Multiparametric Flow Cytometry-Based Immunophenotyping of Mouse Liver Immune Cells
Abstract
:1. Introduction
2. Experimental Design
2.1. Materials
- Debris removal solution (Miltenyi Biotec, Bergisch Gladbach, Germany; Cat. no.: 130-109-398; store protected from light at 4 °C, do not freeze)
- DMEM High Glucose w/stable glutamine, w/sodium pyruvate (Biowest, Riverside, MO, USA; Cat. no.: L0103-500; store protected from light at 4 °C, do not freeze)
- Aerrane (Isoflurane UPC, Baxter, Deerfield, IL, USA; Cat. no.: FDG9623; store protected from light at room temperature (RT))
- Liver dissociation kit, mouse (Miltenyi Biotec, Bergisch Gladbach, Germany; Cat. no.: 130-105-807; individual components store at 4 °C, reconstituted components store for max. 6 months at −20 °C, avoid freeze/thaw cycles)
- Phosphate buffered saline w/o calcium, w/o magnesium (Biowest, Riverside, MO, USA; Cat. no.: P0750; store at 4 °C)
- Red blood cell lysis buffer (RBL; store at RT; see Reagent Setup)
- Trypan blue solution (Sigma-Aldrich, Burlington, MA, USA; Cat. no.: T8154, store at RT)
- Fixation buffer (BD Biosciences, Franklin Lakes, NJ, USA; Cat. no.: 554655, store protected from light at 4 °C, do not freeze), optional reagent
- Flow cytometry (FC) staining buffer (store for max. 1 month at 4 °C, do not freeze; see Reagent Setup)
- Fluorescently labeled antibodies for FC purposes (see Table 1; store protected from light at 4 °C)
2.2. Equipment
- 25 mL tissue sample vessel (Carl Roth, Karlsruhe, Germany; Cat. no.: AYX2.1)
- Blunt dissecting scissors (VWR®, Radnor, PA, USA; Cat. no.: HAMMHSB120-14)
- Cotton pads (Batist Medical a.s., Cerveny Kostelec, Czech Republic; Cat. no.: 5670)
- Dry bath incubator (Major Science, Saratoga, CA, USA; Cat. no.: MD-02N)
- 15 mL conical centrifuge tubes (VWR®, Radnor, PA, USA; Cat. no.: 525-1084)
- 1.5 mL microcentrifuge tubes (VWR®, Radnor, PA, USA; Cat. no.: 89000-028)
- Forceps with round blade (VWR®, Radnor, PA, USA; Cat. no.: 232-0106)
- Forceps with straight blade (VWR®, Radnor, PA, USA; Cat. no.: BSNC00DSA)
- gentleMACS C-tube (Miltenyi Biotec, Bergisch Gladbach, Germany; Cat. no.: 130-093-237)
- gentleMACS Octo dissociator with heaters (Miltenyi Biotec, Bergisch Gladbach, Germany; Cat. no.: 130-096-427)
- Ismatec IPC pump (Ismatec, Wertheim, Germany; Cat. no.: ISM 930)
- Luna-II automated cell counter (Logos Biosystems, Anyang-si, Gyeonggi-do, Korea; Cat. no.: L40002)
- Cell counting slides (Logos Biosystems, Anyang-si, Gyeonggi-do, Korea; Cat. no.: L12003)
- MACS SmartStrainer, 100 µm (Miltenyi Biotec, Bergisch Gladbach, Germany; Cat. no.: 130-098-463)
- Neo Delta Ven T cannula 24G (Delta Med, Viadana, Lombardia, Italy; Cat. no.: 3113122)
- R540 Enhanced Anesthesia Machine (RWD, Baltimore, MD, USA; Cat. no.: R540IE)
- Refrigerated centrifuge with swinging buckets (ThermoFisher Scientific, Waltham, MA, USA; Cat. no.: 15253457)
- Sharp scissors (VWR®, Radnor, PA, USA; Cat. no.: 233-1104)
- Extension tubes (Gama group, Ceske Budejovice, Czech Republic; Cat. no.: 606301-ND)
- Pump tubing (Tygon®, Ismatec, Wertheim, Germany; Cat. no.: ISMCSC0024T, ISMCSC0048T)
- Tweezers (VWR®, Radnor, PA, USA; Cat. no.: 229-0374)
- Water bath (Polysciences, Warrington, PA, USA; Cat. no.: WBE20A12E)
- BD LSRFortessa flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA) or any multiparametric flow cytometer with at least 13-fluorescence detectors)
2.3. Software
- Diva software (Becton Dickinson, Franklin Lakes, NJ, USA, v8.0.1. or later, BD FACSDiva™ Software www.bdbiosciences.com, accessed on 4 August 2022) or any equivalent
