar] [CrossRef]
- Carmichael, L.E.; Clark, W.; Strobeck, C. Development and Characterization of Microsatellite Loci from Lynx (Lynx canadensis), and Their Use in Other Felids. Mol. Ecol. 2000, 9, 2197–2199. [Google Scholar] [CrossRef] [PubMed]
- Krojerová-Prokešová, J.; Turbaková, B.; Jelenčič, M.; Bojda, M.; Kutal, M.; Skrbinšek, T.; Koubek, P.; Bryja, J. Genetic Constraints of Population Expansion of the Carpathian Lynx at the Western Edge of Its Native Distribution Range in Central Europe. Heredity 2019, 122, 785–799. [Google Scholar] [CrossRef] [PubMed]
- Gajdárová, B.; Belotti, E.; Bufka, L.; Volfová, J.; Wölfl, S.; Mináriková, T.; Hollerbach, L.; Duľa, M.; Kleven, O.; Kutal, M.; et al. Long-Term Genetic Monitoring of a Reintroduced Eurasian Lynx Population Does Not Indicate an Ongoing Loss of Genetic Diversity. Glob. Ecol. Conserv. 2023, 42, e02399. [Google Scholar] [CrossRef]
- Janečka, J.E.; Blankenship, T.L.; Hirth, D.H.; Tewes, M.E.; Kilpatrick, C.W.; Grassman, L.I. Kinship and Social Structure of Bobcats (Lynx rufus) Inferred from Microsatellite and Radio-telemetry Data. J. Zool. 2006, 269, 494–501. [Google Scholar] [CrossRef]
- Palormes, F.; Godoy, J.A.; López-Bao, J.V.; Rodríguez, A.; Roques, S.; Casas-Marce, M.; Revilla, E.; Delibes, M. Possible Extinction Vortex for a Population of Iberian Lynx on the Verge of Extirpation. Conserv. Biol. 2012, 26, 689–697. [Google Scholar] [CrossRef]
