HiChIP and Hi-C Protocol Optimized for Primary Murine T Cells
Abstract
:1. Introduction
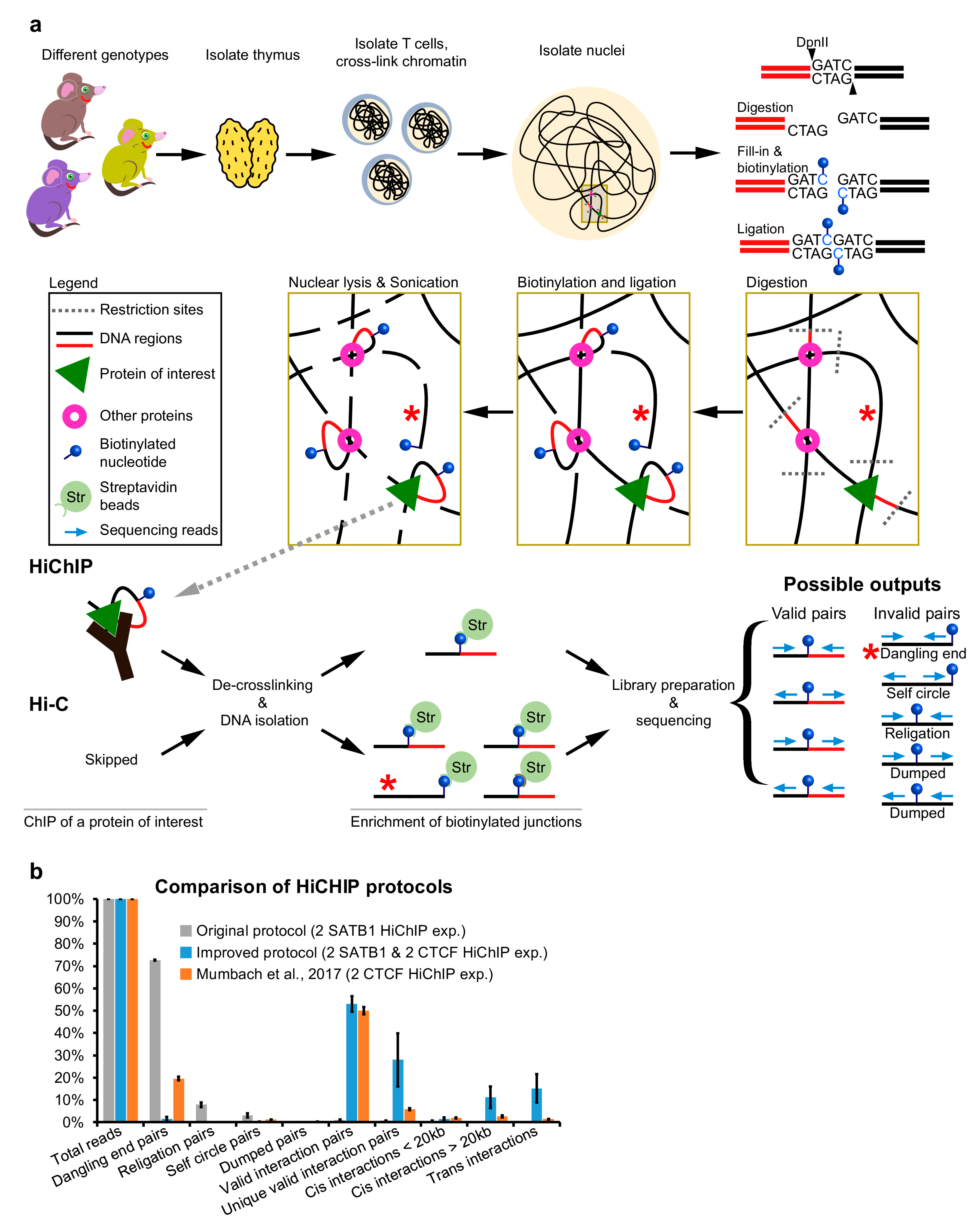
2. Experimental Design
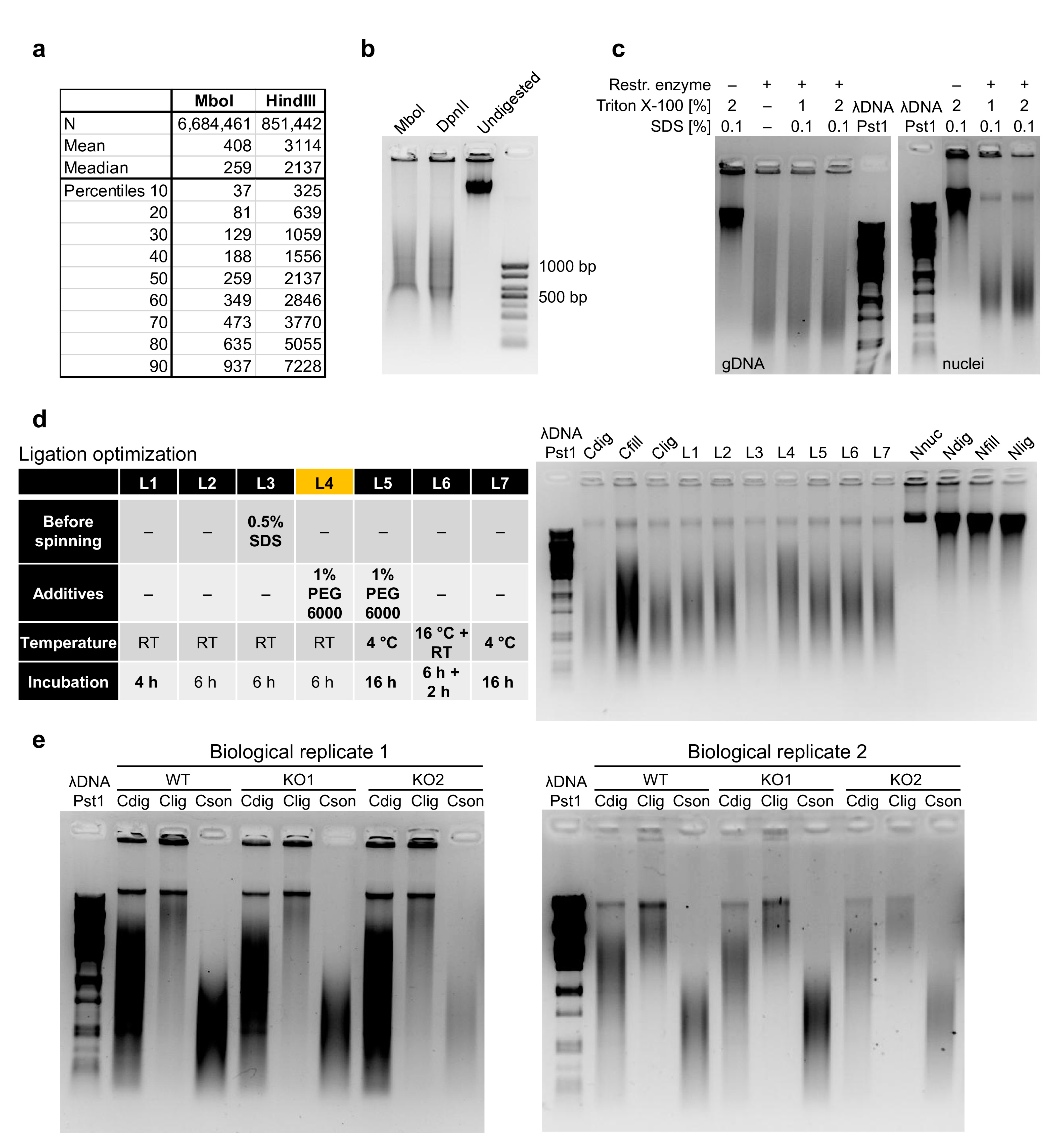
2.1. Cell Lysis and Digestion
2.2. Fill-in, Biotinylation and Ligation
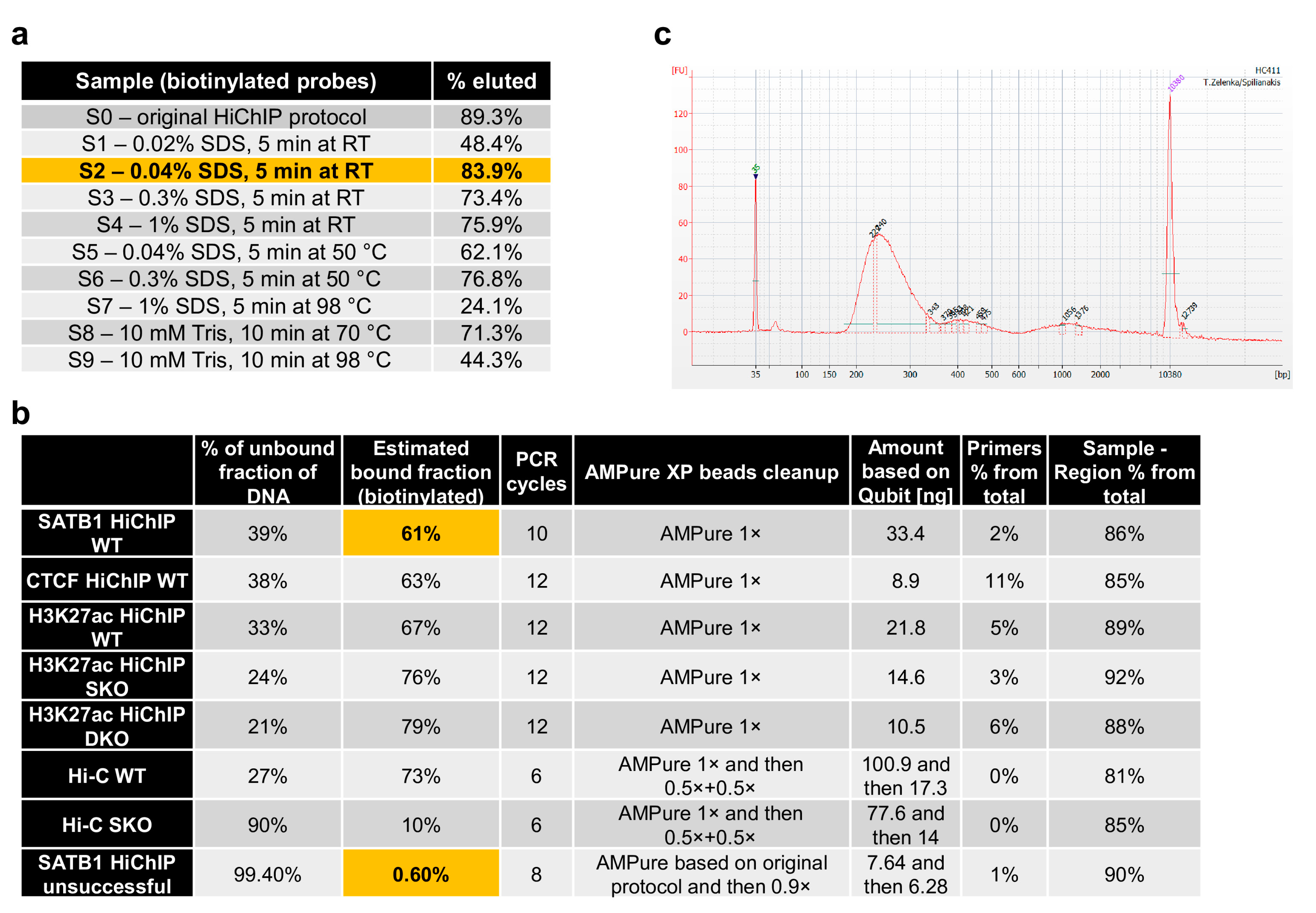
2.3. Optimization of DNA Library Preparation
2.4. Chemicals and Enzymes
- PBS (Sigma-Aldrich, St. Louis, MO, USA, cat. no. P4417)
- 16% paraformaldehyde aqueous solution (Electron Microscopy Sciences, Hatfield, PA, USA, cat. no. 15710)
- NaCl (Merck, Kenilworth, NJ, USA, cat. no. 106404)
- EDTA (Sigma-Aldrich, St. Louis, MO, USA, cat. no. E6511)
- EGTA (Sigma-Aldrich, St. Louis, MO, USA, cat. no. E8145)
- Hepes (Sigma-Aldrich, St. Louis, MO, USA, cat. no. 54457)
- Glycine (PanReac AppliChem, Darmstadt, Germany, cat. no. A1067)
- Tris (PanReac AppliChem, Darmstadt, Germany, cat. no. A1086)
- NP-40 (Sigma-Aldrich, St. Louis, MO, USA, cat. no. 74385)
- Protease inhibitors (custom-made mixture, n/a)
- Phosphatase inhibitors (custom-made mixture, n/a)
- Trypan blue solution (Merck, Kenilworth, NJ, USA, cat. no. 93595)
- SDS (prepare 20% stock) (Sigma-Aldrich, St. Louis, MO, USA, cat. no. L5750)
- Triton X-100 (prepare 20% stock) (Merck, Kenilworth, NJ, USA, cat. no. 108603)
- PEG 6000 (prepare 30% stock) (Merck, Kenilworth, NJ, USA, cat. no. 528877)
- DpnII restriction enzyme (NEB, Ipswich, MA, USA, cat. no. R0543M)
- Biotin-16-dCTP (Jena Bioscience, Jena, Germany, cat. no. NU-809-BIO16-L)
- 10 mM dGTP (Promega, Madison, WI, USA, cat. no. U1240)
- 10 mM dTTP (Promega, Madison, WI, USA, cat. no. U1240)
- 10 mM dATP (Promega, Madison, WI, USA, cat. no. U1240)
- DNA Polymerase I, Large (Klenow) Fragment (NEB, Ipswich, MA, USA, cat. no. M0210L)
- 10× NEB T4 DNA Ligase Buffer with 10 mM ATP (NEB, Ipswich, MA, USA, cat. no. B0202)
- 20 mg/mL BSA (NEB, Ipswich, MA, USA, cat. no. B9000S)
- 400 U/μL T4 DNA Ligase (NEB, Ipswich, MA, USA, cat. no. M0202L)
- RNase A (Qiagen, Hilden, Germany, cat. no. 1007885)
- Proteinase K (Sigma-Aldrich, St. Louis, MO, USA, cat. no. P2308)
- Phenol solution (Merck, Kenilworth, NJ, USA, cat. no. P4557)