- FlowJo analysis software (BD Biosciences, Franklin Lakes, NJ, USA, v10 or later, www.bdbiosciences.com, accessed on 4 August 2022) or any equivalent
3. Procedure
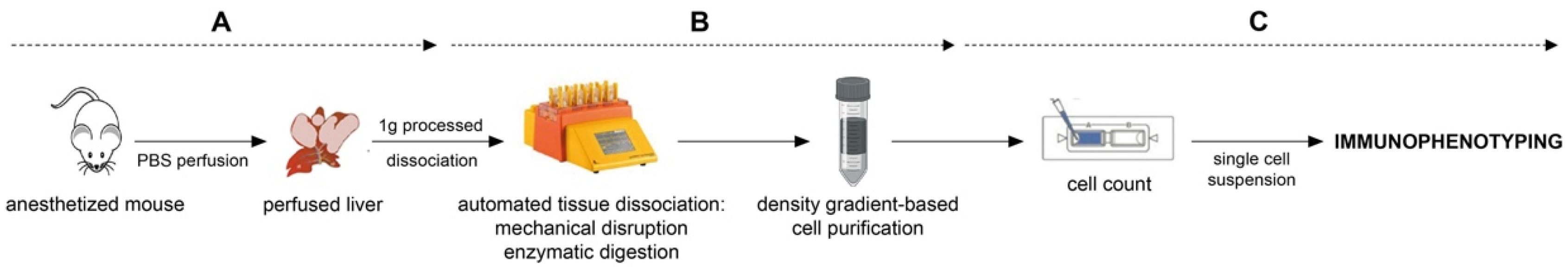
3.1. Liver Perfusion
- Set up of instruments: prime peristaltic pump with tempered PBS, set the flow rate to 2.5 mL/min.
- Anesthetize a mouse using 2–5% isoflurane in air or oxygen mixture until the deep loss of sensitivity.
- Place the fully anesthetized mouse to a supine position in a breathing mask, attach paws to the pad to stretch the mouse.
- Disinfect the abdomen with 70% ethanol. Lift the skin with tweezers. Using blunt dissecting scissors, cut the skin and peritoneum horizontally in the lower abdomen. Continue with a lateral cut on both sides of the abdomen, up to the lower rib cage.
CRITICAL STEP Continuously observe breathing rate to be low and deep without any sign of choking. Be sure not to cut any of the organs or diaphragm.
- Use forceps to grab the abdominal skin and peritoneum and roll the skin up to the rib cage to reveal the abdominal cavity. Move intestines to the side to expose the portal vein.
CRITICAL STEP For optimal procedure, no bleeding should occur.
- Straighten the vein. Place the needle of the cannula in parallel to the portal vein with the bevel up (Figure 2A). Inject the lower part of the vein with the cannula needle, then pull out the needle from the cannula, and move the polymer part further into the vein. A blood backflow should be visible.Note: It is not necessary to immobilize the cannula by a vein ligation.
- Adjust the pump flow rate to 2.5 mL/min, ensure there are no bubbles in the tubing, place the tubing into the cannula, cut one kidney, and immobilize the tubing onto the pad.
CRITICAL STEP Observe an immediate liver color change as a proof of correct perfusion setting (Figure 2B).
- Wash the liver with 30–40 mL of PBS until the liver completely lightens and no blood appears in the wash volume.
- Remove the cannula and turn off the pump. Carefully remove the gallbladder and harvest the liver into 50 mL sample vial with PBS (Figure 2C).
- OPTIONAL STEP Carefully remove the gallbladder to prevent bile contamination of the liver; if contaminated, thoroughly wash the liver with PBS.
3.2. Liver Dissociation
- Cut off 1 g of liver tissue (weight should not exceed 1.2 g of tissue per one dissociation [30]).
- Wash the liver with preheated cell medium and place it into C-tube with liver dissociation mix (according to the manufacturer’s instruction). After attaching C-tube onto the dissociator with heater, launch a 37C_m_LIDK_1 program predefined by the manufacturer (Figure 2D,E).
CRITICAL STEP Aliquoted components should be thawed right before use, repeated freeze-thaw cycles should be strictly avoided.
- Detach C-tube from the dissociator when the program terminates. Gently resuspend obtained liver homogenate and filter it through a pre-wetted 100 µm cell strainer into a 15 mL falcon tube. To avoid loss of cells within the C-tube, wash the tube and strainer with 5 mL of cell culture medium (Figure 2F,G).
- Centrifuge the homogenate sample for 10 min at 300× g, RT. Discard supernatant and resuspend the pellet in PBS (RT).
3.3. Liver Homogenate Processing to Prepare Single Cell Suspension
- 5.
- Centrifuge the obtained cell suspension for 10 min at 300× g, 4 °C and discard the supernatant.
- 6.
- Resuspend the pellet in pre-cooled PBS, add the debris removal solution and overlay gently with the pre-cooled PBS (according to the manufacturer’s instruction).
