A Versatile Protocol for Efficient Transformation and Regeneration in Mega Indica Rice Cultivar MTU1010: Optimization through Hormonal Variables
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Procedure
2.2.1. Surface Sterilization of Mature Rice Seeds
2.2.2. Optimization of Efficient Callus Induction and Regeneration in Rice cv. MTU1010
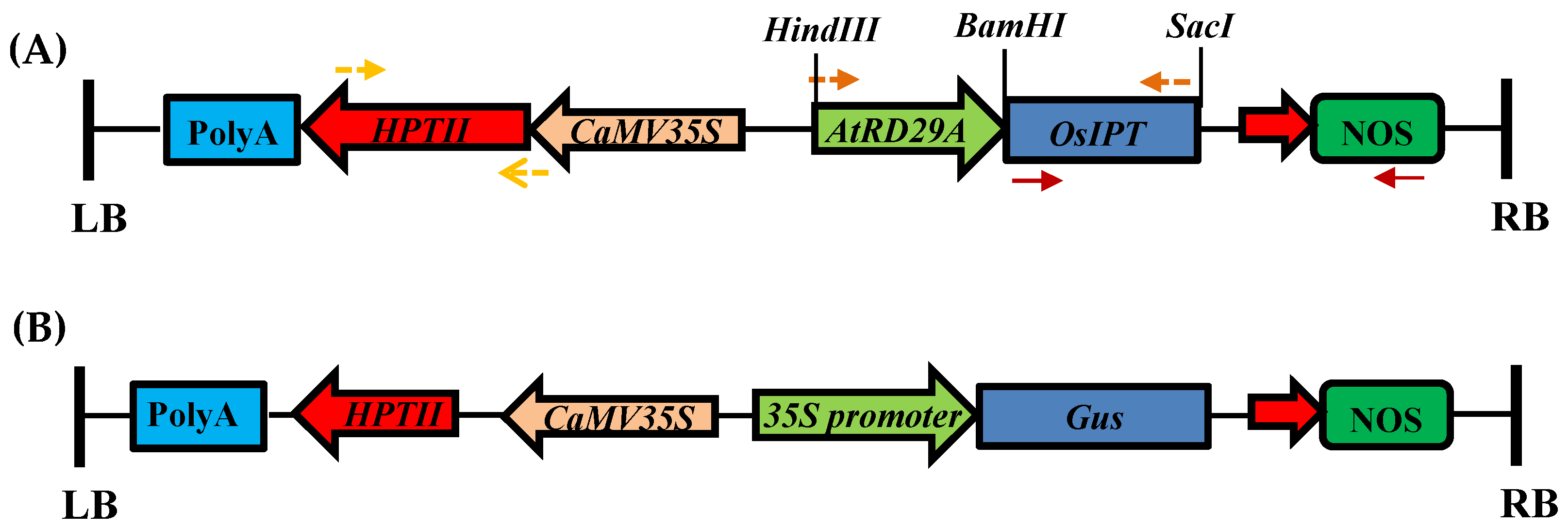
2.2.3. Gene Constructs and Agrobacterium Strain Used for Rice Transformation
2.2.4. Preparation of Primary and Secondary Agrobacterium Culture
2.2.5. Transformation and Co-Cultivation
2.2.6. Washing of Calli and Resting Medium
2.2.7. Selection of Transformed Calli
2.2.8. Regeneration of Transformed Calli
2.2.9. Rooting Medium and Root Hardening
2.2.10. Molecular Confirmation of Putative Transgenic Plants
2.2.11. Southern Blotting
2.2.12. β-Glucuronidase Staining (GUS) Assay
2.2.13. Statistical Analysis
3. Results and Discussion
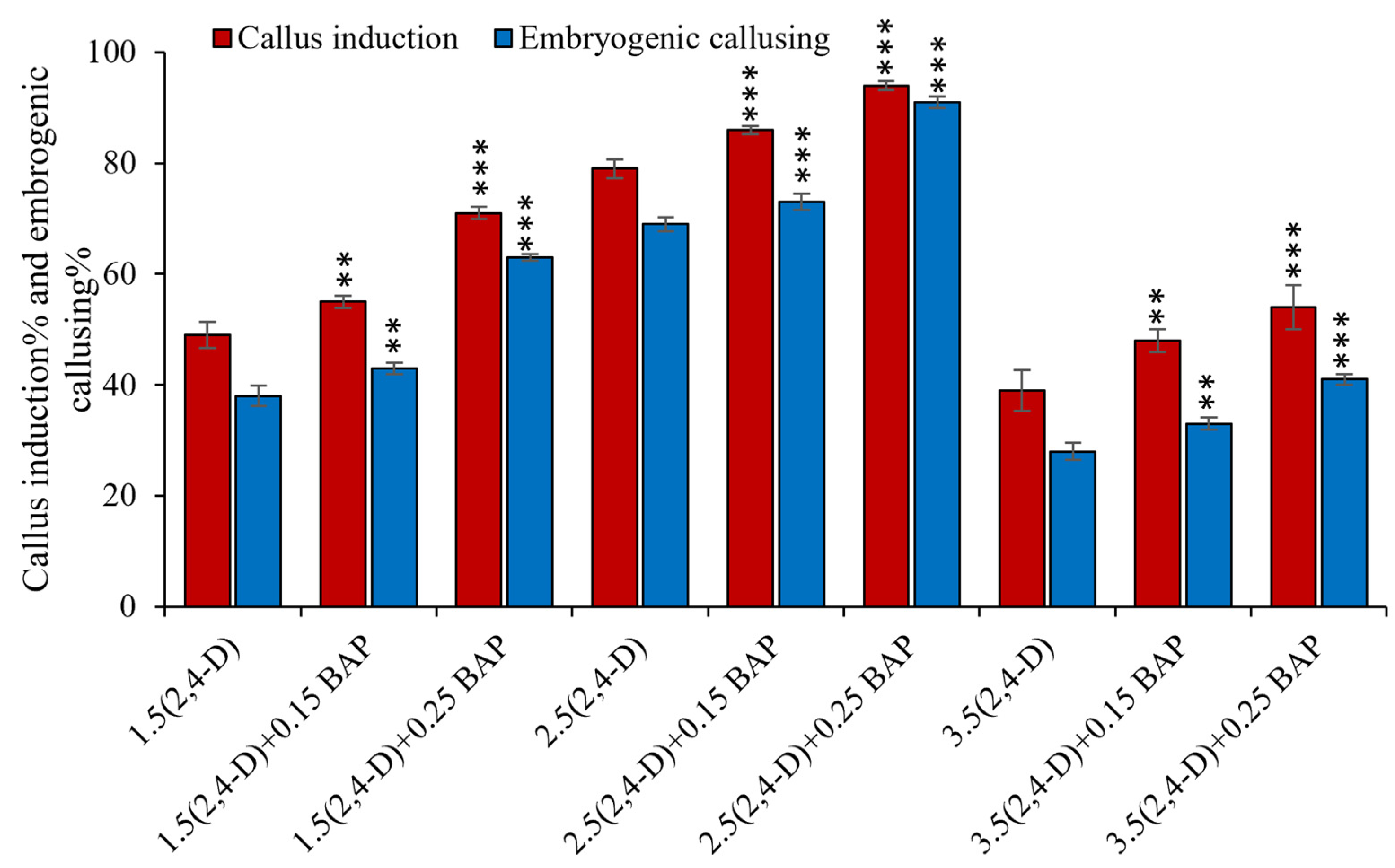
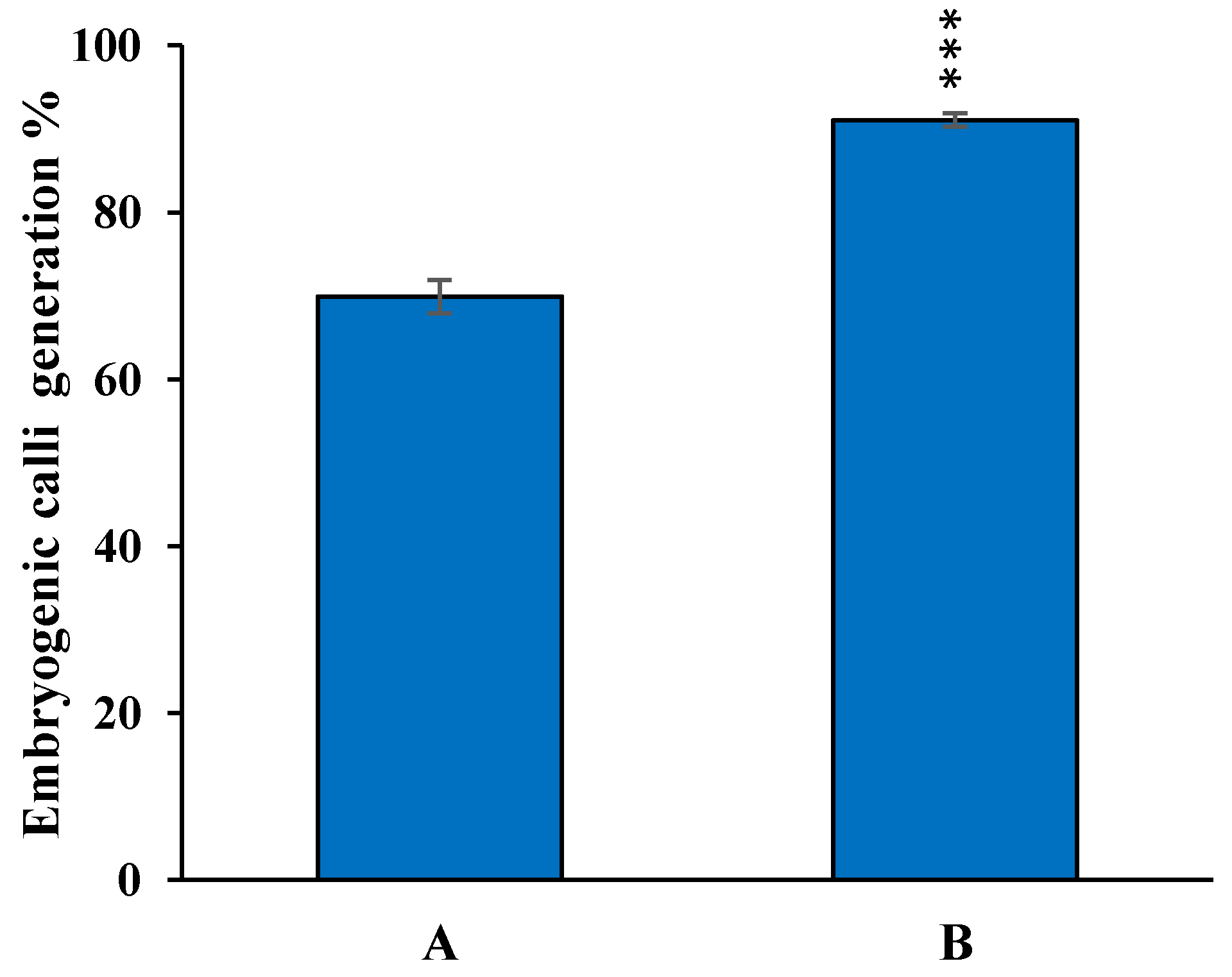
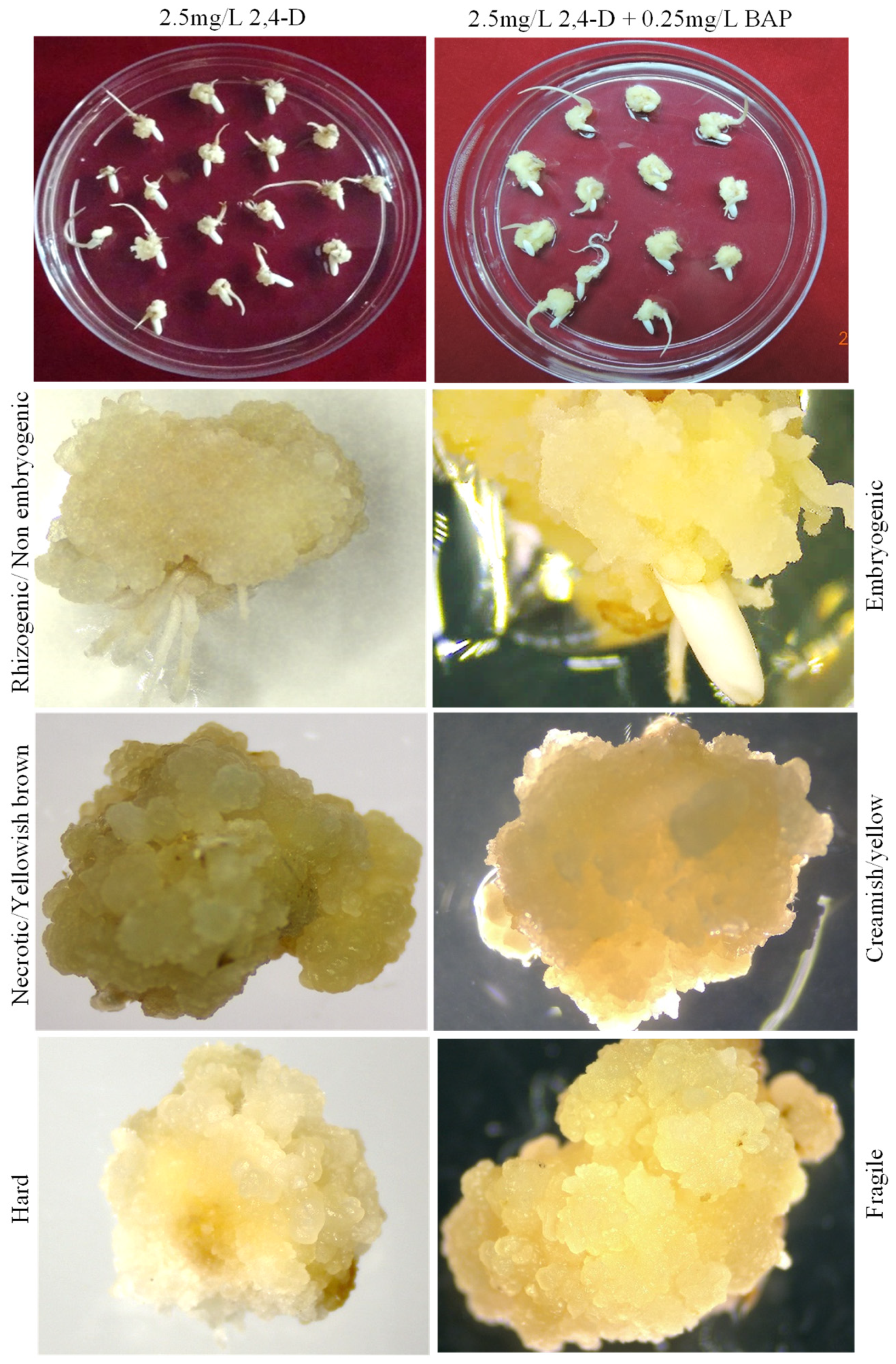
3.1. Optimization of In Vitro Embryogenic Callus Induction in Rice cv. MTU1010
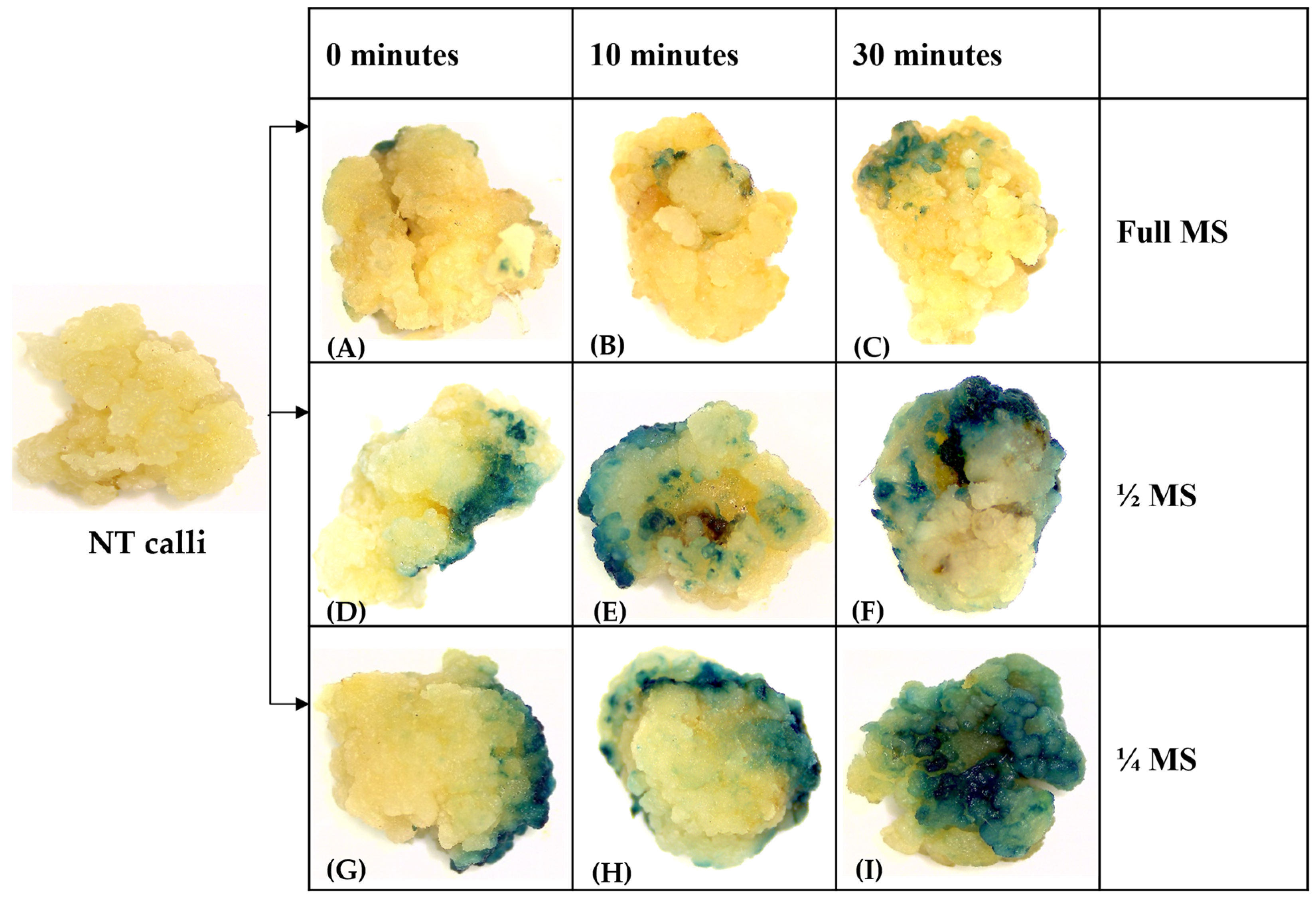
3.2. Downscaling Salt Concentration during Infection and Co-Cultivation Can Enhance the T-DNA Delivery and High Transient Expression Efficiency in Rice cv. MTU1010
3.3. Effect of Pre-Incubation on Transformation Efficiency
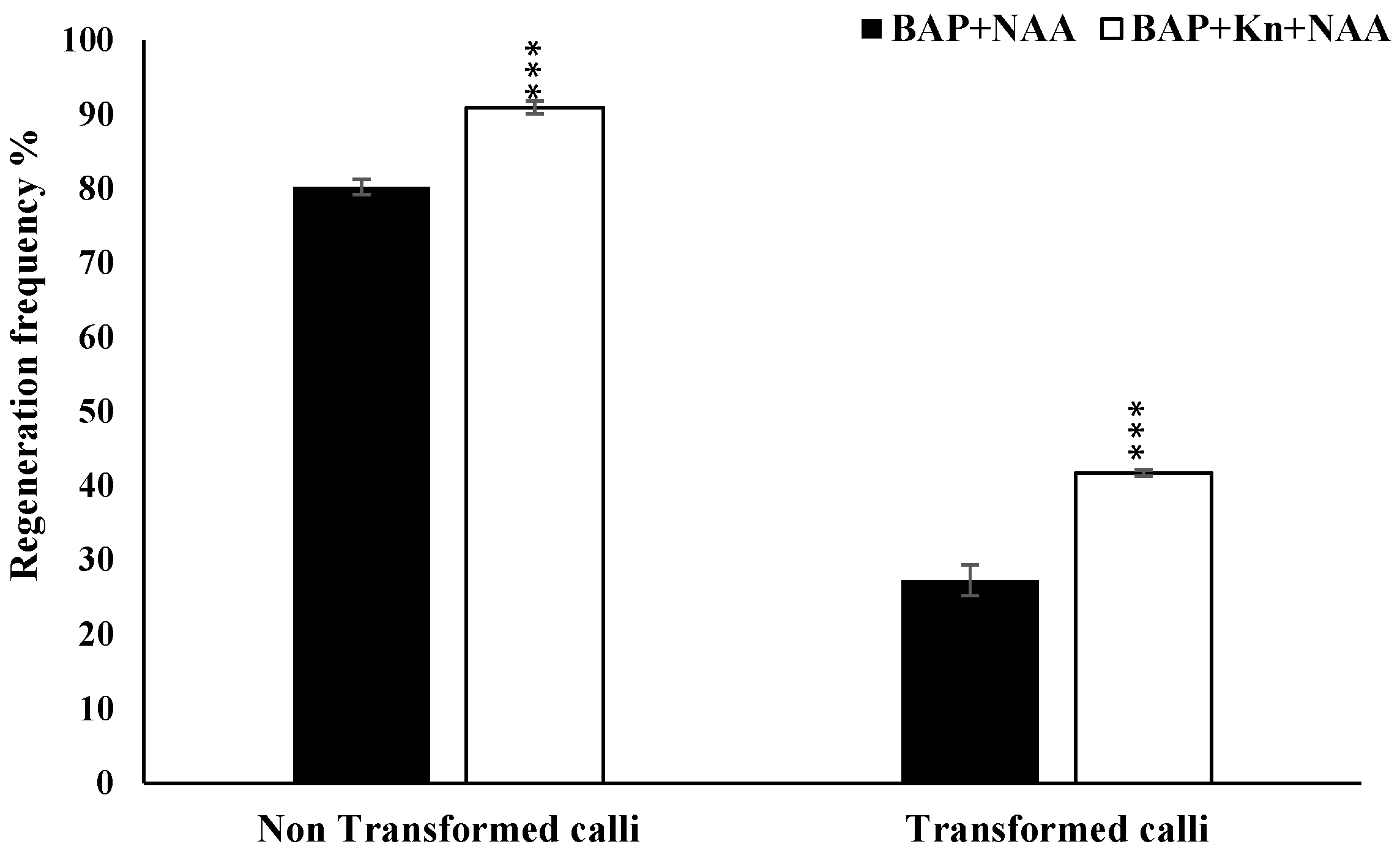
3.4. Optimization of Regeneration Medium and Measurement of Regeneration Frequency
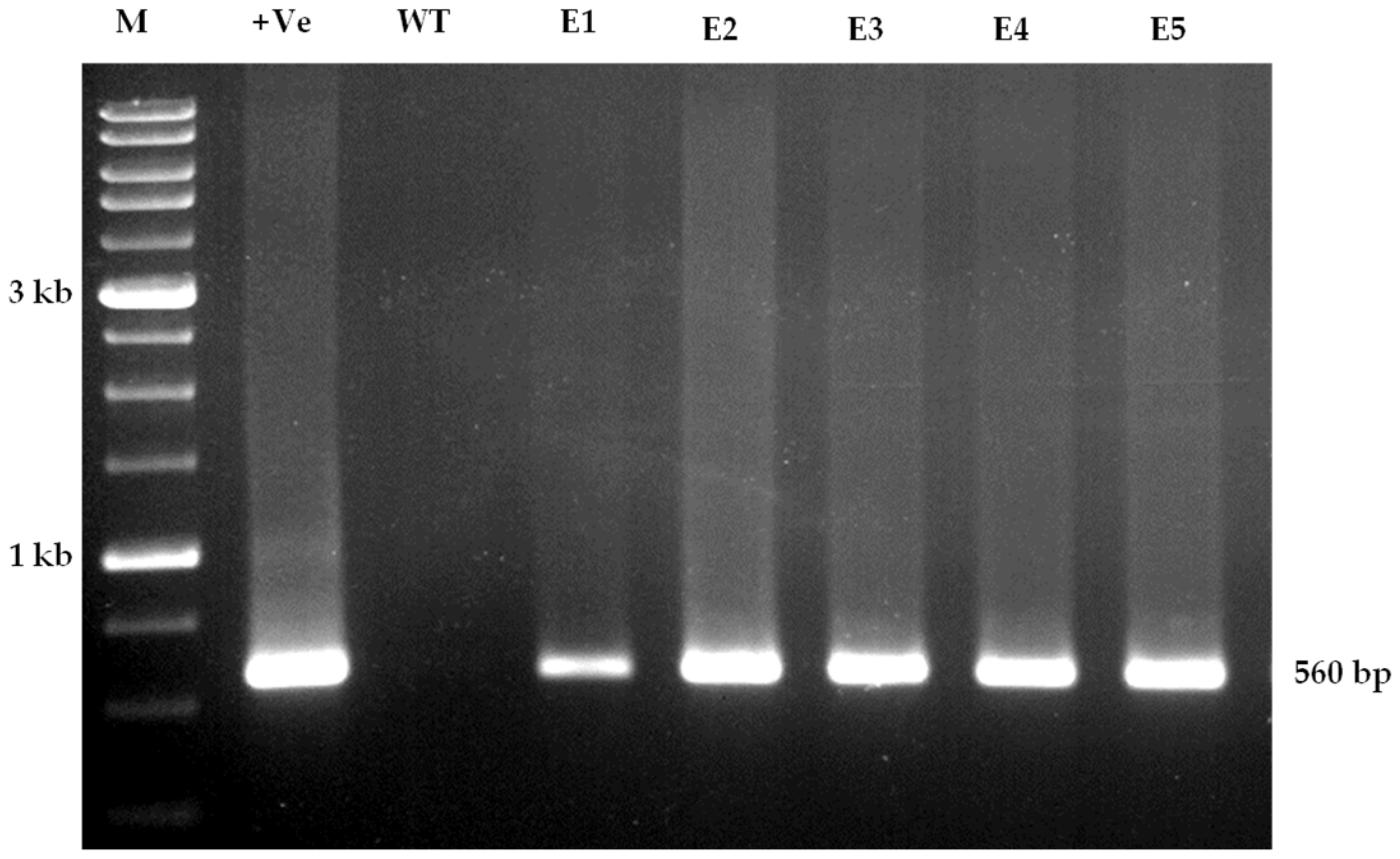
3.5. Molecular Confirmation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviation
References
- Sasaki, T. Rice genome analysis to understand the rice plant as an assembly of genetic codes. Photosynt. Res. 2001, 70, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Pazuki, A.; Sohani, M.M. Phenotypic evaluation of scutellum derived calluses in ‘Indica’ rice cultivars. Acta Agric. Sloven. 2013, 101, 239–247. [Google Scholar]
- Noor, W.; Lone, R.; Kamili, A.N.; Husaini, A.M. Callus induction and regeneration in high-altitude himalayan rice genotype SR4 via seed explant. Biotechnol. Rep. 2022, 36, e00762. [Google Scholar]
