A Light Wand to Untangle the Myocardial Cell Network
Abstract
1. Introduction of Optogenetics
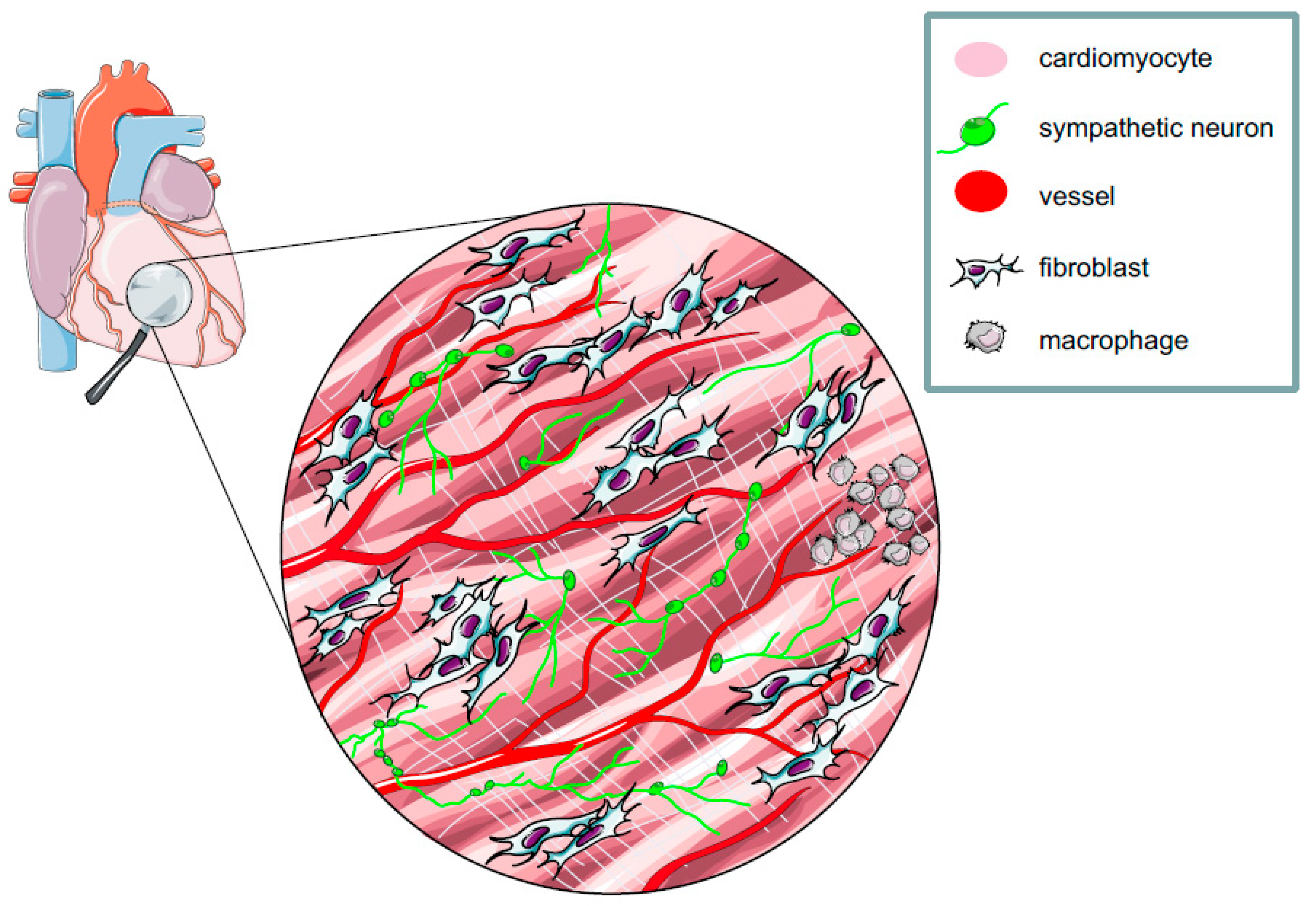
2. The Myocardium: A Complex Network of Excitable and Non-excitable Cells
3. The Heart Goes to Optogenetics
4. All-Optical: Optogenetics, Optical Mapping, and Optoelectronics
5. Optogenetics Targets Specific Heart Cells
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviation
| α-MyHC | alpha-myosin heavy chain |
| ArchT | archeorhodopsin |
| AVN | atrioventricular node |
| APD | action potential duration |
| β-AR | β-adrenoceptors |
| ChR2 | Channelrhodopsin-2 |
| CF | cardiac fibroblast |
| CM | cardiomyocyte |
| cx | connexin |
| EC | endothetial cell |
| ECG | electrocardiography |
| EP | electrophysiology |
| ERP | effective refractory period |
| ET-1 | endothelin-1 |
| MΦ | macrophage |
| NE | noradrenaline |
| NO | nitric oxide |
| NpHR | N. pharaonis HaloRhodopsin |
| PF | Purkinje fiber |
| RVSAN | right ventriclesino atrial node |
| SMC | smooth muscle cell |
| SNs | sympathetic neurons |
| TCU | tandem cell unit |
| TH | tyrosine hydroxylase |
| TxA2 | thromboxane-A2. |
References
- Mattis, J.; Tye, K.M.; Ferenczi, E.A.; Ramakrishnan, C.; O’Shea, D.J.; Prakash, R.; Gunaydin, L.A.; Hyun, M.; Fenno, L.E.; Gradinaru, V.; et al. Principles for applying optogenetic tools derived from direct comparative analysis of microbial opsins. Nat. Methods 2011, 9, 159–172. [Google Scholar] [CrossRef] [PubMed]
- Kopton, R.A.; Baillie, J.S.; Rafferty, S.A.; Moss, R.; Zgierski-Johnston, C.M.; Prykhozhij, S.V.; Stoyek, M.R.; Smith, F.M.; Kohl, P.; Quinn, T.A.; et al. Cardiac Electrophysiological Effects of Light-Activated Chloride Channels. Front. Physiol. 2018, 9, 1806. [Google Scholar] [CrossRef]
- Kleinlogel, S.; Feldbauer, K.; Dempski, R.E.; Fotis, H.; Wood, P.G.; Bamann, C.; Bamberg, E. Ultra light-sensitive and fast neuronal activation with the Ca(2)+-permeable channelrhodopsin CatCh. Nat. Neurosci. 2011, 14, 513–518. [Google Scholar] [CrossRef]
- Gunaydin, L.A.; Yizhar, O.; Berndt, A.; Sohal, V.S.; Deisseroth, K.; Hegemann, P. Ultrafast optogenetic control. Nat. Neurosci. 2010, 13, 387–392. [Google Scholar] [CrossRef]
- Berndt, A.; Yizhar, O.; Gunaydin, L.A.; Hegemann, P.; Deisseroth, K. Bi-stable neural state switches. Nat. Neurosci. 2009, 12, 229–234. [Google Scholar] [CrossRef]
- Zhang, F.; Vierock, J.; Yizhar, O.; Fenno, L.E.; Tsunoda, S.; Kianianmomeni, A.; Prigge, M.; Berndt, A.; Cushman, J.; Polle, J.; et al. The microbial opsin family of optogenetic tools. Cell 2011, 147, 1446–1457. [Google Scholar] [CrossRef] [PubMed]
- Deisseroth, K.; Hegemann, P. The form and function of channelrhodopsin. Science 2017, 357. [Google Scholar] [CrossRef]