CRITICAL STEP Observe phase formation to control the step (Figure 2H).
- 7.
- Centrifuge for 10 min at 3000× g, 4 °C.
CRITICAL STEP Reduce the centrifugation break as well as acceleration rate (level 4 out of 9 applied on centrifuge used in this protocol), 3 phases have to be well defined.
- 8.
- Aspirate the two upper phases and add up to 15 mL of pre-cooled PBS, mix the suspension by gentle inverting the tubes.
- 9.
- Centrifuge for 10 min at 1000× g, 4 °C and discard the supernatant (Figure 2I).
- 10.
- Resuspend pellet in 1 mL of RBL to remove remaining red blood cells and incubate for 5 min at RT.
- 11.
- Fill the tube with PBS, mix the suspension by gentle inverting the tube.
- 12.
- Centrifuge for 5 min at 500× g, RT and discard the supernatant (Figure 2J).
- 13.
- OPTIONAL STEP Repeat the steps C.10–C.12 if pelleted cells are still contaminated with red blood cells, eventually platelets.
- 14.
- Resuspend the pellet in at least 1 mL of PBS to count the cells.
- 15.
- Use any cell counter to determine cell concentration, viability on the basis of Trypan blue exclusion, size distribution and clustering.
- 16.
- OPTIONAL STEP Using the LUNA cell counter, follow the steps below (17–19).
- 17.
- Prepare a 1:1 mixture of cells and Trypan blue (10 µL + 10 µL) to determine the cell count (concentration), viability, distribution, and clustering. Apply 10 µL of the mixture into a cell counting slide chamber, wait until the equilibrium is established.
- 18.
- Set the counting protocol to the following settings: dilution factor 2, min. cell size 3 µm, max. cell size 30 µm, size gating 3–30 µm, live cell sensitivity 7, roundness 60%, declustering level medium.
- 19.
- Apply the loaded protocol on a sample, verify the autofocus and count the cells, verify the gating strategy of the program (Figure 4).
3.4. FC Based Immunophenotyping
- Centrifuge the obtained cell suspension for 10 min at 300× g, 4 °C and discard the supernatant. Wash with an excessive volume of PBS and centrifuge for 5 min at 500× g, RT. Discard the supernatant.
- Resuspend cells in 40 µL of PBS.
- To distinguish live and dead cells, add the live/dead Zombie NIR marker at a pre-determined dilution (Table 1) and incubate for 20 min at RT, avoid light.Note: Any other viability dye can be used. The used Zombie viability kit is a fixable (paraformaldehyde or methanol), an amine-reactive fluorescent dye that is non-permeant to live cells.
- In parallel, prepare a positive control of dead cells by boiling 0.3–0.5 × 106 cells in 40 µL PBS for 5 min at 65 °C, cool the sample down to RT, perform staining as in D.3 step. This sample is also used as a single stain control to create a compensation matrix.
- Wash the cells by adding 150 µL of PBS (RT) and centrifuge for 5 min at 500× g, RT. Discard the supernatant.
- Perform specific staining by resuspending the pellets in 40 µL of FC staining buffer (see Reagent Setup), incubate with the staining antibody mixture or relevant isotype controls at a pre-determined concentration (Table 1) for 30 min at 4 °C, avoid light.
- Wash the cells by adding 150 µL of FC buffer and centrifuge for 5 min at 500× g, RT, discard the supernatant.
- OPTIONAL STEP Fix the cells by resuspending the pellets in 80 µL of Fixation buffer, incubate for 15–45 min at RT, avoid light; wash the cells by adding 150 µL of FC buffer and centrifuge for 5 min at 500× g, RT, discard the supernatant.
- Resuspend the cells in 250 µL of FC buffer and transfer cell suspension into a FC tube through its cell strainer snap cap. Samples without the fixation step are intended for immediate analysis; however, fixed samples can be stored at 4 °C for up to one week and then assayed.
- For a compensation matrix set up, prepare single stained samples using a drop of both types of compensation beads (anti-rat/hamster and negative particle set, Table 1) into 30 µL of FC buffer (1 drop is approximately of 50 µL equivalent). Perform staining directly in FC tubes to minimize potential losses.
- Add the specific staining antibody in the same dilution as for the immunophenotyping (count sample volume as a composition: 50 µL drop of specific + 50 µL negative beads + 30 µL of FC buffer), incubate under the same conditions as in step 6.
- Wash the beads by adding 2 mL of FC buffer and centrifuge for 10 min at 200× g, RT, discard the supernatant.
- Resuspend the pelleted beads in 250 µL of FC buffer, vortex thoroughly.
- Launch a calibration procedure at the flow cytometer, create the compensation matrix using the unstained and single stained samples, and calculate the compensations.
- OPTIONAL STEP If required, define the acquisition mode in terms of cells or beads used for the compensation set up depending on the flow cytometer available.