- Khush, G.S. What it will take to feed 5.0 billion rice consumers in 2030. Plant Mol. Biol. 2005, 59, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Zeigler, R.S.; Barclay, A. The Relevance of Rice; Springer: New York, NY, USA, 2008. [Google Scholar]
- Zhao, M.; Lin, Y.; Chen, H. Improving nutritional quality of rice for human health. Theor Appl Genet. 2020, 133, 1397–1413. [Google Scholar] [CrossRef]
- Yadav, S.K.; Yadav, P.; Chinnusamy, V. Nutrigenomics in Cereals. In Biofortification in Cereals; Deshmukh, R., Nadaf, A., Ansari, W.A., Singh, K., Sonah, H., Eds.; Springer: Singapore, 2023. [Google Scholar]
- Azmi, M.; Azlan, S.; Yim, K.M.; George, T.V.; Chew, S.E. Control of weedy rice in direct-seeded rice using the clearfield production system in Malaysia. Pak. J. Weed Sci. Res. 2012, 18, 49–53. [Google Scholar]
- Bzour, M.I.; Zuki, F.M.; Mispan, M.S. Introduction of imidazolinone herbicide and clearfield rice between weedy rice-control efficiency and environmental concerns. Environ. Rev. 2018, 26, 181–198. [Google Scholar] [CrossRef]
- Zulkarnain, W.M.; Ismail, M.R.; Saud, H.M.; Othman, R.; Habib, S.H.; Kausar, H. Growth and yield response to water availability at different growth stages of rice. J. Food Agric. Environ. 2013, 11, 540–544. [Google Scholar]
- Oo, K.S.; Lang, N.T. Developing salt tolerance in rice by mutagenesis. Omonrice 2005, 13, 126–134. [Google Scholar]
- Miah, M.A.A.; Pathan, M.S.; Quayum, H.A. Production of salt tolerant rice breeding line via doubled haploid. Euphytica 1996, 91, 285–288. [Google Scholar] [CrossRef]
- Sathish, P.; Gamborg, O.L.; Nabors, M.W. Establishment of stable NaCl-resistant rice plant lines from anther culture: Distribution pattern of K+/Na+ in callus and plant cells. Theor. Appl Genet. 1997, 95, 1203–1209. [Google Scholar] [CrossRef]
- Hoque, A.; Haque, M.A.; Sarker, M.R.A.; Rahman, M.A. Marker-assisted introgression of saltol locus into genetic background of BRRI Dhan-49. Int. J. Biosci. 2015, 6, 71–80. [Google Scholar]