- Jun, J.; Han, S.H.; Jeong, T.-J.; Park, H.C.; Lee, B.; Kwak, M. Wildlife Forensics Using Mitochondrial DNA Sequences: Species Identification Based on Hairs Collected in the Field and Confiscated Tanned Felidae Leathers. Genes Genom. 2011, 33, 721–726. [Google Scholar] [CrossRef]
- Waits, L.P.; Paetkau, D. Non-Invasive Genetic Sampling Tools for Wildlife Biologists: A Review of Applications and Recommendations for Accurate Data Collection. J. Wildl. Manag. 2005, 69, 1419–1433. [Google Scholar] [CrossRef]
- Simbolo, M.; Gottardi, M.; Corbo, V.; Fassan, M.; Mafficini, A.; Malpeli, G.; Lawlor, R.T.; Scarpa, A. DNA Qualification Workflow for Next Generation Sequencing of Histopathological Samples. PLoS ONE 2013, 8, e62692. [Google Scholar] [CrossRef]
- Lee, S.B.; McCord, B.; Buel, E. Advances in Forensic DNA Quantification: A Review. Electrophoresis 2014, 35, 3044–3052. [Google Scholar] [CrossRef]
- Červený, J.; Koubek, P.; Bufka, L. Eurasian Lynx (Lynx lynx) and Its Chance for Survival in Central Europe: The Case of the Czech Republic. Acta Zool. Litu. 2002, 12, 428–432. [Google Scholar] [CrossRef]
- Schmidt, K.; Ratkiewicz, M.; Konopinski, M.K. The Importance of Genetic Variability and Population Differentiation in the Eurasian Lynx Lynx lynx for Conservation, in the Context of Habitat and Climate Change. Mammal Rev. 2011, 41, 112–124. [Google Scholar] [CrossRef]