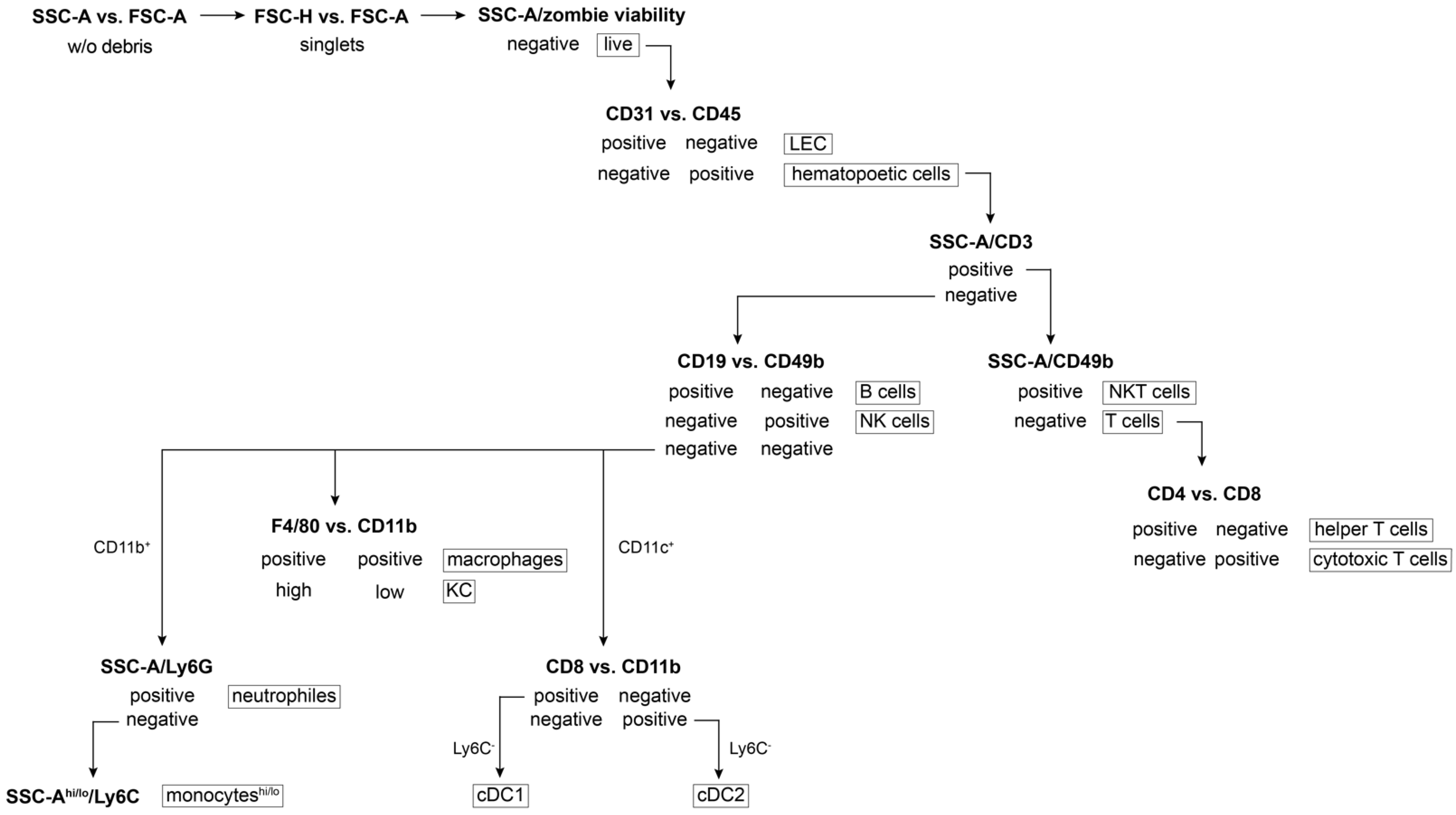
- Formulate the gating strategy (Figure 3) to monitor cell subsets of interest, respect subset hierarchy and marker exclusivity.
- Run samples for the immunophenotyping purposes by gating on a rare population (either KC or neutrophils).
- Acquire and record data by collecting at least 10,000 events of the population of interest.
- OPTIONAL STEP If necessary, record the same sample several times by gating on various immune populations to then easily define and characterize any population.
- Export fsc files to evaluate the data in FlowJo software or any equivalent.
3.5. Data Analysis
- Start the gating strategy first by the debris exclusion, looking at forward and side scatter, followed by a doublet and dead cell exclusion.
- Gate the particular immune population by exclusion of non-desired cells and the selection of those specific cells (Table 2, Figure 3 and Figure 5).Note: Zombie viability dye is permeant only to cells with compromised membranes, therefore a negative population needs to be gated as live subset.