- Xu, R.; Li, H.; Qin, R.; Wang, L.; Li, L.; Wei, P. Gene targeting using the Agrobacterium tumefaciens mediated CRISPR-Cas system in rice. Rice 2014, 7, 5. [Google Scholar] [CrossRef] [PubMed]
- Mostafiz, S.; Wagiran, A. Efficient callus induction and regeneration in selected indica rice. Agronomy 2018, 8, 77. [Google Scholar] [CrossRef]
- Magar, M.M.; Rani, C.V.D.; Anuradha, G. Marker Assisted Selection for Bacterial Leaf Blight Resistance in Segregating Populations of Cottondora Sannalu. Int. J. Appl. Sci. Biotechnol. 2014, 2, 229–237. [Google Scholar] [CrossRef]
- Sahoo, K.K.; Tripathi, A.K.; Pareek, A.; Sopory, S.K.; and Singla-Pareek, S.L. An improved protocol for efficient transformation and regeneration of diverse indica rice cultivars. Plant Methods 2011, 7, 49. [Google Scholar] [CrossRef]
- Sahoo, R.K.; Tuteja, N. Development of Agrobacterium-mediated transformation technology for mature seed-derived callus tissues of indica rice cultivar IR64. GM Crops Food 2012, 3, 123–128. [Google Scholar] [CrossRef]
- Lin, Y.J.; and Zhang, Q. Optimising the tissue culture conditions for high efficiency transformation of indica rice. Plant Cell Rep. 2005, 23, 540–547. [Google Scholar] [CrossRef]
- Yookongkaew, N.; Srivatanakul, M.; Narangajavana, J. Development of genotype-independent regeneration system for transformation of rice (Oryza sativa ssp. indica). J. Plant Res. 2007, 120, 237–245. [Google Scholar] [CrossRef]
- Jain, R.K.; Khehra, G.S.; Lee, S.H.; Blackhall, N.W.; Marchant, R.; Davey, M.R.; Power, J.B.; Cocking, E.C.; Gosal, S.S. An improved procedure for plant regeneration from indica and japonica rice protoplasts. Plant Cell Rep. 1995, 14, 515–519. [Google Scholar] [CrossRef]
- Kumar, K.K.; Maruthasalam, S.; Loganathan, M.; Sudhakar, D.; Balasubramanian, P. An improved Agrobacterium mediated transformation protocol for recalcitrant elite indica rice cultivars. Plant Mol. Biol. Rep. 2005, 23, 67–73. [Google Scholar] [CrossRef]