- Sommer, R.S.; Benecke, N. Late Pleistocene and Holocene Development of the Felid Fauna (Felidae) of Europe: A Review. J. Zool. 2006, 269, 7–19. [Google Scholar] [CrossRef]
- Arlettaz, R.; Chapron, G.; Kéry, M.; Klaus, E.; Mettaz, S.; Roder, S.; Vignali, S.; Zimmermann, F.; Braunisch, V. Poaching Threatens the Establishment of a Lynx Population, Highlighting the Need for a Centralized Judiciary Approach. Front. Conserv. Sci. 2021, 2, 665000. [Google Scholar] [CrossRef]
- Menotti-Raymond, M.; David, V.A.; Lyons, L.A.; Schäffer, A.A.; Tomlin, J.F.; Hutton, M.K.; O’Brien, S.J. A Genetic Linkage Map of Microsatellites in the Domestic Cat (Felis catus). Genomics 1999, 57, 9–23. [Google Scholar] [CrossRef]
- Pilgrim, K.L.; McKeley, K.S.; Riddle, A.E.; Schwartz, M.K. Felid Sex Identification Based on Noninvasive Genetic Samples. Mol. Ecol. Notes 2005, 5, 60–61. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A Tool to Design Target-Specific Primers for Polymerase Chain Reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef]
- Ng, K.K.S.; Lee, S.L.; Tnah, L.H.; Nurul-Farhanah, Z.; Ng, C.H.; Lee, C.T.; Tani, N.; Diway, B.; Lai, P.S.; Khoo, E. Forensic Timber Identification: A Case Study of a CITES Listed Species, Gonystylus bancanus (Thymelaeaceae). Forensic Sci. Int. Genet. 2016, 23, 197–209. [Google Scholar] [CrossRef]