- Boyden, E.S.; Zhang, F.; Bamberg, E.; Nagel, G.; Deisseroth, K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 2005, 8, 1263–1268. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, J.G.; Boyden, E.S. Optogenetic tools for analyzing the neural circuits of behavior. Trends Cogn. Sci. 2011, 15, 592–600. [Google Scholar] [CrossRef]
- Deisseroth, K.; Feng, G.; Majewska, A.K.; Miesenbock, G.; Ting, A.; Schnitzer, M.J. Next-generation optical technologies for illuminating genetically targeted brain circuits. J. Neurosci. 2006, 26, 10380–10386. [Google Scholar] [CrossRef]
- Fenno, L.; Yizhar, O.; Deisseroth, K. The development and application of optogenetics. Annu. Rev. Neurosci. 2011, 34, 389–412. [Google Scholar] [CrossRef]
- Zhang, F.; Aravanis, A.M.; Adamantidis, A.; de Lecea, L.; Deisseroth, K. Circuit-breakers: Optical technologies for probing neural signals and systems. Nat. Rev. Neurosci. 2007, 8, 577–581. [Google Scholar] [CrossRef]
- Aravanis, A.M.; Wang, L.P.; Zhang, F.; Meltzer, L.A.; Mogri, M.Z.; Schneider, M.B.; Deisseroth, K. An optical neural interface: In vivo control of rodent motor cortex with integrated fiberoptic and optogenetic technology. J. Neural Eng. 2007, 4, S143–S156. [Google Scholar] [CrossRef] [PubMed]
- Kravitz, A.V.; Freeze, B.S.; Parker, P.R.; Kay, K.; Thwin, M.T.; Deisseroth, K.; Kreitzer, A.C. Regulation of parkinsonian motor behaviours by optogenetic control of basal ganglia circuitry. Nature 2010, 466, 622–626. [Google Scholar] [CrossRef]
- Tonnesen, J.; Parish, C.L.; Sorensen, A.T.; Andersson, A.; Lundberg, C.; Deisseroth, K.; Arenas, E.; Lindvall, O.; Kokaia, M. Functional integration of grafted neural stem cell-derived dopaminergic neurons monitored by optogenetics in an in vitro Parkinson model. PLoS ONE 2011, 6, e17560. [Google Scholar] [CrossRef]
- G, N.; Tan, A.; Farhatnia, Y.; Rajadas, J.; Hamblin, M.R.; Khaw, P.T.; Seifalian, A.M. Channelrhodopsins: Visual regeneration and neural activation by a light switch. New Biotechnol. 2013, 30, 461–474. [Google Scholar] [CrossRef][Green Version]
- Yizhar, O.; Fenno, L.E.; Davidson, T.J.; Mogri, M.; Deisseroth, K. Optogenetics in neural systems. Neuron 2011, 71, 9–34. [Google Scholar] [CrossRef]
- Wanjare, M.; Huang, N.F. Regulation of the microenvironment for cardiac tissue engineering. Regen. Med. 2017, 12, 187–201. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, I.; Fuseler, J.W.; Price, R.L.; Borg, T.K.; Baudino, T.A. Determination of cell types and numbers during cardiac development in the neonatal and adult rat and mouse. Am. J. Physiol. Heart Circ. Physiol. 2007, 293, H1883–H1891. [Google Scholar] [CrossRef] [PubMed]
- LeGrice, I.; Pope, A.; Smaill, B. The Architecture of the Heart: Myocyte Organization and the Cardiac Extracellular Matrix. Dev. Cardiovasc. Med. 2005, 253, 3–22. [Google Scholar]