- Rance, I.M.; Tian, W.; Mathews, H.; de Kochko, A.; Beachy, R.N.; and Fauquet, C. Partial desiccation of mature embryo-derived calli, a simple treatment that dramatically enhances the regeneration ability of indica rice. Plant Cell Rep. 1994, 13, 647–651. [Google Scholar] [CrossRef] [PubMed]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Yoshida, S.; Forno, D.A.; Cock, J.A.; Gomez, K.A. Routine procedures for growing rice plants I culture solution. In Laboratory Manual for Physiological Studies in Rice; IRRI: Los Baños, Philippines, 1976; pp. 61–66. [Google Scholar]
- Murray, M.G.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4326. [Google Scholar] [CrossRef]
- Rowland, L.J.; Nguyen, B. Use of polyethylene glycol for purification of DNA from leaf tissue of woody plants. Biotechniques 1993, 14, 734–736. [Google Scholar] [PubMed]
- Jefferson, R.A. Assaying chimeric genes in plants: The GUS gene fusion system. Plant Mol. Biol. Rep. 1987, 5, 387–405. [Google Scholar] [CrossRef]
- Strickland, S.G.; Nichol, J.W.; McCaU, C.M.; Stuart, D.A. Effect of carbohydrate source on alfalfa somatic embryogenesis. Plant Sci. 1987, 48, 113–121. [Google Scholar] [CrossRef]
- Chu, C.C.; Hill, R.D.; Brule-Babel, A.I. High frequency of pollen embryoid formation and plant regeneration in Triticum aestivum L. on monosaccharide containing media. Plant Sci. 1990, 66, 255–262. [Google Scholar] [CrossRef]
- Verma, R.K.; Santosh Kumar, V.V.; Yadav, S.K.; Pushkar, S.; Rao, M.V.; and Chinnusamy, V. Overexpression of ABA Receptor PYL10 Gene Confers Drought and Cold Tolerance to Indica Rice. Front. Plant Sci. 2019, 10, 1488. [Google Scholar] [CrossRef]
- Santosh Kumar, V.V.; Verma, R.K.; Yadav, S.K.; Yadav, P.; Watts, A.; Rao, M.V.; and Chinnusamy, V. CRISPR-Cas9 mediated genome editing of drought and salt tolerance (OsDST) gene in indica mega rice cultivar MTU1010. Physiol. Mol. Biol. Plants Int. J. Funct. Plant Biol. 2020, 26, 1099–1110. [Google Scholar] [CrossRef]
- Fry, J.; Barnason, A.; Horsch, R.B. Transformation of Brassica napus with Agrobacterium tumefaciens based vectors. Plant Cell Rep. 1987, 6, 321–325. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.; Fry, J.E.; Pang, S.; Zhou, H.; Hironaka, C.M.; Duncan, D.R.; Conner, T.W.; Wan, Y. Genetic transformation of wheat mediated by Agrobacterium tumefaciens. Plant Physiol. 1997, 115, 971–980. [Google Scholar] [CrossRef] [PubMed]