- Chloroform (Merck, Kenilworth, NJ, USA, cat. no. 102445)
- Isoamylalcohol (Merck, Kenilworth, NJ, USA, cat. no. 100979)
- Dynabeads™ Protein A (Invitrogen, Waltham, MA, USA, cat. no. 10002D)
- Dynabeads™ Protein G (Invitrogen, Waltham, MA, USA, cat. no. 10004D)
- ChIP DNA Clean and Concentrator kit (Zymo, Irvine, CA, USA, cat. no. D5205)
- Custom-made anti-SATB1 (Davids Biotechnologie, Regensburg, Germany, cat. no. n/a)
- Anti-H3K27ac (Abcam, Cambridge, United Kingdom, cat. no. ab4729)
- Anti-CTCF (Abcam, Cambridge, United Kingdom, cat. no. ab70303)
- Sodium deoxycholate (Sigma-Aldrich, St. Louis, MO, USA, cat. no. D6750)
- LiCl (Sigma-Aldrich, St. Louis, MO, USA, cat. no. L7026)
- Qubit™ dsDNA HS Assay Kit (Invitrogen, Waltham, MA, USA, cat. no. Q32851)
- Streptavidin C-1 beads (Invitrogen, Waltham, MA, USA, cat. no. 65002)
- Tween-20 (Merck, Kenilworth, NJ, USA, cat. no. 822184)
- N,N-Dimethylformamide (Sigma-Aldrich, St. Louis, MO, USA, cat. no. D4551)
- MgCl2 (Ambion, Austin, TX, USA, cat. no. AM9530G)
- Nextera DNA Sample Preparation Kit (Illumina, San Diego, CA, USA, cat. no. FC-121-1030)
- Nextera DNA Sample Preparation INDEX Kit (Illumina, San Diego, CA, USA, cat. no. FC-121-1011)
- Phusion® High-Fidelity PCR Master Mix with HF Buffer (NEB, Ipswich, MA, USA, cat. no. M0531S)
- AMPure XP beads (Beckman Coulter Genomics, Chaska, MN, USA, cat. no. A63880)
3. Procedure
3.1. Cell Preparation (Required Time: 1.5 h)
- Isolate the thymus from freshly euthanized animals and homogenize it to prepare a single-cell suspension in 10 mL of 1× PBS. Homogenization can be achieved, for example, by rubbing the tissue by a syringe pestle against a 40 µm cell strainer (Falcon, 352340) to simultaneously separate and filter the cells.
- Spin down the cells at 500× g at 4 °C for 5 min and wash them twice with 10 mL of 1× PBS.
- Count the cells (for example, using a hemocytometer or an automated cell counter) and use 10 million cells per HiChIP experiment or 5 million cells per Hi-C experiment. Bring up the volume to 10 mL with 1× PBS and resuspend the cells well.