- Zak, R. Development and proliferative capacity of cardiac muscle cells. Circ. Res. 1974, 35, 17–26. [Google Scholar]
- Nag, A.C. Study of non-muscle cells of the adult mammalian heart: A fine structural analysis and distribution. Cytobios 1980, 28, 41–61. [Google Scholar] [PubMed]
- Tian, Y.; Morrisey, E.E. Importance of myocyte-nonmyocyte interactions in cardiac development and disease. Circ. Res. 2012, 110, 1023–1034. [Google Scholar] [CrossRef]
- Pinto, A.R.; Ilinykh, A.; Ivey, M.J.; Kuwabara, J.T.; D’Antoni, M.L.; Debuque, R.; Chandran, A.; Wang, L.; Arora, K.; Rosenthal, N.A.; et al. Revisiting Cardiac Cellular Composition. Circ. Res. 2016, 118, 400–409. [Google Scholar] [CrossRef]
- Ongstad, E.; Kohl, P. Fibroblast-myocyte coupling in the heart: Potential relevance for therapeutic interventions. J. Mol. Cell. Cardiol. 2016, 91, 238–246. [Google Scholar] [CrossRef]
- Rog-Zielinska, E.A.; Norris, R.A.; Kohl, P.; Markwald, R. The Living Scar—Cardiac Fibroblasts and the Injured Heart. Trends Mol. Med. 2016, 22, 99–114. [Google Scholar] [CrossRef]
- Mollmann, H.; Nef, H.M.; Kostin, S.; von Kalle, C.; Pilz, I.; Weber, M.; Schaper, J.; Hamm, C.W.; Elsasser, A. Bone marrow-derived cells contribute to infarct remodelling. Cardiovasc. Res. 2006, 71, 661–671. [Google Scholar] [CrossRef]
- Endo, J.; Sano, M.; Fujita, J.; Hayashida, K.; Yuasa, S.; Aoyama, N.; Takehara, Y.; Kato, O.; Makino, S.; Ogawa, S.; et al. Bone marrow derived cells are involved in the pathogenesis of cardiac hypertrophy in response to pressure overload. Circulation 2007, 116, 1176–1184. [Google Scholar] [CrossRef]
- Tillmanns, J.; Hoffmann, D.; Habbaba, Y.; Schmitto, J.D.; Sedding, D.; Fraccarollo, D.; Galuppo, P.; Bauersachs, J. Fibroblast activation protein alpha expression identifies activated fibroblasts after myocardial infarction. J. Mol. Cell. Cardiol. 2015, 87, 194–203. [Google Scholar] [CrossRef]
- Chilton, L.; Giles, W.R.; Smith, G.L. Evidence of intercellular coupling between co-cultured adult rabbit ventricular myocytes and myofibroblasts. J. Physiol. 2007, 583, 225–236. [Google Scholar] [CrossRef]
- Camelliti, P.; Green, C.R.; LeGrice, I.; Kohl, P. Fibroblast network in rabbit sinoatrial node: Structural and functional identification of homogeneous and heterogeneous cell coupling. Circ. Res. 2004, 94, 828–835. [Google Scholar] [CrossRef] [PubMed]
- Kohl, P.; Camelliti, P. Fibroblast-myocyte connections in the heart. Heart Rhythm 2012, 9, 461–464. [Google Scholar] [CrossRef]
- De Maziere, A.M.; van Ginneken, A.C.; Wilders, R.; Jongsma, H.J.; Bouman, L.N. Spatial and functional relationship between myocytes and fibroblasts in the rabbit sinoatrial node. J. Mol. Cell. Cardiol. 1992, 24, 567–578. [Google Scholar] [CrossRef]