- Paz, M.M.; Shou, H.; Guo, Z.; Zhang, Z.; Banerjee, A.K.; Wang, K. Assessment of conditions affecting Agrobacterium-mediated soybean transformation using the cotyledonary node explant. Euphytica 2004, 136, 167–179. [Google Scholar] [CrossRef]
- Sheng, J.; Citovsky, V. Agrobacterium-plant cell DNA transport: Have virulence proteins, will travel. Plant Cell 1996, 8, 1699–1710. [Google Scholar]
- Rana, M.M.; Han, Z.X.; Song, D.P.; Liu, G.F.; Li, D.X.; Wan, X.C.; Karthikeyan, A.; and Wei, S. Effect of Medium Supplements on Agrobacterium rhizogenes Mediated Hairy Root Induction from the Callus Tissues of Camellia sinensis var. sinensis. Int. J. Mol. Sci. 2016, 17, 1132. [Google Scholar] [CrossRef] [PubMed]
- Shrawat, A.K.; Lorz, H. Agrobacterium-mediated transformation of cereals: A promising approach crossing barriers. Plant Biotechnol. J. 2006, 4, 575–603. [Google Scholar] [CrossRef]
- Hiei, Y.; Komari, T. Agrobacterium-mediated transformation of rice using immature embryos or calli induced from mature seed. Nat. Protocols 2008, 3, 824–834. [Google Scholar] [CrossRef]
- Karthikeyan, A.; Pandian, S.K.; Ramesh, M. Agrobacterium-mediated transformation of leaf base derived callus tissues of popular indica rice (Oryza sativa L. sub sp indica cv. ADT 43). Plant Sci. Int. J. Exp. Plant Biol. 2011, 181, 258–268. [Google Scholar] [CrossRef]
- Vennapusa, A.R.; Vemanna, R.S.; Reddy, B.H.R.; Babitha, K.C.; Kiranmai, K.; Nareshkumar, A.; Sudhakar, C. An Efficient Callus Induction and Regeneration Protocol for a Drought Tolerant Rice Indica Genotype AC39020. J. Plant Sci. 2015, 3, 248–254. [Google Scholar] [CrossRef]
- Achary, V.M.M.; Reddy, M.K. CRISPR-Cas9 mediated mutation in GRAIN WIDTH and WEIGHT2 (GW2) locus improves aleurone layer and grain nutritional quality in rice. Sci. Rep. 2021, 11, 21941. [Google Scholar] [CrossRef]
- Greenwood, J.R.; Glaus, A.N. Optimized rice transformation protocol for transformation of the blast susceptible Indica rice accession CO39. CABI Agric Biosci. 2022, 3, 35. [Google Scholar] [CrossRef]




| Name of the Media | Composition |
|---|---|