- Add 1 mL of Fixation Buffer (11% Formaldehyde (methanol free), 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA and 50 mM Hepes pH 8.0).
- Incubate the samples for 10 min at room temperature while rocking.
- Quench the reaction by adding Glycine to 0.2 M final concentration (870 μL, 2.5 M stock), mix well and incubate at room temperature for 5 min.
- Spin down the cells at 1000× g at 4 °C for 5 min.
- Discard the supernatant, resuspend the cell pellet in 1 mL ice-cold 1× PBS and transfer the solution into a fresh Eppendorf tube.
- Spin down the cells at 1000× g at 4 °C for 5 min.
- Repeat the wash (steps 8–9) one more time and discard the supernatant.
 . PAUSE STEP Either proceed with the protocol or flash-freeze the cell pellets in liquid nitrogen (or a dry ice/ethanol bath) and store them at –80 °C. We have compared both approaches, and freezing the cells did not affect the efficiency of subsequent steps.
. PAUSE STEP Either proceed with the protocol or flash-freeze the cell pellets in liquid nitrogen (or a dry ice/ethanol bath) and store them at –80 °C. We have compared both approaches, and freezing the cells did not affect the efficiency of subsequent steps.3.2. Lysis and Restriction Enzyme Digestion (Required Time: 20 h)
- 11.
- Resuspend the pellets of 10 million cells per HiChIP sample (or 5 million cells for Hi-C) in 500 μL of ice-cold Hi-C Lysis Buffer (10 mM Tris-HCl pH 8.0, 10 mM NaCl, 0.2% NP-40 and 1× Protease Inhibitors) and rotate them at 4 °C for 1–1.5 h.
 . CRITICAL STEP Efficient preparation of nuclei is critical, so we advise monitoring cell lysis under a microscope upon Trypan blue staining, which should not stain the cells with an intact cellular membrane. Extended cellular lysis up to 4 h did not negatively affect the experiment, unlike the insufficient lysis.
. CRITICAL STEP Efficient preparation of nuclei is critical, so we advise monitoring cell lysis under a microscope upon Trypan blue staining, which should not stain the cells with an intact cellular membrane. Extended cellular lysis up to 4 h did not negatively affect the experiment, unlike the insufficient lysis.- 12.