- OPTIONAL STEP To quantify individual populations and their subsets, export the frequency of various subsets as a percentage in either the live cells or the parent population regarding the data representation.
4. Expected Results and Discussion
5. Reagents Setup
- RBL buffer
- 0.1 mM EDTA
- 12 mM NaHCO3
- 155 mM NH4Cl
- Store at RT, no contamination should appear.
- FC staining buffer
- 0.5% BSA (w/v)
- 2 mM EDTA
- Prepare a solution in 1x PBS. Store at 4 °C up to 1 month without any preservative such as 0.02% (v/v) thimerosal or 0.02–0.05% (w/v) sodium azide.
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bogdanos, D.P.; Gao, B.; Gershwin, M.E. Liver Immunology. Compr. Physiol. 2013, 3, 567–598. [Google Scholar] [CrossRef] [PubMed]
- Heymann, F.; Tacke, F. Immunology in the Liver—From Homeostasis to Disease. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 88–110. [Google Scholar] [CrossRef] [PubMed]
- Freitas-Lopes, M.A.; Mafra, K.; David, B.A.; Carvalho-Gontijo, R.; Menezes, G.B. Differential Location and Distribution of Hepatic Immune Cells. Cells 2017, 6, 48. [Google Scholar] [CrossRef] [PubMed]
- Blom, K.G.; Qazi, M.R.; Matos, J.B.N.; Nelson, B.D.; DePierre, J.W.; Abedi-Valugerdi, M. Isolation of Murine Intrahepatic Immune Cells Employing a Modified Procedure for Mechanical Disruption and Functional Characterization of the B, T and Natural Killer T Cells Obtained. Clin. Exp. Immunol. 2009, 155, 320–329. [Google Scholar] [CrossRef] [PubMed]
- Tacke, F.; Zimmermann, H.W. Macrophage Heterogeneity in Liver Injury and Fibrosis. J. Hepatol. 2014, 60, 1090–1096. [Google Scholar] [CrossRef] [PubMed]
- Klugewitz, K.; Adams, D.H.; Emoto, M.; Eulenburg, K.; Hamann, A. The Composition of Intrahepatic Lymphocytes: Shaped by Selective Recruitment? Trends Immunol. 2004, 25, 590–594. [Google Scholar] [CrossRef]
- Lian, Z.-X.; Li, L. The Liver as a Lymphoid Organ. In Liver Immunology, 3rd ed.; Gerswin, M.E., Vierling, J.M., Tanaka, A., Manns, M.P., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 17–33. ISBN 978-3-030-51709-0. [Google Scholar]
- Kuipery, A.; Gehring, A.J.; Isogawa, M. Mechanisms of HBV Immune Evasion. Antivir. Res. 2020, 179, 104816. [Google Scholar] [CrossRef]
- You, Q.; Cheng, L.; Kedl, R.M.; Ju, C. Mechanism of T Cell Tolerance Induction by Murine Hepatic Kupffer Cells. Hepatology 2008, 48, 978–990. [Google Scholar] [CrossRef]
- Diehl, L.; Schurich, A.; Grochtmann, R.; Hegenbarth, S.; Chen, L.; Knolle, P.A. Tolerogenic Maturation of Liver Sinusoidal Endothelial Cells Promotes B7-Homolog 1-Dependent CD8+ T Cell Tolerance. Hepatology 2008, 47, 296–305. [Google Scholar] [CrossRef]
- Xu, L.; Yin, W.; Sun, R.; Wei, H.; Tian, Z. Kupffer Cell-Derived IL-10 Plays a Key Role in Maintaining Humoral Immune Tolerance in Hepatitis B Virus-Persistent Mice. Hepatology 2014, 59, 443–452. [Google Scholar] [CrossRef]
- Zhou, J.; Peng, H.; Li, K.; Qu, K.; Wang, B.; Wu, Y.; Ye, L.; Dong, Z.; Wei, H.; Sun, R.; et al. Liver-Resident NK Cells Control Antiviral Activity of Hepatic T Cells via the PD-1-PD-L1 Axis. Immunity 2019, 50, 403–417.e4. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Tian, Z. Liver-Mediated Adaptive Immune Tolerance. Front. Immunol. 2019, 10, 2525. [Google Scholar] [CrossRef]
- Wu, L.; Peng, W.-H.; Wu, H.-L.; Miaw, S.-C.; Yeh, S.-H.; Yang, H.-C.; Liao, P.-H.; Lin, J.-S.; Chen, Y.; Hong, Y.-T.; et al. Lymphocyte Antigen 6 Complex, Locus C+ Monocytes and Kupffer Cells Orchestrate Liver Immune Responses against Hepatitis B Virus in Mice. Hepatology 2019, 69, 2364–2380. [Google Scholar] [CrossRef] [PubMed]
- David, B.A.; Rubino, S.; Moreira, T.G.; Freitas-Lopes, M.A.; Araújo, A.M.; Paul, N.E.; Rezende, R.M.; Menezes, G.B. Isolation and High-Dimensional Phenotyping of Gastrointestinal Immune Cells. Immunology 2017, 151, 56–70. [Google Scholar] [CrossRef]