| Callus induction media (CIM) | MS Salts with B5 vitamins, 300 mg/L Casein hydrolysate, 560 mg/L L-Proline, 36 g/L Maltose monohydrate, pH-5.8, 4 g/L Phytagel. Autoclave the media and allow them to cool to room temperature and then add 2.5 mg/L 2,4-D and 0.25 mg/L 6-BAP |
| Yeast extract Mannitol medium (YEM) | Yeast extract 1 g/L, Mannitol 10 g/L, NaCl 1 g/L, MgSO4.7H2O 0.2 g/L, K2HPO4 0.5 g/L, pH-6.8–7. Autoclave and store the media at room temperature |
| Yeast extract Mannitol Agar medium (YEMA) | To YEM medium, add 1.5% Agar |
| Resuspension medium (RSM-1) | MS salts with 1.5% sucrose and adjust the pH to 5.4 autoclave and store at room temperature |
| Resuspension medium (RSM-2) | ½ MS or ¼ MS salts with 1.5% sucrose and adjust the pH to 5.4 autoclave and store at room temperature |
| Co-cultivation medium (CCM-1) | MS Salts with B5 vitamins, 300 mg/L Casein hydrolysate, 560 mg/L L-Proline, 36 g/L Maltose monohydrate, pH-5.8, 4 g/L Phytagel. Autoclave and then add 2.5 mg/L 2,4-D, 0.25 mg/L 6-BAP and 150 µM acetosyringone (freshly prepared) |
| Co-cultivation medium (CCM-2) | ½ MS Salts with B5 vitamins, 300 mg/L Casein hydrolysate, 560 mg/L L-Proline, 36 g/L Maltose monohydrate, pH-5.8, 4 g/L Phytagel. Autoclave and then add 2.5 mg/L 2,4-D, 0.25 mg/L 6-BAP and 150 µM acetosyringone (freshly prepared) |
| Resting Media | To the CIM medium, add 300 mg/L Cefotaxime, 200 mg/L Timentin. |
| Selection medium-1 (SM-1) | To the CIM medium, add 300 mg/L Cefotaxime, 200 mg/L Timentin, 50 mg/L Hygromycin |
| Selection medium-2 (SM-2) | To the CIM medium, add 250 mg/L Cefotaxime, 50 mg/L Hygromycin |
| Regeneration medium (RM-1) | MS salts, 30 g/L Maltose monohydrate, pH-5.8, 1% Agarose. Autoclave the media and then add 2.5 mg/L 6-BAP, 1 mg/L Kinetin, 0.5 mg/L NAA, 250 mg/L cefotaxime and 35 mg/L Hygromycin |
| Regeneration medium (RM-2) | MS salts, 30 g/L Maltose monohydrate, pH-5.8, 0.8% Agarose. Autoclave the media and add 2.5 mg/L 6-BAP, 1 mg/L Kinetin and 0.5 mg/L NAA, 30 mg/L Hygromycin |
| Rooting medium | ½ MS salts, 20 g/L Sucrose, 10 g/L Glucose, pH-5.8 then add 2.5 g/L Phytagel. Autoclave the media and then add 0.05 mg/L NAA |