- Spin down the nuclei at 2500× g at 4 °C for 5 min and discard the supernatant.
- 13.
- Wash the pellet once with 500 μL of ice-cold Hi-C Lysis Buffer.
- 14.
- Remove the supernatant and gently resuspend the pellet by pipetting up and down in 100 μL of 0.5% SDS.
- 15.
- Incubate at 62 °C for 10 min and then add 300 μL of ddH2O and 50 μL of 20% Triton X-100 to quench the SDS.
- 16.
- Mix by pipetting and incubate at 37 °C for 15 min.
- 17.
- Add 50 μL of 10× DpnII Buffer and check the pH with 4 μL spotted on a pH-indicator paper. If it is in the range of pH 6–7, you can proceed; otherwise, fix the pH with the addition of Tris-HCl pH 8.0.
 . CRITICAL STEP We noticed that older batches of Triton X-100 tend to have low pH, which negatively affects the enzymatic reactions.
. CRITICAL STEP We noticed that older batches of Triton X-100 tend to have low pH, which negatively affects the enzymatic reactions.- 18.
- Add 4 μL (200 U) of DpnII restriction enzyme (NEB, R0543M—50 U/μL), and digest chromatin for 16 h at 37 °C, while shaking (300 rpm).
- 19.
- Heat inactivate DpnII at 62 °C, for 20 min and let it cool down to room temperature.
- 20.
- Resuspend the nuclei and split each HiChIP sample into two halves, each in a different tube. Do not split the Hi-C samples. Keep them as they are.
- 21.
- Spin down the nuclei at 2500× g at 4 °C for 5 min and discard the supernatant.
3.3. Fill-in Reaction and Proximity Ligation (Required Time: 7.5 h)
- 22.
- Resuspend the nuclei in 300 μL of Fill-in Master Mix according to the Table 1 below.
- 23.
- Mix by pipetting and incubate at 37 °C for 30 min while shaking (300 rpm).
- 24.
- Merge the two halves of each HiChIP sample back together (does not apply to Hi-C), mix it carefully by pipetting and keep 15 μL as the control C1.
- 25.
- Add 15 μL of 20% SDS (final 0.5%) to inactivate the Klenow enzyme and mix carefully by pipetting.
- 26.
- Add 32 μL of 20% Triton X-100 (final 1%) to quench the SDS. This makes the nuclear pellet visible (translucent).
- 27.
- Incubate at 37 °C for 5 min.
- 28.
- Split the HiChIP samples into two halves again, before ligation (do not split the Hi-C sample).
- 29.
- Spin down the nuclei at 2500× g at 4 °C for 10 min and discard the supernatant.
- 30.
- Resuspend the nuclei in 1200 μL of Ligation Master Mix according to Table 2 below.
- 31.
- Incubate the reaction for 6 h at room temperature with rotation. In the meanwhile, for the HiChIP experiment, start the incubation of antibodies with the magnetic Protein A/G DynaBeads (needed for step 41).
- 32.
- Merge the two halves of the HiChIP samples back together (does not apply to Hi-C), mix carefully by pipetting and take 120 μL as the control C2.
- 33.
- Spin down the nuclei at 2400× g at 4 °C for 15 min and discard the supernatant.
- 34.
- To further decrease the number of dangling ends, we recommend implementing the removal of unligated, biotinylated ends [17]. For HiChIP libraries, it can either be performed right after the ligation in T4 ligation buffer (step 32) or after nuclei spin down (at this step, in its dedicated T4 ligase reaction buffer). For the reaction, add only dGTP and dATP nucleotides and T4 DNA polymerase, which will naturally manifest its 3′-5′ exonuclease activity and remove the unligated 3′ biotin-dCTP. Alternatively, this step can be performed after DNA purification (step 67).
 PAUSE STEP Either proceed with the protocol or flash-freeze the pellets of nuclei in liquid nitrogen (or dry ice/ethanol bath) and store them at –80 °C.
PAUSE STEP Either proceed with the protocol or flash-freeze the pellets of nuclei in liquid nitrogen (or dry ice/ethanol bath) and store them at –80 °C.3.4. Sonication (Required Time: 1 h)
- 35.
- Resuspend the HiChIP pellets in 60 μL (and Hi-C pellets in 30 μL) of Sonication Buffer (1% SDS, 50 mM Tris-HCl pH 8.0, 20 mM EDTA and 1× Protease Inhibitors), pipette up and down 20 times to mix and incubate at room temperature for 15 min. Try to avoid air bubbles.
- 36.
- Add 540 μL of cold 1× TE buffer (pH 8.0) into HiChIP samples (270 μL for Hi-C) and keep close to ice.
 CRITICAL STEP SDS will start precipitating when on ice, so try to avoid it by controlling the sample and putting it on and off the ice.
CRITICAL STEP SDS will start precipitating when on ice, so try to avoid it by controlling the sample and putting it on and off the ice.- 37.
- Sonicate with a Labsonic M—Tip sonicator (or any other sonicator, based on your optimized conditions) for 1.5 min (3 cycles with 30 s ON/OFF, 40% Power). Sonication time and power may be optimized based on the cell type and instrument used.
- 38.
- Transfer the sample in a 1.5 mL Eppendorf tube and spin at 20,000× g at 4 °C for 20 min. Carefully collect the supernatant into a clean tube.
- 39.
- In HiChIP experiments, transfer 50 μL into a new tube as the control C3.
- 40.
- For solely Hi-C experiments, use the entire volume to purify the DNA in the Elution section (step 60). Alternatively, use 50 μL for Hi-C and use the rest for the HiChIP experiment.
3.5. HiChIP Samples—Immunoprecipitation (Required Time: 16 h)
- 41.
- For HiChIP samples, start the incubation of the magnetic Protein A/G DynaBeads with antibodies earlier, optimally during the ligation step (step 31).
- 42.
- Wash aliquots of 60 μL (or 30 μL + 30 μL, when bought separately) of Protein A/G DynaBeads for each sample, three times with 0.5 mL cold BSA/1×PBS (1 mg/mL BSA in 1× PBS).
- 43.
- Perform the washes as follows: Add the solution. Remove the tubes from the magnet and invert them several times to resuspend the beads. Place the tubes on the magnet and collect the beads for 2 min. Remove the supernatant. Ideally, perform all steps with the magnetic rack on ice.
- 44.
- Resuspend the beads in 1000 μL of BSA/1×PBS + 1× Protease Inhibitors in Protein LoBind tubes.