- Fang, X.; Du, P.; Liu, Y.; Tang, J. Efficient Isolation of Mouse Liver NKT Cells by Perfusion. PLoS ONE 2010, 5, e10288. [Google Scholar] [CrossRef]
- Finlon, J.M.; Burchill, M.A.; Tamburini, B.A.J. Digestion of the Murine Liver for a Flow Cytometric Analysis of Lymphatic Endothelial Cells. J. Vis. Exp. 2019, 143, e58621. [Google Scholar] [CrossRef]
- Li, P.; Li, J.; Li, M.; Gong, J.; He, K. An Efficient Method to Isolate and Culture Mouse Kupffer Cells. Immunol. Lett. 2014, 158, 52–56. [Google Scholar] [CrossRef] [PubMed]
- Lynch, R.W.; Hawley, C.A.; Pellicoro, A.; Bain, C.C.; Iredale, J.P.; Jenkins, S.J. An Efficient Method to Isolate Kupffer Cells Eliminating Endothelial Cell Contamination and Selective Bias. J. Leukoc. Biol. 2018, 104, 579–586. [Google Scholar] [CrossRef]
- Shi, W.; Wang, Y.; Zhang, C.; Jin, H.; Zeng, Z.; Wei, L.; Tian, Y.; Zhang, D.; Sun, G. Isolation and Purification of Immune Cells from the Liver. Int. Immunopharmacol. 2020, 85, 106632. [Google Scholar] [CrossRef]
- Mohar, I.; Brempelis, K.J.; Murray, S.A.; Ebrahimkhani, M.R.; Crispe, I.N. Isolation of Non-Parenchymal Cells from the Mouse Liver. In Malaria Vaccines. Methods in Molecular Biology; Vaughan, A., Ed.; Humana Press: New York, NY, USA, 2015; Volume 1325, pp. 3–17. ISBN 9781493928149. [Google Scholar]
- Cabral, F.; Miller, C.M.; Kudrna, K.M.; Hass, B.E.; Daubendiek, J.G.; Kellar, B.M.; Harris, E.N. Purification of Hepatocytes and Sinusoidal Endothelial Cells from Mouse Liver Perfusion. J. Vis. Exp. 2018, 132, 56993. [Google Scholar] [CrossRef]
- Aparicio-Vergara, M.; Tencerova, M.; Morgantini, C.; Barreby, E.; Aouadi, M. Isolation of Kupffer Cells and Hepatocytes from a Single Mouse Liver. In Alpha-1 Antitrypsin Deficiency. Methods in Molecular Biology; Borel, F., Mueller, C., Eds.; Humana Press: New York, NY, USA, 2017; Volume 1639, pp. 161–171. ISBN 9781493971633. [Google Scholar]
- Sulen, A. Liver Macrophage Isolation by Flow Cytometry Sorting. In Kupffer Cells; Aouadi, M., Azzimato, V., Eds.; Humana Press: New York, NY, USA, 2020; Volume 2164, pp. 15–20. ISBN 9781071607046. [Google Scholar]
- Wei, C.; Ni, C.; Song, T.; Liu, Y.; Yang, X.; Zheng, Z.; Jia, Y.; Yuan, Y.; Guan, K.; Xu, Y.; et al. The Hepatitis B Virus X Protein Disrupts Innate Immunity by Downregulating Mitochondrial Antiviral Signaling Protein. J. Immunol. 2010, 185, 1158–1168. [Google Scholar] [CrossRef] [PubMed]
- Gondois-Rey, F.; Granjeaud, S.; Kieu, S.L.T.; Herrera, D.; Hirsch, I.; Olive, D. Multiparametric Cytometry for Exploration of Complex Cellular Dynamics. Cytom. Part A 2012, 81, 332–342. [Google Scholar] [CrossRef] [PubMed]
- Ferrer-Font, L.; Pellefigues, C.; Mayer, J.U.; Small, S.J.; Jaimes, M.C.; Price, K.M. Panel Design and Optimization for High-Dimensional Immunophenotyping Assays Using Spectral Flow Cytometry. Curr. Protoc. Cytom. 2020, 92, e70. [Google Scholar] [CrossRef]
- Giladi, A.; Paul, F.; Herzog, Y.; Lubling, Y.; Weiner, A.; Yofe, I.; Jaitin, D.; Cabezas-Wallscheid, N.; Dress, R.J.; Ginhoux, F.; et al. Single-Cell Characterization of Haematopoietic Progenitors and Their Trajectories in Homeostasis and Perturbed Haematopoiesis. Nat. Cell Biol. 2018, 20, 836–846. [Google Scholar] [CrossRef] [PubMed]
- Jaitin, D.A.; Kenigsberg, E.; Keren-Shaul, H.; Elefant, N.; Paul, F.; Zaretsky, I.; Mildner, A.; Cohen, N.; Jung, S.; Tanay, A.; et al. Massively Parallel Single-Cell RNA-Seq for Marker-Free Decomposition of Tissues into Cell Types. Science 2014, 343, 776–779. [Google Scholar] [CrossRef]
- Miltenyi Biotec Liver Dissociation Kit Mouse Protocol. Available online: https://www.miltenyibiotec.com/US-en/products/liver-dissociation-kit-mouse.html?countryRedirected=1#gref (accessed on 17 February 2021).