- Kakkar, R.; Lee, R.T. Intramyocardial fibroblast myocyte communication. Circ. Res. 2010, 106, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Ottaviano, F.G.; Yee, K.O. Communication signals between cardiac fibroblasts and cardiac myocytes. J. Cardiovasc. Pharmacol. 2011, 57, 513–521. [Google Scholar] [CrossRef] [PubMed]
- Kofron, C.M.; Mende, U. In vitro models of the cardiac microenvironment to study myocyte and non-myocyte crosstalk: Bioinspired approaches beyond the polystyrene dish. J. Physiol. 2017, 595, 3891–3905. [Google Scholar] [CrossRef]
- Zhang, Y.; Kanter, E.M.; Laing, J.G.; Aprhys, C.; Johns, D.C.; Kardami, E.; Yamada, K.A. Connexin43 expression levels influence intercellular coupling and cell proliferation of native murine cardiac fibroblasts. Cell Commun. Adhes. 2008, 15, 289–303. [Google Scholar] [CrossRef]
- Louault, C.; Benamer, N.; Faivre, J.F.; Potreau, D.; Bescond, J. Implication of connexins 40 and 43 in functional coupling between mouse cardiac fibroblasts in primary culture. Biochim. Biophys. Acta 2008, 1778, 2097–2104. [Google Scholar] [CrossRef]
- Rook, M.B.; van Ginneken, A.C.; de Jonge, B.; el Aoumari, A.; Gros, D.; Jongsma, H.J. Differences in gap junction channels between cardiac myocytes, fibroblasts, and heterologous pairs. Am. J. Physiol 1992, 263, C959–C977. [Google Scholar] [CrossRef]
- He, K.; Shi, X.; Zhang, X.; Dang, S.; Ma, X.; Liu, F.; Xu, M.; Lv, Z.; Han, D.; Fang, X.; et al. Long-distance intercellular connectivity between cardiomyocytes and cardiofibroblasts mediated by membrane nanotubes. Cardiovasc. Res. 2011, 92, 39–47. [Google Scholar] [CrossRef]
- Wang, X.; Gerdes, H.H. Long-distance electrical coupling via tunneling nanotubes. Biochim. Biophys. Acta 2012, 1818, 2082–2086. [Google Scholar] [CrossRef]
- Nguyen, T.P.; Xie, Y.; Garfinkel, A.; Qu, Z.; Weiss, J.N. Arrhythmogenic consequences of myofibroblast-myocyte coupling. Cardiovasc. Res. 2012, 93, 242–251. [Google Scholar] [CrossRef] [PubMed]
- Goshima, K. Formation of nexuses and electrotonic transmission between myocardial and FL cells in monolayer culture. Exp. Cell Res. 1970, 63, 124–130. [Google Scholar] [CrossRef]
- Nagel, G.; Ollig, D.; Fuhrmann, M.; Kateriya, S.; Musti, A.M.; Bamberg, E.; Hegemann, P. Channelrhodopsin-1: A light-gated proton channel in green algae. Science 2002, 296, 2395–2398. [Google Scholar] [CrossRef] [PubMed]
- Nagel, G.; Szellas, T.; Huhn, W.; Kateriya, S.; Adeishvili, N.; Berthold, P.; Ollig, D.; Hegemann, P.; Bamberg, E. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc. Natl. Acad. Sci. USA 2003, 100, 13940–13945. [Google Scholar] [CrossRef]