- 45.
- Add the antibodies of interest based on your experimental needs. The amounts range based on the type and quality of the antibody and other factors as well, so we recommend running some optimization experiments first. We used 8 μg of custom-made anti-SATB1, 2 µg of anti-H3K27ac (Abcam, Cambridge, United Kingdom, cat. no. ab4729) and 7 µg of anti-CTCF (Abcam, Cambridge, United Kingdom, cat. no. ab70303) antibodies.
- 46.
- Incubate the beads with the antibodies for 4–6 h at 4 °C with rotation.
- 47.
- Wash the antibody-coupled beads three times with 0.5 mL of ice-cold BSA/1×PBS, and if needed, you can leave them on ice until ready.
- 48.
- After the sonication step, add 20% Triton X-100 into each tube. Choose the volume depending on the sample volume to obtain a final concentration of 1%.
- 49.
- Mix well and incubate at 37 °C for 15 min.
- 50.
- Add an equal volume (sample + Triton) of 2× ChIP Binding Buffer (20 mM Tris-pH 8, 2 mM EDTA, 0.2% sodium deoxycholate and 2× Protease + Phosphatase Inhibitors).
- 51.
- Combine the chromatin samples with the washed antibody-coupled beads and transfer everything into DNA LoBind tubes.
- 52.
- Incubate at 4 °C for 12–16 h with rotation.
- 53.
- OPTIONAL STEP You can also prepare 15 μL of Protein A/G DynaBeads to preclear chromatin before the immunoprecipitation step. In this case, combine washed preclearing beads with chromatin in ChIP Binding Buffer in DNA LoBind tubes and rotate the samples at 4 °C for 1 h. Collect the preclearing beads toward a magnet and combine the supernatant (with the precleared chromatin) with the washed antibody-coupled beads. Transfer everything into a new DNA LoBind tube and incubate at 4 °C for 12–16 h with rotation.
- 54.
- Wash the beads with the precipitated chromatin 4–7 times (depending on how much background you are getting, start with four washes) with ice-cold RIPA Buffer (50 mM Hepes pH 8.0, 1% NP-40, 0.70% sodium deoxycholate, 0.5 M LiCl, 1 mM EDTA and 1× Protease Inhibitors). Perform the washes as follows:
- 55.
- Place the tubes on a magnetic rack on ice. Once the beads have been collected toward the magnet, rotate the tube swiftly and let the beads travel from one side of the tube to the other. Repeat a few times, then aspirate the supernatant without disturbing the beads and repeat the washes as required.
- 56.
- Wash once with 1 mL ice-cold TE buffer (pH 8.0).
- 57.
- Resuspend the beads in 1 mL TE buffer (pH 8.0) and transfer the samples into a NEW tube.
- 58.
- Collect the beads toward the magnetic stand, discard the supernatant and continue with the next section.
3.6. Elution of Immune Complexes and DNA Isolation (Required Time: 20 h)
- 59.
- There are three control samples: C1 (15 μL), C2 (120 μL) and C3 (50 μL) previously collected. Bring the volume up to 120 μL with 1× TE buffer (pH 8.0); i.e., 105, 0 and 70 μL, respectively.
- 60.
- Split the Hi-C samples into two tubes, containing ~120–140 μL each. If you have lower volumes, bring it up to 120 μL with 1× TE buffer (pH 8.0).
- 61.
- Add into each Hi-C sample, onto the HiChIP beads and also in the control samples, 125 μL of Hi-C Elution Buffer (10 mM Tris-HCl pH 8.0, 5 mM EDTA, 300 mM NaCl and 1% SDS), mix by pipetting and incubate at 65 °C for 6–16 h.
- 62.
- Add to HiChIP samples 110 μL of TE Buffer (add 115–95 μL into Hi-C samples for a total volume of 235 μL), mix by pipetting and collect the beads toward a magnetic stand. Transfer the supernatant into a fresh tube.
- 63.
- Add, into both control and HiChIP samples, 10 μL of RNase A (10 mg/mL) and incubate at 37 °C for 30 min.
- 64.
- Add 8 μL of Proteinase K (10 mg/mL stock solution) and incubate for 2–3 h at 55 °C.
- 65.
- Clean up the samples (~250 μL) using the ChIP DNA Clean and Concentrator kit (Zymo, D5205) or by the traditional Phenol-Chloroform-Isoamyl alcohol (25:24:1) method.
- 66.
- When using the kit, follow the manufacturer’s instructions. Briefly, apply five volumes of ChIP DNA Binding Buffer and re-load the column 2–3 times since it has a smaller capacity. Elute the DNA by applying 6 μL (12 μL for Hi-C) of pre-warmed Elution Buffer directly onto the center of the column matrix and incubate at room temperature for 1 minute. Spin down the tube and repeat the elution step one more time to obtain 12 μL (24 μL for Hi-C) in total.
- 67.
- Use 2 μL of each sample to quantify DNA on Qubit using the dsDNA HS Assay Kit and load 800–1000 ng of control samples on 1.5% TBE agarose gel to qualitatively review the prepared DNA libraries (see Figure 2c).
- 68.
- Additionally, you can use a part of the C3 control sample to set up a control PCR reaction. Ligation of two DpnII blunt DNA ends produces a novel restriction site recognized by the restriction enzyme ClaI [17]. Hence primers designed for two adjacent restriction fragments with a high likelihood to be ligated together can be used to amplify the ligated molecule. If the fragments were properly digested, filled in and re-ligated, then they should be digested by ClaI. The ratio between the digested and undigested fragments provides another piece of evidence about the protocol’s efficiency.
 PAUSE STEP Purified DNA can be stored long-term at –20 °C.
PAUSE STEP Purified DNA can be stored long-term at –20 °C.3.7. Biotin Pull-Down and Preparation of DNA Libraries for Illumina Sequencing (Required Time: 1.5 h)
- 69.
- Use all the material left for HiChIP samples and 100 ng of Hi-C samples.
- 70.
- Bring up the volume of all the samples to 25 μL with ddH2O.
- 71.
- Prepare for biotin pull-down, for HiChIP, by washing 5 μL of Streptavidin C-1 beads with 500 μL of Tween Wash Buffer (5 mM Tris-HCl pH 7.5, 0.5 mM EDTA, 1 M NaCl and 0.05% Tween-20).
- 72.
- Resuspend the beads in 25 μL of 2× Biotin Binding Buffer (10 mM Tris-HCl pH 7.5, 1 mM EDTA and 2M NaCl) and add them to 25 μL of DNA samples.