| Root hardening | ½ strength Yoshida Medium or ½ strength Hoagland medium |
| Acclimatization | Soil rite mixture |
| Transplanting | Soil |
| Media | Composition | [19] | [18] | [41] | [42] | [43] | [44] |
|---|---|---|---|---|---|---|---|
| Genotypes | IR64 | CSR10, IR64, PB1, Swarna | ADT 43 | AC39020 | MTU1010 | CO39 | |
| CIM (Callus Induction Media) |
| 4.4 g/L 0.03 g/L 0.065 g/L --- 30 g/L --- 4 g/L 2 g/L 2.5 mg/L 0.15 mg/L 5.8 | 4.4 g/L 0.3 g/L 0.60 g/L 30 g/L --- 10 g/L 3 g/L --- 3 mg/L 0.25 mg/L 5.2 | --- --- --- --- --- --- 3 g/L --- 2.5 mg/L --- 5.8 | --- --- --- --- --- --- --- --- 2 mg/L --- 5.8 | 4.4 g/L 0.4 g/L 0.7 g/L 30 g/L --- --- 4 g/L --- 2.5 mg/L --- 5.8 | 4.3 g/L 0.3 g/L 2.8 g/L 30 g/L --- --- 3 g/L --- 3 mg/L --- 5.8 |
| Resuspension medium |
| 4.4 g/L 68 g/L 36 g/L 3 g/L 4 g/L 150 µM 5.2 | 4.4 g/L 68 g/L 36 g/L 3 g/L 4 g/L 150 μM 5.2 | Not Mentioned | Not Mentioned | Not Mentioned | 4.3 g/L 68 g/L 36 g/L 3 g/L 4 g/L 150 µM 5.2 |
| CO-C (Co-Cultivation Media) |
| 4.4 g/L 0.03 g/L 0.065 g/L --- 30 g/L --- 4 g/L 2 g/L 2.5 mg/L 0.15 mg/L 150 µM 5.8 | 4.4 g/L 0.3 g/L 0.60 g/L 30 g/L --- 10 g/L 3 g/L --- 3 mg/L 0.25 mg/L 150 µM 5.2 | --- --- --- --- --- --- 3 g/L --- 2.5 mg/L --- 100 µM 5.8 | Not Mentioned | 4.4 g/L --- 0.5 g/L --- 20 g/L 10 g/L 4 g/L --- 1.5 mg/L --- 200 µM 5.2 | 4.3 g/L 0.3 g/L 0.60 g/L --- --- 10 g/L 4 g/L --- 3 mg/L --- 150 µM 5.8 |
| Selection Media |
| 4.4 g/L 0.03 g/L 0.065 g/L --- 30 g/L --- --- 4 g/L 2 g/L 2.5 mg/L 0.15 mg/L 50 mg/L 300 mg/L --- --- 5.8 | 4.4 g/L 0.3 g/L 0.60 g/L 30 g/L --- 10 g/L --- 3 g/L --- 3 mg/L 0.25 mg/L 50 mg/L 250 mg/L --- --- 5.2 | --- --- --- --- --- --- --- 3 g/L --- 2.5 mg/L --- --- --- --- --- 5.8 | Not Mentioned | 4.4 g/L 0.4 g/L 0.7 g/L --- 30 g/L --- 1 g/L 4 g/L --- 3 mg/L --- 50 mg/L --- --- 600 mg/L 5.8 | 4.3 g/L 0.3 g/L 2.8 g/L --- 30 g/L --- --- 3 g/L --- 3 mg/L --- 50 mg/L --- 250 mg/L --- 5.8 |
| Regeneration Media (I) |