- Li, X.; Gutierrez, D.V.; Hanson, M.G.; Han, J.; Mark, M.D.; Chiel, H.; Hegemann, P.; Landmesser, L.T.; Herlitze, S. Fast noninvasive activation and inhibition of neural and network activity by vertebrate rhodopsin and green algae channelrhodopsin. Proc. Natl. Acad. Sci. USA 2005, 102, 17816–17821. [Google Scholar] [CrossRef]
- Nagel, G.; Szellas, T.; Kateriya, S.; Adeishvili, N.; Hegemann, P.; Bamberg, E. Channelrhodopsins: Directly light-gated cation channels. Biochem. Soc. Trans. 2005, 33, 863–866. [Google Scholar] [CrossRef]
- Method of the Year 2010. Nat. Method 2011, 8, 1. [CrossRef]
- Bruegmann, T.; Malan, D.; Hesse, M.; Beiert, T.; Fuegemann, C.J.; Fleischmann, B.K.; Sasse, P. Optogenetic control of heart muscle in vitro and in vivo. Nat. Methods 2010, 7, 897–900. [Google Scholar] [CrossRef] [PubMed]
- Arrenberg, A.B.; Stainier, D.Y.; Baier, H.; Huisken, J. Optogenetic control of cardiac function. Science 2010, 330, 971–974. [Google Scholar] [CrossRef]
- Jia, Z.; Valiunas, V.; Lu, Z.; Bien, H.; Liu, H.; Wang, H.Z.; Rosati, B.; Brink, P.R.; Cohen, I.S.; Entcheva, E. Stimulating cardiac muscle by light: Cardiac optogenetics by cell delivery. Circ. Arrhythm. Electrophysiol. 2011, 4, 753–760. [Google Scholar] [CrossRef]
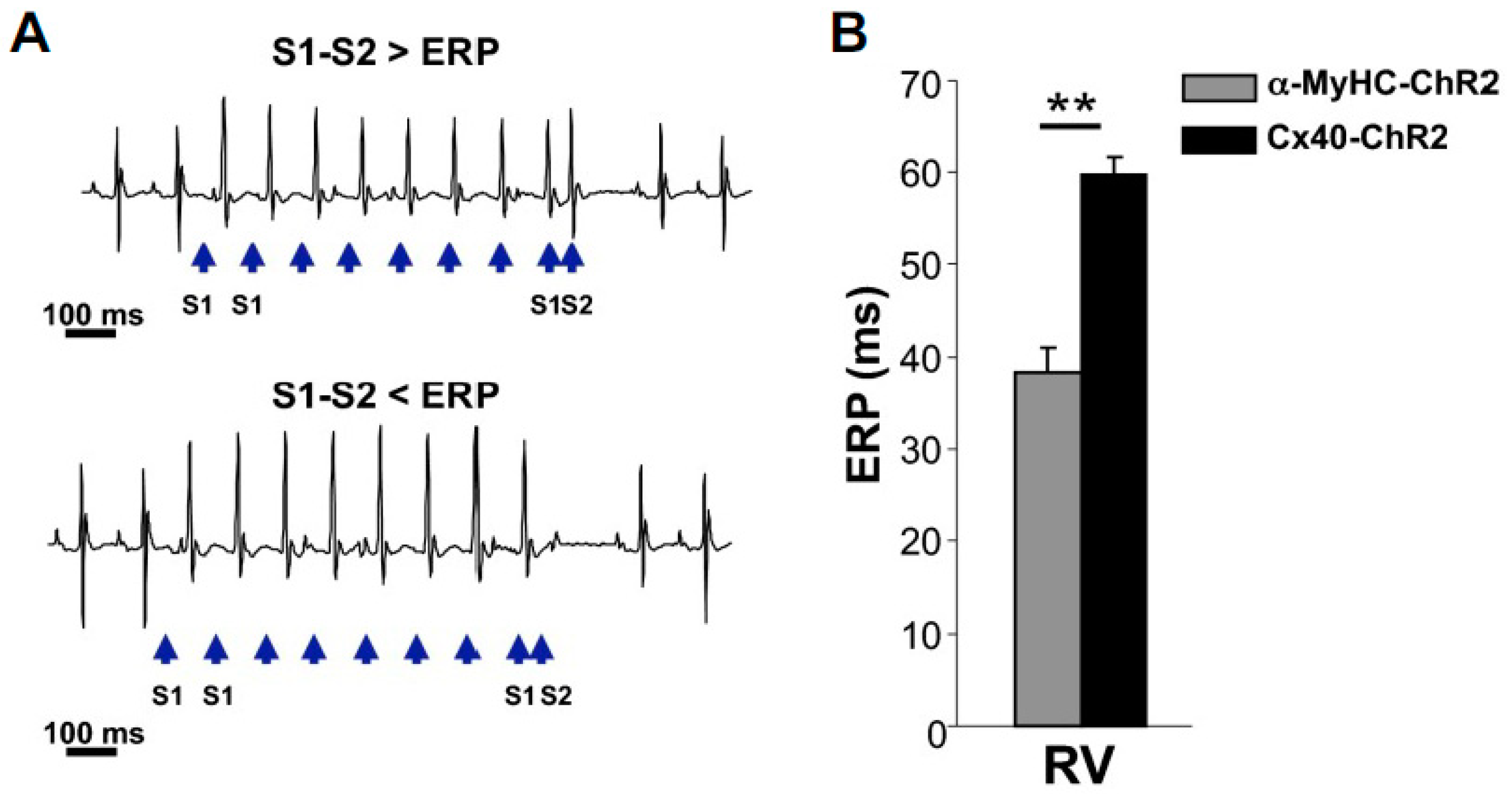
- Zaglia, T.; Pianca, N.; Borile, G.; Da Broi, F.; Richter, C.; Campione, M.; Lehnart, S.E.; Luther, S.; Corrado, D.; Miquerol, L.; et al. Optogenetic determination of the myocardial requirements for extrasystoles by cell type-specific targeting of ChannelRhodopsin-2. Proc. Natl. Acad. Sci. USA 2015, 112, E4495–E4504. [Google Scholar] [CrossRef]
- Bruegmann, T.; Boyle, P.M.; Vogt, C.C.; Karathanos, T.V.; Arevalo, H.J.; Fleischmann, B.K.; Trayanova, N.A.; Sasse, P. Optogenetic defibrillation terminates ventricular arrhythmia in mouse hearts and human simulations. J. Clin. Investig. 2016, 126, 3894–3904. [Google Scholar] [CrossRef]
- Boyle, P.M.; Karathanos, T.V.; Trayanova, N.A. Cardiac Optogenetics: 2018. JACC Clin. Electrophysiol. 2018, 4, 155–167. [Google Scholar] [CrossRef]
- Williams, J.C.; Entcheva, E. Optogenetic versus Electrical Stimulation of Human Cardiomyocytes: Modeling Insights. Biophys. J. 2015, 108, 1934–1945. [Google Scholar] [CrossRef]
- Karathanos, T.V.; Bayer, J.D.; Wang, D.; Boyle, P.M.; Trayanova, N.A. Opsin spectral sensitivity determines the effectiveness of optogenetic termination of ventricular fibrillation in the human heart: A simulation study. J. Physiol. 2016, 594, 6879–6891. [Google Scholar] [CrossRef]
- Entcheva, E.; Bub, G. All-optical control of cardiac excitation: Combined high-resolution optogenetic actuation and optical mapping. J. Physiol. 2016, 594, 2503–2510. [Google Scholar] [CrossRef]