- 73.
- Incubate at room temperature for 20 min, resuspending from time to time by pipetting up and down.
- 74.
- Separate the beads on a magnet and transfer the supernatant into a fresh tube.
- 75.
- Use 20 μL of the supernatant to measure DNA concentration using Qubit as quality control.
- 76.
- Wash the beads twice with 400 μL of Tween Wash Buffer and resuspend them at room temperature.
- 77.
- Wash the beads with 100 μL of 1× TD Buffer (10 mM Tris-HCl pH 7.5, 5 mM MgCl2 and 10% dimethylformamide).
- 78.
- Resuspend the beads in 25 μL of 2× TD Buffer, add 20–24 μL ddH2O (depending on the amount of Tn5 used, the total volume should be 50 μL) and add the appropriate amount of Tn5 enzyme from the Nextera DNA Sample Preparation Kit (Illumina, FC-121-1030) based on the amount of your precipitated material.
- 79.
- Adjust the amount of Tn5 linearly for different amounts of post-ChIP DNA. Use 1 μL for the lowest yields of ~5 ng of DNA and up to 5 μL for 100 ng of DNA.
- 80.
- Incubate at 55 °C for 10 min with interval pipetting up and down to resuspend.
- 81.
- Place samples on the magnet and remove the supernatant.
- 82.
- Resuspend in 300 μL of Strip Buffer (0.15% SDS, 10 mM Tris-HCl pH 8.0 and 50 mM EDTA) and incubate at room temperature for 5 min to strip off and deactivate Tn5.
- 83.
- Wash the beads once with 400 μL of Tween Wash Buffer.
- 84.
- Wash the beads once with 500 μL of 10 mM Tris-HCl (pH 8.0).
3.8. PCR and Post-PCR Size Selection (Required Time: 1.5 h)
- 85.
- Resuspend the beads in 50 μL of PCR master mix prepared according to Table 3. Due to different Nextera barcodes for each sample, it needs to be pipetted individually.
- 86.
- Run the PCR program according to the conditions described in Table 4 with the number of repeated cycles estimated based on the amount of post-ChIP DNA determined by Qubit. An accurate estimation of the number of PCR cycles is never an easy task. One option is to follow the recommendation from the original HiChIP paper: “Greater amount than 50 ng run for 5 cycles, approximately 50 ng for 6 cycles, 25 ng for 7 cycles, 12.5 ng for 8 cycles, etc” [9]. However, the quality of libraries is not reflected in the quantification, so this approach is really just an estimation (although often sufficient). In our laboratory, we tend to amplify a bit more, ranging between 8–14 cycles for 50–3 ng of post-ChIP DNA, respectively, in order to obtain a sufficient amount of DNA.
- 87.
- Place the amplified DNA libraries on a magnet and transfer the supernatant into new tubes.
- 88.
- Size selection should be performed based on your technical availability. In our laboratory, we use AMPure XP beads with success; hence, here, we describe their use.
- 89.
- Bring AMPure XP beads to room temperature.
- 90.
- There are usually no long DNA fragments in HiChIP libraries, so we usually perform only one-sided size selection. In some libraries, there are also long DNA fragments present. In that case, perform double-sided size selection. The ratios given here are approximate and based on our empirical experience. Each batch of beads may behave differently, so you may test out your own sample-to-bead ratios for one-sided and double-sided size selection on a 100 bp DNA ladder for agarose gels.
- 91.
- Use a 1× ratio of AMPure XP beads to the volume of your sample, i.e., put 50 μL of AMPure XP beads into 50 μL of DNA solution.
- 92.
- Mix it well on a vortex mixer or by pipetting up and down at least 10 times.
- 93.
- Incubate at room temperature for 5 min.
- 94.
- Place the tube on a magnetic rack to separate beads and remove the supernatant.
- 95.
- OPTIONAL STEP For double-sided size selection, first incubate with 0.5× sample volume of beads (25 μL) and keep the supernatant (discard the beads). Add to the supernatant 0.4× of the original sample volume (20 μL) of beads and incubate again. This time discard the supernatant and continue with bead wash.
- 96.
- Add 200 µL of 80% freshly prepared ethanol to the tube with the beads while on the magnetic stand.
- 97.
- Incubate at room temperature for 30 s and then carefully remove and discard the supernatant.
- 98.
- Repeat the previous washing step one more time.
- 99.
- Air-dry the beads with the cap open for a maximum of ~5 min. Do not overdry the beads/DNA as it may result in lower recovery of target DNA.
- 100.
- Elute DNA from the beads by adding 15 μL of pre-warmed Elution Buffer (Tris-HC pH 8.0).
- 101.
- Remove the tube from the magnet. Mix it well on a vortex mixer or by pipetting up and down.
- 102.
- Incubate at room temperature for 2 min.
- 103.
- Put the tube in a magnetic rack until the solution is clear, approximately 3–5 min.
- 104.
- Transfer the supernatant to a clean DNA LoBind tube.
- 105.
- Quantify 2 μL of the DNA libraries with Qubit and check 1 μL on an Agilent Bioanalyzer System.
4. Expected Results
5. Reagents Setup
5.1. Custom-Made Protease and Phosphatase
| Inhibitor of | Name | Time | Stock (c) |
| Proteases | Aprotinin | 300 nM | 300,000 |
| Proteases | Leupeptin | 10 μM | 10,000 |
| Proteases | Pepstatin A | 1 μM | 1000 |
| Proteases | EDTA | 10 mM | 500 |
| Proteases | PMSF | 1 mM | 100 |
| Phosphatases | NaF | 10 mM | 500 |
| Phosphatases | Na3VO4 (pH 10.0) | 2 mM | 200 |
5.2. Solutions
- Fixation buffer (11% formaldehyde, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 50 mM Hepes pH 8.0);
- Hi-C lysis buffer (10 mM Tris-HCl pH 8, 10 mM NaCl, 0.2% NP40, 1× Protease Inhibitors);
- 0.5% SDS;
- Sonication buffer (1% SDS, 50 mM Tris-HCl pH 8, 20 mM EDTA, 1× Protease Inhibitors);
- 1× TE (pH 8);
- Hi-C elution buffer (10 mM Tris-HCl pH 8, 5 mM EDTA, 300 mM NaCl, 1% SDS);
- BSA/1×PBS (1 mg/mL BSA (0.1%) in 1× PBS);
- 2× ChIP binding buffer (20 mM Tris-HCl pH 8, 2 mM EDTA, 0.2% sodium deoxycholate, 2× Protease + Phosphatase Inhibitors);
- RIPA buffer (50 mM Hepes pH 8, 1% NP-40, 0.70% sodium deoxycholate, 0.5 M LiCl, 1 mM EDTA, 1× Protease Inhibitors);
- Tween wash buffer (5 mM Tris-HCl pH 7.5, 0.5 mM EDTA, 1 M NaCl, 0.05% Tween-20);
- 2× Biotin binding buffer (10 mM Tris-HCl pH 7.5, 1 mM EDTA, 2M NaCl);
- 1× TD buffer (10 mM Tris-HCl pH 7.5, 5 mM MgCl2, 10% dimethylformamide);
- 2× TD buffer (20 mM Tris-HCl pH 7.5, 10 mM MgCl2, 20% dimethylformamide);
- Strip buffer (0.15% SDS, 10 mM Tris-HCl pH 8, 50 mM EDTA).