- Potoczniak, M.J.; Chermak, M.; Quarino, L.; Tobe, S.S.; Conte, J. Development of a Multiplex, PCR-Based Genotyping Assay for African and Asian Elephants for Forensic Purposes. Int. J. Leg. Med. 2020, 134, 55–62. [Google Scholar] [CrossRef]
- Gill, P.; Whitaker, J.; Flaxman, C.; Brown, N.; Buckleton, J. An Investigation of the Rigor of Interpretation Rules for STRs Derived from Less than 100 Pg of DNA. Forensic Sci. Int. 2000, 112, 17–40. [Google Scholar] [CrossRef] [PubMed]
- Schrader, C.; Schielke, A.; Ellerbroek, L.; Johne, R. PCR Inhibitors—Occurrence, Properties and Removal. J. Appl. Microbiol. 2012, 113, 1014–1026. [Google Scholar] [CrossRef] [PubMed]
- Sidstedt, M.; Jansson, L.; Nilsson, E.; Noppa, L.; Forsman, M.; Rådström, P.; Hedman, J. Humic Substances Cause Fluorescence Inhibition in Real-Time Polymerase Chain Reaction. Anal. Biochem. 2015, 487, 30–37. [Google Scholar] [CrossRef]
- Sidstedt, M.; Rådström, P.; Hedman, J. PCR Inhibition in QPCR, DPCR and MPS—Mechanisms and Solutions. Anal. Bioanal. Chem. 2020, 412, 2009–2023. [Google Scholar] [CrossRef]
- Polner, M.; Moell, D. Interagency Collaboration and Combating Wildlife Crime. In Environmental Crime and Collaborative State Intervention; Palgrave Macmillan: London, UK, 2016; pp. 59–75. [Google Scholar]