- Vogt, C.C.; Bruegmann, T.; Malan, D.; Ottersbach, A.; Roell, W.; Fleischmann, B.K.; Sasse, P. Systemic gene transfer enables optogenetic pacing of mouse hearts. Cardiovasc. Res. 2015, 106, 338–343. [Google Scholar] [CrossRef]
- Crocini, C.; Ferrantini, C.; Coppini, R.; Scardigli, M.; Yan, P.; Loew, L.M.; Smith, G.; Cerbai, E.; Poggesi, C.; Pavone, F.S.; et al. Optogenetics design of mechanistically-based stimulation patterns for cardiac defibrillation. Sci. Rep. 2016, 6, 35628. [Google Scholar] [CrossRef]
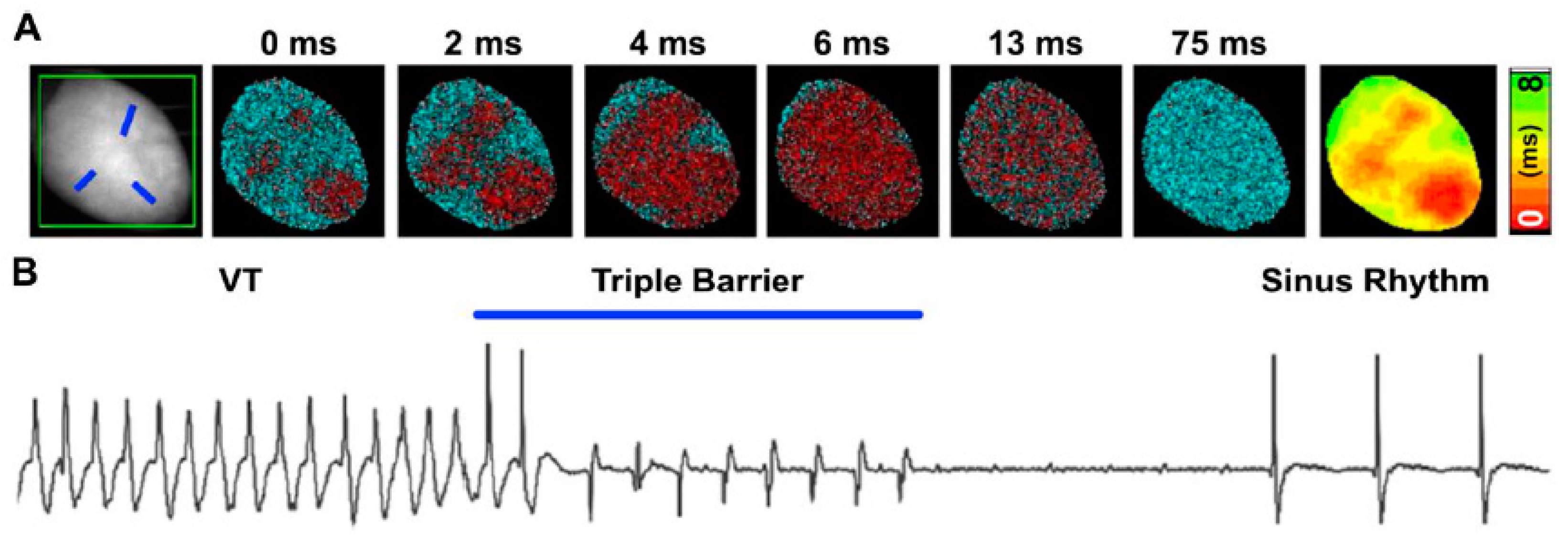
- Watanabe, M.; Feola, I.; Majumder, R.; Jangsangthong, W.; Teplenin, A.S.; Ypey, D.L.; Schalij, M.J.; Zeppenfeld, K.; de Vries, A.A.; Pijnappels, D.A. Optogenetic manipulation of anatomical re-entry by light-guided generation of a reversible local conduction block. Cardiovasc. Res. 2017, 113, 354–366. [Google Scholar] [CrossRef]
- Nyns, E.C.A.; Kip, A.; Bart, C.I.; Plomp, J.J.; Zeppenfeld, K.; Schalij, M.J.; de Vries, A.A.F.; Pijnappels, D.A. Optogenetic termination of ventricular arrhythmias in the whole heart: Towards biological cardiac rhythm management. Eur. Heart J. 2017, 38, 2132–2136. [Google Scholar] [CrossRef]
- Wang, S.; Kugelman, T.; Buch, A.; Herman, M.; Han, Y.; Karakatsani, M.E.; Hussaini, S.A.; Duff, K.; Konofagou, E.E. Non-invasive, Focused Ultrasound-Facilitated Gene Delivery for Optogenetics. Sci. Rep. 2017, 7, 39955. [Google Scholar] [CrossRef] [PubMed]
- Klimas, A.; Ambrosi, C.M.; Yu, J.; Williams, J.C.; Bien, H.; Entcheva, E. OptoDyCE as an automated system for high-throughput all-optical dynamic cardiac electrophysiology. Nat. Commun. 2016, 7, 11542. [Google Scholar] [CrossRef]
- Colatsky, T.; Fermini, B.; Gintant, G.; Pierson, J.B.; Sager, P.; Sekino, Y.; Strauss, D.G.; Stockbridge, N. The Comprehensive in Vitro Proarrhythmia Assay (CiPA) initiative—Update on progress. J. Pharmacol. Toxicol. Methods 2016, 81, 15–20. [Google Scholar] [CrossRef]
- Dempsey, G.T.; Chaudhary, K.W.; Atwater, N.; Nguyen, C.; Brown, B.S.; McNeish, J.D.; Cohen, A.E.; Kralj, J.M. Cardiotoxicity screening with simultaneous optogenetic pacing, voltage imaging and calcium imaging. J. Pharmacol. Toxicol. Methods 2016, 81, 240–250. [Google Scholar] [CrossRef]
- Lapp, H.; Bruegmann, T.; Malan, D.; Friedrichs, S.; Kilgus, C.; Heidsieck, A.; Sasse, P. Frequency-dependent drug screening using optogenetic stimulation of human iPSC-derived cardiomyocytes. Sci. Rep. 2017, 7, 9629. [Google Scholar] [CrossRef]
- Richter, C.; Christoph, J.; Lehnart, S.E.; Luther, S. Optogenetic Light Crafting Tools for the Control of Cardiac Arrhythmias. Methods Mol. Biol. 2016, 1408, 293–302. [Google Scholar] [CrossRef]