- Choi, W.-M.; Eun, H.S.; Lee, Y.-S.; Kim, S.J.; Kim, M.-H.; Lee, J.-H.; Shim, Y.-R.; Kim, H.-H.; Kim, Y.E.; Yi, H.-S.; et al. Experimental Applications of in Situ Liver Perfusion Machinery for the Study of Liver Disease. Mol. Cells 2019, 42, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Rogers, A.B. Stress of Strains: Inbred Mice in Liver Research. Gene Expr. 2018, 19, 61–67. [Google Scholar] [CrossRef]
- Sellers, R.S.; Clifford, C.B.; Treuting, P.M.; Brayton, C. Immunological Variation between Inbred Laboratory Mouse Strains: Points to Consider in Phenotyping Genetically Immunomodified Mice. Vet. Pathol. 2012, 49, 32–43. [Google Scholar] [CrossRef]
- Medina-Montano, C.; Cacicedo, M.L.; Svensson, M.; Limeres, M.J.; Zeyn, Y.; Chaves-Giraldo, J.E.; Röhrig, N.; Grabbe, S.; Gehring, S.; Bros, M. Enrichment Methods for Murine Liver Non-Parenchymal Cells Differentially Affect Their Immunophenotype and Responsiveness towards Stimulation. Int. J. Mol. Sci. 2022, 23, 6543. [Google Scholar] [CrossRef]
- Canè, S.; Ugel, S.; Trovato, R.; Marigo, I.; De Sanctis, F.; Sartoris, S.; Bronte, V. The Endless Saga of Monocyte Diversity. Front. Immunol. 2019, 10, 1786. [Google Scholar] [CrossRef] [Green Version]
- Lee, P.Y.; Wang, J.-X.; Parisini, E.; Dascher, C.C.; Nigrovic, P.A. Ly6 Family Proteins in Neutrophil Biology. J. Leukoc. Biol. 2013, 94, 585–594. [Google Scholar] [CrossRef] [PubMed]
- Deniset, J.F.; Kubes, P. Neutrophil Heterogeneity: Bona Fide Subsets or Polarization States? J. Leukoc. Biol. 2018, 103, 829–838. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Liu, X.; Li, Z.; Chai, Y.; Jiang, Y.; Wang, Q.; Ji, Y.; Zhu, Z.; Wan, Y.; Yuan, Z.; et al. CD8+NKT-like Cells Regulate the Immune Response by Killing Antigen-Bearing DCs. Sci. Rep. 2015, 5, 14124. [Google Scholar] [CrossRef]
- Li, Z.; Wu, Y.; Wang, C.; Zhang, M. Mouse CD8+NKT-like Cells Exert Dual Cytotoxicity against Mouse Tumor Cells and Myeloid-Derived Suppressor Cells. Cancer Immunol. Immunother. 2019, 68, 1303–1315. [Google Scholar] [CrossRef] [PubMed]
- Pimkova Polidarova, M.; Brehova, P.; Dejmek, M.; Birkus, G.; Brazdova, A. STING Agonist-Mediated Cytokine Secretion Is Accompanied by Monocyte Apoptosis. ACS Infect. Dis. 2022, 8, 463–471. [Google Scholar] [CrossRef] [PubMed]
- Pfeiffer, E.; Kegel, V.; Zeilinger, K.; Hengstler, J.G.; Nüssler, A.K.; Seehofer, D.; Damm, G. Featured Article: Isolation, Characterization, and Cultivation of Human Hepatocytes and Non-Parenchymal Liver Cells. Exp. Biol. Med. 2015, 240, 645–656. [Google Scholar] [CrossRef] [Green Version]



| Cell Staining (cat. no.) | Clone | Dilution | Isotype Controls (cat. no.) | Manufacturer | Staining Buffer | FC Compensations |
|---|---|---|---|---|---|---|
| rat anti-mouse CD3 BV605 (564009) | 17A2 | 1/100 | BV605 Rat IgG2b, κ (563145) | BD Biosciences | FC | CompBead Anti-Rat and Anti-Hamster Ig κ/Negative Control Compensation Particles Set (552845) |
| rat anti-mouse CD4 BV421(562891) | GK1.5 | 1/50 | BV421 Rat IgG2b, κ (562603) | BD Biosciences | FC | |