- Van Asch, E. Exploring the Effectiveness of International Cooperation to Combat Transnational Organized Wildlife Crime: Lessons Learned from Initiatives in Asia. Ph.D. Thesis, University of Sheffield, Sheffield, UK, 2017. [Google Scholar]
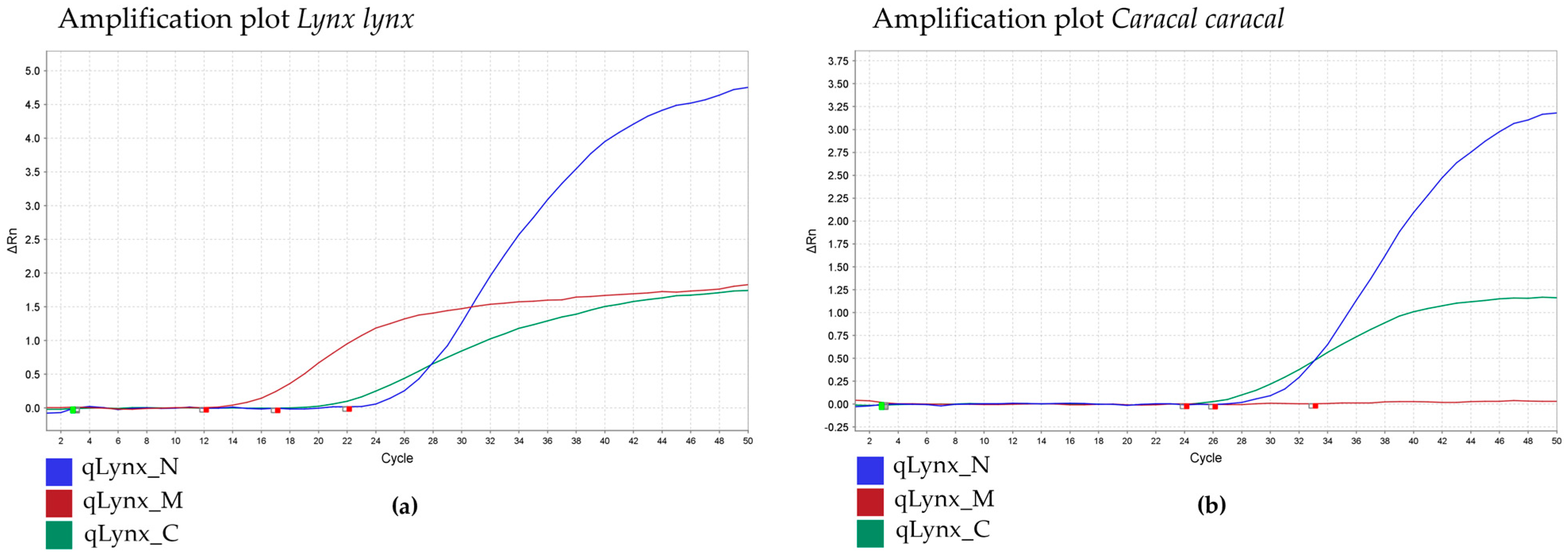
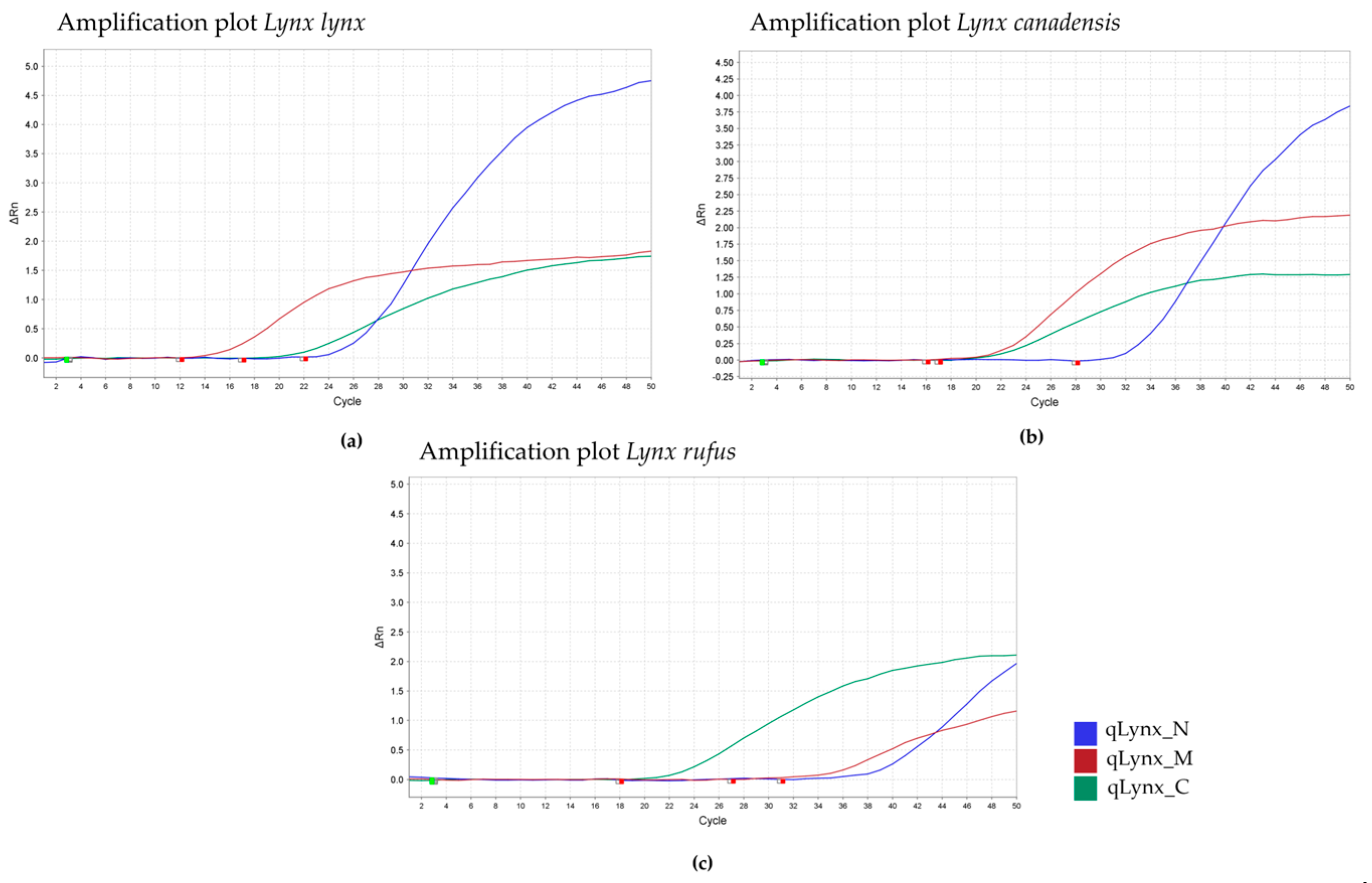
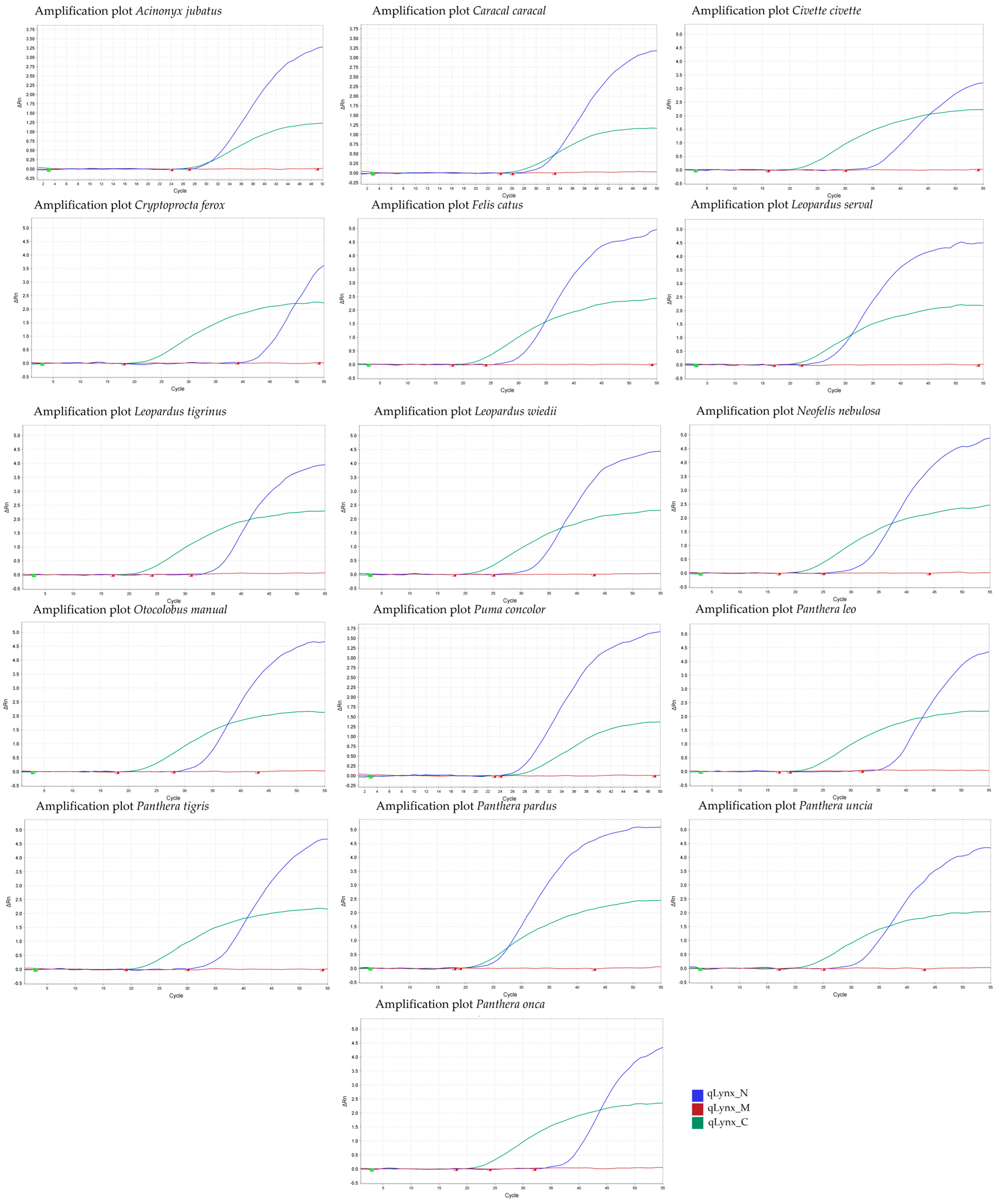
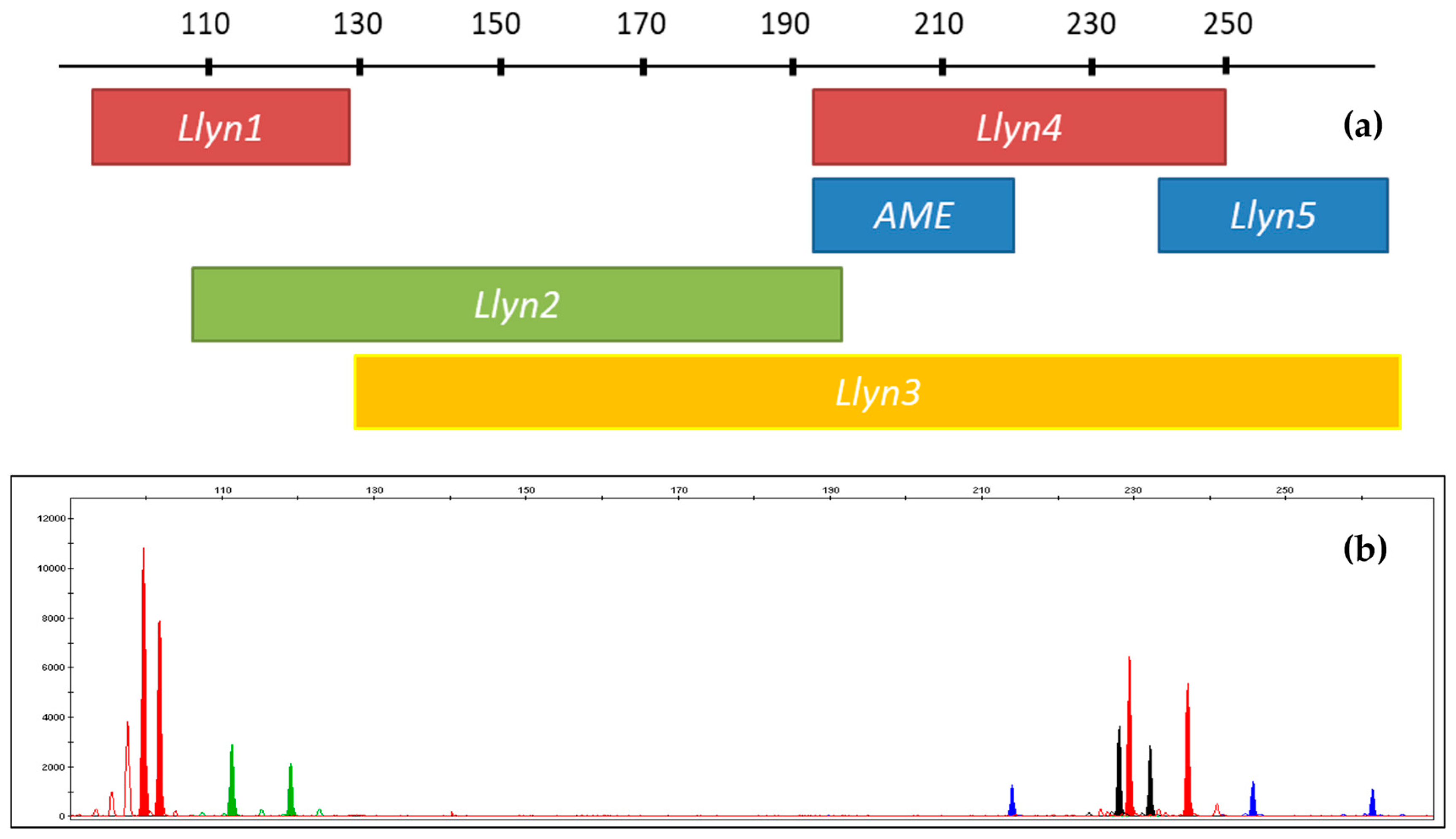
| Primer/Probe Name | Final Concentration (mM) | Sequence (5′-3′) | PCR Product Size (bp = Base Pairs) | Specificity | TaqMan Probe Fluorescent Label |
|---|---|---|---|---|---|
| qLynxM_F | 5 | GTCCCCTTCCACCCATACTAT | 139 bp | Cyt b (mtDNA) | --- |
| qLynxM_R | 5 | ACTTAGGGGGTTAGCGGGGATATAA | --- | ||
| qLynxM_probe | 1.7 | CTCACCAGACCTGTTAGGA | probe | VIC | |
| qLynxC_F | 5 | CTGCTAGGTTTAGCGCGTGAC | 261 bp | IPC | --- |
| qLynxC_R | 5 | GGGGACCATGCTTGCG | --- | ||
| qLynx_probe | 1.7 | TGCACGATTCAAGCACGAT | probe | NED | |
| qLynxN_F | 3.3 | AGTCCACTTCTCATTGCCCCTT | 132 bp | PLP (nDNA) | --- |
| qLynxN_R | 3.3 | ACCTTCCCTGAGTTCTCCATACC | --- | ||
| qLynxN_probe | 1.7 | CTCACCAGACCTGTTAGGA | probe | 6-FAM |
| STR Marker | Label | Size (bp) | Repeat Motif | Primer Design | Primer Concentration (µM) |
|---|---|---|---|---|---|
| Amelogenin | FAM | 193; 214 | [59] | 2.75 | |