- Majumder, R.; Feola, I.; Teplenin, A.S.; de Vries, A.A.; Panfilov, A.V.; Pijnappels, D.A. Optogenetics enables real-time spatiotemporal control over spiral wave dynamics in an excitable cardiac system. eLife 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Moreno, A.; Endicott, K.; Skancke, M.; Dwyer, M.K.; Brennan, J.; Efimov, I.R.; Trachiotis, G.; Mendelowitz, D.; Kay, M.W. Sudden Heart Rate Reduction Upon Optogenetic Release of Acetylcholine From Cardiac Parasympathetic Neurons in Perfused Hearts. Front. Physiol. 2019, 10, 16. [Google Scholar] [CrossRef]
- Nyns, E.C.A.; Poelma, R.H.; Volkers, L.; Plomp, J.J.; Bart, C.I.; Kip, A.M.; van Brakel, T.J.; Zeppenfeld, K.; Schalij, M.J.; Zhang, G.Q.; et al. An automated hybrid bioelectronic system for autogenous restoration of sinus rhythm in atrial fibrillation. Sci. Transl. Med. 2019, 11. [Google Scholar] [CrossRef]
- Ambrosi, C.M.; Klimas, A.; Yu, J.; Entcheva, E. Cardiac applications of optogenetics. Prog. Biophys. Mol. Biol. 2014, 115, 294–304. [Google Scholar] [CrossRef][Green Version]
- Cunningham, J.G.; Klein, B.A. Textbook of Veterinary Physiology; Elsevier Saunders: St. Louis, MO, USA, 2013. [Google Scholar]
- Boyden, P.A.; Dun, W.; Robinson, R.B. Cardiac Purkinje fibers and arrhythmias; The GK Moe Award Lecture 2015. Heart Rhythm 2016, 13, 1172–1181. [Google Scholar] [CrossRef]
- Scheinman, M.M. Role of the His-Purkinje system in the genesis of cardiac arrhythmia. Heart Rhythm 2009, 6, 1050–1058. [Google Scholar] [CrossRef]
- Cerrone, M.; Noujaim, S.F.; Tolkacheva, E.G.; Talkachou, A.; O’Connell, R.; Berenfeld, O.; Anumonwo, J.; Pandit, S.V.; Vikstrom, K.; Napolitano, C.; et al. Arrhythmogenic mechanisms in a mouse model of catecholaminergic polymorphic ventricular tachycardia. Circ. Res. 2007, 101, 1039–1048. [Google Scholar] [CrossRef] [PubMed]
- Weiss, J.N.; Nivala, M.; Garfinkel, A.; Qu, Z. Alternans and arrhythmias: From cell to heart. Circ. Res. 2011, 108, 98–112. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Sato, D.; Garfinkel, A.; Qu, Z.; Weiss, J.N. So little source, so much sink: Requirements for afterdepolarizations to propagate in tissue. Biophys. J. 2010, 99, 1408–1415. [Google Scholar] [CrossRef]
- Franzoso, M.; Zaglia, T.; Mongillo, M. Putting together the clues of the everlasting neuro-cardiac liaison. Biochim. Biophys. Acta 2016, 1863, 1904–1915. [Google Scholar] [CrossRef]
- Yang, J.; Liu, Y.; Fan, X.; Li, Z.; Cheng, Y. A pathway and network review on beta-adrenoceptor signaling and beta blockers in cardiac remodeling. Heart Fail. Rev. 2014, 19, 799–814. [Google Scholar] [CrossRef]
- Vilar, S.; Sobarzo-Sanchez, E.; Santana, L.; Uriarte, E. Molecular Docking and Drug Discovery in beta-Adrenergic Receptors. Curr. Med. Chem. 2017, 24, 4340–4359. [Google Scholar] [CrossRef]