| rat anti-mouse CD8 BV510 (563068) | 53-6.8 | 1/50 | BV510 Rat IgG2a, κ (562952) | BD Biosciences | FC | |
| rat anti-mouse CD11b APC-R700 (564985) | M1/71 | 1/100 | APC-R700 Rat IgG2b, κ (564984) | BD Biosciences | FC | |
| hamster anti-mouse CD11c PE-CF594 (565591) | N418 | 1/50 | PE-CF594 Hamster IgG2, λ1 | BD Biosciences | FC | |
| rat anti-mouse CD19 BUV395 (563557) | 1D3 | 1/100 | BUV395 Rat IgG2a, κ (563556) | BD Biosciences | FC | |
| rat anti-mouse Ly-6C PE (560592) | AL-22 | 1/50 | PE Rat IgM, κ (553943) | BD Biosciences | FC | |
| rat anti-mouse Ly-6G APC (560599) | 1A8 | 1/50 | APC Rat IgG2a κ (553932) | BD Biosciences | FC | |
| rat anti-mouse CD45 PerCP (561047) | 30-F11 | 1/100 | PerCP Rat IgG2b, κ (552991) | BD Biosciences | FC | |
| rat anti-mouse CD49b FITC (553857) | DX5 | 1/100 | FITC Rat IgM, κ (553942) | BD Biosciences | FC | |
| rat anti-mouse CD31 BUV496 (741084) | 390 | 1/100 | BUV496 Rat IgG2a, κ (564663) | BD Biosciences | FC | |
| rat anti-mouse F4/80 BV650 (743282) | T45-2342 | 1/50 | BV650 Rat IgG2a, κ (563236) | BD Biosciences | FC | |
| live/dead marker Zombie NIR (423106) | n/a * | 1/200 | n/a * | Biolegend | PBS | cells |
| Immune Population | Immunophenotype |
|---|---|
| liver endothelial cells | CD31+ CD45− |
| hematopoietic cells (leukocytes) | CD31− CD45+ |
| T cells | CD31− CD45+ CD3+ CD49b− |
| helper T cells | CD31− CD45+ CD3+ CD49b− CD4+ CD8− |
| cytotoxic T cells | CD31− CD45+ CD3+ CD49b− CD4− CD8+ |
| B cells | CD31− CD45+ CD3− CD49b− CD19+ |
| NK cells | CD31− CD45+ CD3− CD49b+ |
| NKT cells | CD31− CD45+ CD3+ CD49b+ |
| neutrophils | CD31− CD45+ CD3− CD19− CD49b− CD11b+ Ly6G+ |
| reparative monocytes | CD31− CD45+ CD3− CD19− CD49b− CD11b+ Ly6G− Ly6Clo |
| inflammatory monocytes | CD31− CD45+ CD3− CD19− CD49b− CD11b+ Ly6G− Ly6Chi |
| CD8 cDC1 | CD31− CD45+ CD3− CD19− CD49b− CD11c+ CD8+ CD11b− Ly6C− |
| CD11b cDC2 | CD31− CD45+ CD3− CD19− CD49b− CD11c+ CD8− CD11b+ Ly6C− |
| KC | CD31− CD45+ CD3− CD19− CD49b− CD11blo F4/80hi |
| macrophages | CD31− CD45+ CD3− CD19− CD49b− CD11b+ F4/80+ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vanekova, L.; Polidarova, M.P.; Veverka, V.; Birkus, G.; Brazdova, A. Multiparametric Flow Cytometry-Based Immunophenotyping of Mouse Liver Immune Cells. Methods Protoc. 2022, 5, 70. https://doi.org/10.3390/mps5050070
Vanekova L, Polidarova MP, Veverka V, Birkus G, Brazdova A. Multiparametric Flow Cytometry-Based Immunophenotyping of Mouse Liver Immune Cells. Methods and Protocols. 2022; 5(5):70. https://doi.org/10.3390/mps5050070
Chicago/Turabian StyleVanekova, Lenka, Marketa Pimkova Polidarova, Vaclav Veverka, Gabriel Birkus, and Andrea Brazdova. 2022. "Multiparametric Flow Cytometry-Based Immunophenotyping of Mouse Liver Immune Cells" Methods and Protocols 5, no. 5: 70. https://doi.org/10.3390/mps5050070
APA StyleVanekova, L., Polidarova, M. P., Veverka, V., Birkus, G., & Brazdova, A. (2022). Multiparametric Flow Cytometry-Based Immunophenotyping of Mouse Liver Immune Cells. Methods and Protocols, 5(5), 70. https://doi.org/10.3390/mps5050070