| Lc110 (alias Llyn1) | ATTO565 | 91–120 | (T)3(GT)14 | [45] | 0.75 |
| FCA559 (alias Llyn2) | YAKYE | 100–195 | (GAAA)n | [58] | 2.75 |
| FCA391 (alias Llyn3) | ATTO550 | 129–273 | (GATA)n | [58] | 1 |
| F115 (alias Llyn4) | ATTO565 | 193–250 | (GAA)n | [58] | 1.25 |
| FCA723 (alias Llyn5) | FAM | 243–317 | (AAAG)n | [58] | 0.75 |
| Species | Sample | Amelogenin | Lc110 (Alias Llyn1) | FCA559 (Alias Llyn2) | FCA391 (Alias Llyn3) | F115 (Alias Llyn4) | FCA723 (Alias Llyn5) |
|---|---|---|---|---|---|---|---|
| Lynx lynx | LL57 | F | 18,19 | 14,14 | 13,13 | 34,36 | 10,10 |
| Lynx lynx | LL56 | M | 19,19 | 12,13 | 13,13 | 34,42 | 10,11 |
| Lynx lynx | LL40 | F | 19,20 | 12,12 | 12,13 | 40,42 | 10,13 |
| Lynx lynx | LL41 | M | 19,19 | 15,15 | 13,13 | 30,34 | 8,11 |
| Lynx lynx | LL55 | F | 18,18 | 11,13 | 11,12 | 33,40 | 8,10 |
| Lynx lynx | LL61 | M | 18,22 | 11,11 | 11,13 | 36,37 | 10,14 |
| Lynx lynx | LL54 | M | 16,20 | 11,14 | 13,13 | 34,36 | 8,11 |
| Lynx lynx | LL62 | M | 18,19 | 12,13 | 12,15 | 42,43 | 5,7 |
| Lynx lynx | LL147 | F | 18,19 | 13,15 | 11,13 | 34,37 | 10,10 |
| Lynx lynx | LL65 | M | 18,19 | 11,13 | 11,12 | 32,40 | 7,10 |
| Lynx lynx | LL67 | M | 18,19 | 13,13 | 13,15 | 32,34 | 9,10 |
| Lynx lynx | LL68 | M | 18,19 | 13,15 | 11,12 | 34,38 | 10,14 |
| Lynx lynx | LL148 | F | 15,16 | 13,14 | 13,13 | 30,36 | 10,10 |