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A

References
- Dekker, J.; Rippe, K.; Dekker, M.; Kleckner, N. Capturing Chromosome Conformation. Science 2002, 295, 1306–1311. [Google Scholar] [CrossRef] [Green Version]
- Kempfer, R.; Pombo, A. Methods for Mapping 3D Chromosome Architecture. Nat. Rev. Genet. 2020, 21, 207–226. [Google Scholar] [CrossRef]
- Sati, S.; Cavalli, G. Chromosome Conformation Capture Technologies and Their Impact in Understanding Genome Function. Chromosoma 2017, 126, 33–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Wit, E.; de Laat, W. A Decade of 3C Technologies: Insights into Nuclear Organization. Genes Dev. 2012, 26, 11–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lieberman-Aiden, E.; van Berkum, N.L.; Williams, L.; Imakaev, M.; Ragoczy, T.; Telling, A.; Amit, I.; Lajoie, B.R.; Sabo, P.J.; Dorschner, M.O.; et al. Comprehensive Mapping of Long-Range Interactions Reveals Folding Principles of the Human Genome. Science 2009, 326, 289–293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fullwood, M.J.; Liu, M.H.; Pan, Y.F.; Liu, J.; Han, X.; Mohamed, Y.B.; Orlov, Y.L.; Velkov, S.; Ho, A.; Mei, P.H.; et al. An Oestrogen Receptor α-Bound Human Chromatin Interactome. Nature 2009, 462, 58–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, G.; Fullwood, M.J.; Xu, H.; Mulawadi, F.H.; Velkov, S.; Vega, V.; Ariyaratne, P.N.; Mohamed, Y.B.; Ooi, H.-S.; Tennakoon, C.; et al. ChIA-PET Tool for Comprehensive Chromatin Interaction Analysis with Paired-End Tag Sequencing. Genome Biol. 2010, 11, R22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, R.; Yu, M.; Li, G.; Chee, S.; Liu, T.; Schmitt, A.D.; Ren, B. Mapping of Long-Range Chromatin Interactions by Proximity Ligation-Assisted ChIP-Seq. Cell Res. 2016, 26, 1345–1348. [Google Scholar] [CrossRef] [Green Version]
- Mumbach, M.R.; Rubin, A.J.; Flynn, R.A.; Dai, C.; Khavari, P.A.; Greenleaf, W.J.; Chang, H.Y. HiChIP: Efficient and Sensitive Analysis of Protein-Directed Genome Architecture. Nat. Methods 2016, 13, 919–922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rao, S.S.P.; Huntley, M.H.; Durand, N.C.; Stamenova, E.K.; Bochkov, I.D.; Robinson, J.T.; Sanborn, A.L.; Machol, I.; Omer, A.D.; Lander, E.S.; et al. A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell 2014, 159, 1665–1680. [Google Scholar] [CrossRef] [Green Version]
- Lajoie, B.R.; Dekker, J.; Kaplan, N. The Hitchhiker’s Guide to Hi-C Analysis: Practical Guidelines. Methods 2015, 72, 65–75. [Google Scholar] [CrossRef] [Green Version]
- Servant, N.; Varoquaux, N.; Lajoie, B.R.; Viara, E.; Chen, C.-J.; Vert, J.-P.; Heard, E.; Dekker, J.; Barillot, E. HiC-Pro: An Optimized and Flexible Pipeline for Hi-C Data Processing. Genome Biol. 2015, 16, 259. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharyya, S.; Chandra, V.; Vijayanand, P.; Ay, F. Identification of Significant Chromatin Contacts from HiChIP Data by FitHiChIP. Nat. Commun. 2019, 10, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rawlings, J.S.; Gatzka, M.; Thomas, P.G.; Ihle, J.N. Chromatin Condensation via the Condensin II Complex Is Required for Peripheral T-Cell Quiescence. EMBO J. 2011, 30, 263–276. [Google Scholar] [CrossRef] [PubMed]
- Mumbach, M.R.; Satpathy, A.T.; Boyle, E.A.; Dai, C.; Gowen, B.G.; Cho, S.W.; Nguyen, M.L.; Rubin, A.J.; Granja, J.M.; Kazane, K.R.; et al. Enhancer Connectome in Primary Human Cells Identifies Target Genes of Disease-Associated DNA Elements. Nat. Genet. 2017, 49, 1602–1612. [Google Scholar] [CrossRef] [Green Version]
- Arrigoni, L.; Richter, A.S.; Betancourt, E.; Bruder, K.; Diehl, S.; Manke, T.; Bönisch, U. Standardizing Chromatin Research: A Simple and Universal Method for ChIP-Seq. Nucleic Acids Res. 2016, 44, e67. [Google Scholar] [CrossRef]
- Belaghzal, H.; Dekker, J.; Gibcus, J.H. Hi-C 2.0: An Optimized Hi-C Procedure for High-Resolution Genome-Wide Mapping of Chromosome Conformation. Methods 2017, 123, 56–65. [Google Scholar] [CrossRef]
- Conlan, L.H.; José, T.J.; Thornton, K.C.; Dupureur, C.M. Modulating Restriction Endonuclease Activities and Specificities Using Neutral Detergents. Biotechniques 1999, 27, 955–960. [Google Scholar] [CrossRef]
- Mizrahi, V.; Benkovic, P.A.; Benkovic, S.J. Mechanism of the Idling-Turnover Reaction of the Large (Klenow) Fragment of Escherichia Coli DNA Polymerase I. Proc. Natl. Acad. Sci. USA 1986, 83, 231–235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, G.; Li, J.; Hu, T.; Wei, H.; Guan, Y. Realizing Directional Cloning Using Sticky Ends Produced by 3′-5′ Exonuclease of Klenow Fragment. J. Biosci. 2013, 38, 857–866. [Google Scholar] [CrossRef] [PubMed]