| 4.4 g/L --- 30 g/L 3 mg/L 0.5 mg/L 1 mg/L --- --- 40 mg/L 10 g/L 5.8 | 4.4 g/L 30 g/L --- --- 0.2 mg/L 2 mg/L 250 mg/L --- 30 mg/L 10 g/L 5.8 | 4.4 g/L 30 g/L --- 1 mg/L 1.5 mg/L 1.2 mg/L --- --- 30 mg/L 10 g/L 5.8 | 4.4 g/L 30 g/L --- 4 mg/L 0.5 mg/L --- --- --- 30 mg/L 10 g/L 5.8 | 4.4 g/L --- 30 g/L 2 mg/L 0.5 mg/L 1 mg/L --- 600 mg/L 30 mg/L 4 g/L 5.8 | 4.3 g/L 30 g/L --- --- 0.2 mg/L 2 mg/L --- --- --- 10 g/L 5.8 |
| Regeneration Media (II) |
| 4.4 g/L --- 30 g/L 3 mg/L 0.5 mg/L 1 mg/L --- 40 mg/L 8 g/L 5.8 | 4.4 g/L 30 g/L --- 2.7 mg/L 0.5 mg/L 1.2 mg/L 250 mg/L 30 mg/L 8 g/L 5.8 | Not Mentioned | Not Mentioned | Not Mentioned | 4.3 g/L 30 g/L --- --- 0.2 mg/L 2 mg/L --- 30 mg/L 8 g/L 5.8 |
| Rooting Media |
| 4.4 g/L 20 g/L 10 g/L --- --- --- 4 g/L | 2.2 g/L 30 g/L --- --- 250 mg/L 30 g/L 3 g/L | Not Mentioned | Not Mentioned | 2.2 g/L 15 g/L --- --- --- --- 4 g/L | 2.15 g/L 30 g/L --- --- --- 30 mg/L 3 g/L |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yadav, P.; Santosh Kumar, V.V.; Priya, J.; Yadav, S.K.; Nagar, S.; Singh, M.; Chinnusamy, V. A Versatile Protocol for Efficient Transformation and Regeneration in Mega Indica Rice Cultivar MTU1010: Optimization through Hormonal Variables. Methods Protoc. 2023, 6, 113. https://doi.org/10.3390/mps6060113
Yadav P, Santosh Kumar VV, Priya J, Yadav SK, Nagar S, Singh M, Chinnusamy V. A Versatile Protocol for Efficient Transformation and Regeneration in Mega Indica Rice Cultivar MTU1010: Optimization through Hormonal Variables. Methods and Protocols. 2023; 6(6):113. https://doi.org/10.3390/mps6060113
Chicago/Turabian StyleYadav, Pragya, V. V. Santosh Kumar, Jyoti Priya, Shashank Kumar Yadav, Shivani Nagar, Meenu Singh, and Viswanathan Chinnusamy. 2023. "A Versatile Protocol for Efficient Transformation and Regeneration in Mega Indica Rice Cultivar MTU1010: Optimization through Hormonal Variables" Methods and Protocols 6, no. 6: 113. https://doi.org/10.3390/mps6060113
APA StyleYadav, P., Santosh Kumar, V. V., Priya, J., Yadav, S. K., Nagar, S., Singh, M., & Chinnusamy, V. (2023). A Versatile Protocol for Efficient Transformation and Regeneration in Mega Indica Rice Cultivar MTU1010: Optimization through Hormonal Variables. Methods and Protocols, 6(6), 113. https://doi.org/10.3390/mps6060113