| Lynx lynx | LL149 | F | 16,19 | 12,16 | 13,13 | 34,36 | 10,10 |
| Lynx lynx | LL63 | M | 18,19 | 13,15 | 11,12 | 34,38 | 10,14 |
| Lynx canadensis | LC52 | M | 16,18 | 21, 24 | 11,13 | 31,41 | 7,10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mahlerová, K.; Alaverdyan, J.; Vaňková, L.; Vaněk, D. Molecular Tools for Lynx spp. qPCR Identification and STR-Based Individual Identification of Eurasian Lynx (Lynx lynx) in Forensic Casework. Methods Protoc. 2025, 8, 47. https://doi.org/10.3390/mps8030047
Mahlerová K, Alaverdyan J, Vaňková L, Vaněk D. Molecular Tools for Lynx spp. qPCR Identification and STR-Based Individual Identification of Eurasian Lynx (Lynx lynx) in Forensic Casework. Methods and Protocols. 2025; 8(3):47. https://doi.org/10.3390/mps8030047
Chicago/Turabian StyleMahlerová, Karolina, Johana Alaverdyan, Lenka Vaňková, and Daniel Vaněk. 2025. "Molecular Tools for Lynx spp. qPCR Identification and STR-Based Individual Identification of Eurasian Lynx (Lynx lynx) in Forensic Casework" Methods and Protocols 8, no. 3: 47. https://doi.org/10.3390/mps8030047
APA StyleMahlerová, K., Alaverdyan, J., Vaňková, L., & Vaněk, D. (2025). Molecular Tools for Lynx spp. qPCR Identification and STR-Based Individual Identification of Eurasian Lynx (Lynx lynx) in Forensic Casework. Methods and Protocols, 8(3), 47. https://doi.org/10.3390/mps8030047