- Clark, J.M.; Joyce, C.M.; Beardsley, G.P. Novel Blunt-End Addition Reactions Catalyzed by DNA Polymerase I of Escherichia Coli. J. Mol. Biol. 1987, 198, 123–127. [Google Scholar] [CrossRef]
- Montecucco, A.; Lestingi, M.; Pedrali-Noy, G.; Spadari, S.; Ciarrocchi, G. Use of ATP, DATP and Their Alpha-Thio Derivatives to Study DNA Ligase Adenylation. Biochem. J. 1990, 271, 265–268. [Google Scholar] [CrossRef] [Green Version]
- Bauer, R.J.; Zhelkovsky, A.; Bilotti, K.; Crowell, L.E.; Evans, T.C., Jr.; McReynolds, L.A.; Lohman, G.J.S. Comparative Analysis of the End-Joining Activity of Several DNA Ligases. PLoS ONE 2017, 12, e0190062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayashi, K.; Nakazawa, M.; Ishizaki, Y.; Hiraoka, N.; Obayashi, A. Regulation of Inter- and Intramolecular Ligation with T4 DNA Ligase in the Presence of Polyethylene Glycol. Nucleic Acids Res. 1986, 14, 7617–7631. [Google Scholar] [CrossRef] [Green Version]
- Teraoka, H.; Tsukada, K. Influence of Polyethylene Glycol on the Ligation Reaction with Calf Thymus DNA Ligases I and II. J. Biochem. 1987, 101, 225–231. [Google Scholar] [CrossRef] [PubMed]
- Pösö, H.; Kuosmanen, M. Spermidine and Spermine Stimulate the Activity of T4-DNA Ligase. Biochem. Biophys. Res. Commun. 1983, 117, 217–222. [Google Scholar] [CrossRef]
- Picelli, S.; Björklund, Å.K.; Reinius, B.; Sagasser, S.; Winberg, G.; Sandberg, R. Tn5 Transposase and Tagmentation Procedures for Massively Scaled Sequencing Projects. Genome Res. 2014, 24, 2033–2040. [Google Scholar] [CrossRef]
- Amini, S.; Pushkarev, D.; Christiansen, L.; Kostem, E.; Royce, T.; Turk, C.; Pignatelli, N.; Adey, A.; Kitzman, J.O.; Vijayan, K.; et al. Haplotype-Resolved Whole Genome Sequencing by Contiguity Preserving Transposition and Combinatorial Indexing. Nat. Genet. 2014, 46, 1343–1349. [Google Scholar] [CrossRef] [PubMed]
- Nagano, T.; Várnai, C.; Schoenfelder, S.; Javierre, B.-M.; Wingett, S.W.; Fraser, P. Comparison of Hi-C Results Using in-Solution versus in-Nucleus Ligation. Genome Biol. 2015, 16, 175. [Google Scholar] [CrossRef] [Green Version]
- Spilianakis, C.G.; Lalioti, M.D.; Town, T.; Lee, G.R.; Flavell, R.A. Interchromosomal Associations between Alternatively Expressed Loci. Nature 2005, 435, 637–645. [Google Scholar] [CrossRef]
- Zelenka, T.; Klonizakis, A.; Tsoukatou, D.; Franzenburg, S.; Tzerpos, P.; Papamatheakis, D.A.; Tzonevrakis, I.R.; Nikolaou, C.; Plewczynski, D.; Spilianakis, C. The 3D Enhancer Network of the Developing T Cell Genome Is Controlled by SATB1. bioRxiv 2021. [Google Scholar] [CrossRef]

| Reagent | Volume Per Reaction |
|---|---|
| 10× NEBuffer 2 (NEB, M0210L) | 30 μL |
| ddH2O | 238.5 μL |
| 1 mM Biotin-16-dCTP (Jena Bioscience, NU-809-BIO16-L) | 15 μL |
| 10 mM dATP (Promega, U1240) | 1.5 μL |
| 10 mM dGTP (Promega, U1240) | 1.5 μL |
| 10 mM dTTP (Promega, U1240) | 1.5 μL |
| 5 U/μL DNA Polymerase I, Klenow Fragment (NEB, M0210L) | 12 μL |
| Reagent | Volume Per Reaction |
|---|---|
| 10× NEB T4 DNA Ligase Buffer with 10 mM ATP (NEB, B0202) | 120 μL |
| 20% Triton X-100 (1% final) | 60 μL |
| 2% (20 mg/mL) BSA (NEB, B9000S) | 6 μL |
| 30% PEG 6000 (1% final) | 40 μL |
| 400 U/μL T4 DNA Ligase (NEB, M0202L) | 5 μL |
| ddH2O | 969 μL |
| Reagent | Volume Per Reaction |
|---|---|
| Phusion HF 2× PCR Master Mix (NEB, M0531S) | 25 μL |
| Nextera Index 1 N7XX | 1–1.5 μL |
| Nextera Index 2 N5XX | 1–1.5 μL |
| ddH2O | 22–23 μL |
| Reaction | Temperature | Time | Cycle Number |
|---|---|---|---|
| Gap-filling required upon tagmentation | 72 °C | 5 min | 1 cycle |
| Initial Denaturation | 98 °C | 1 min | 1 cycle |
| Denaturation | 98 °C | 15 s | 5–15 cycles |
| Annealing | 63 °C | 35 s | |
| Extension | 72 °C | 1 min |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zelenka, T.; Spilianakis, C. HiChIP and Hi-C Protocol Optimized for Primary Murine T Cells. Methods Protoc. 2021, 4, 49. https://doi.org/10.3390/mps4030049
Zelenka T, Spilianakis C. HiChIP and Hi-C Protocol Optimized for Primary Murine T Cells. Methods and Protocols. 2021; 4(3):49. https://doi.org/10.3390/mps4030049
Chicago/Turabian StyleZelenka, Tomas, and Charalampos Spilianakis. 2021. "HiChIP and Hi-C Protocol Optimized for Primary Murine T Cells" Methods and Protocols 4, no. 3: 49. https://doi.org/10.3390/mps4030049
APA StyleZelenka, T., & Spilianakis, C. (2021). HiChIP and Hi-C Protocol Optimized for Primary Murine T Cells. Methods and Protocols, 4(3), 49. https://doi.org/10.3390/mps4030049