- Wengrowski, A.M.; Wang, X.; Tapa, S.; Posnack, N.G.; Mendelowitz, D.; Kay, M.W. Optogenetic release of norepinephrine from cardiac sympathetic neurons alters mechanical and electrical function. Cardiovasc. Res. 2015, 105, 143–150. [Google Scholar] [CrossRef]
- Prando, V.; Da Broi, F.; Franzoso, M.; Plazzo, A.P.; Pianca, N.; Francolini, M.; Basso, C.; Kay, M.W.; Zaglia, T.; Mongillo, M. Dynamics of neuroeffector coupling at cardiac sympathetic synapses. J. Physiol. 2018, 596, 2055–2075. [Google Scholar] [CrossRef]
- Malloy, C.; Sifers, J.; Mikos, A.; Samadi, A.; Omar, A.; Hermanns, C.; Cooper, R.L. Using optogenetics to assess neuroendocrine modulation of heart rate in Drosophila melanogaster larvae. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 2017, 203, 791–806. [Google Scholar] [CrossRef]
- Buckley, U.; Shivkumar, K.; Ardell, J.L. Autonomic Regulation Therapy in Heart Failure. Curr. Heart Fail. Rep. 2015, 12, 284–293. [Google Scholar] [CrossRef]
- Shen, M.J.; Zipes, D.P. Interventional and device-based autonomic modulation in heart failure. Heart Fail. Clin. 2015, 11, 337–348. [Google Scholar] [CrossRef]
- Yu, L.; Zhou, L.; Cao, G.; Po, S.S.; Huang, B.; Zhou, X.; Wang, M.; Yuan, S.; Wang, Z.; Wang, S.; et al. Optogenetic Modulation of Cardiac Sympathetic Nerve Activity to Prevent Ventricular Arrhythmias. J. Am. Coll. Cardiol. 2017, 70, 2778–2790. [Google Scholar] [CrossRef]
- Nussinovitch, U.; Shinnawi, R.; Gepstein, L. Modulation of cardiac tissue electrophysiological properties with light-sensitive proteins. Cardiovasc. Res. 2014, 102, 176–187. [Google Scholar] [CrossRef]
- Nussinovitch, U.; Gepstein, L. Optogenetics for suppression of cardiac electrical activity in human and rat cardiomyocyte cultures. Neurophotonics 2015, 2, 031204. [Google Scholar] [CrossRef]
- Mahoney, V.M.; Mezzano, V.; Mirams, G.R.; Maass, K.; Li, Z.; Cerrone, M.; Vasquez, C.; Bapat, A.; Delmar, M.; Morley, G.E. Connexin43 contributes to electrotonic conduction across scar tissue in the intact heart. Sci. Rep. 2016, 6, 26744. [Google Scholar] [CrossRef]
- Moore-Morris, T.; Cattaneo, P.; Puceat, M.; Evans, S.M. Origins of cardiac fibroblasts. J. Mol. Cell. Cardiol. 2016, 91, 1–5. [Google Scholar] [CrossRef]
- Mikawa, T.; Gourdie, R.G. Pericardial mesoderm generates a population of coronary smooth muscle cells migrating into the heart along with ingrowth of the epicardial organ. Dev. Biol. 1996, 174, 221–232. [Google Scholar] [CrossRef]
- Vasquez, C.; Morley, G.E. The origin and arrhythmogenic potential of fibroblasts in cardiac disease. J. Cardiovasc. Transl. Res. 2012, 5, 760–767. [Google Scholar] [CrossRef][Green Version]
- Kohl, P.; Kamkin, A.G.; Kiseleva, I.S.; Streubel, T. Mechanosensitive cells in the atrium of frog heart. Exp. Physiol. 1992, 77, 213–216. [Google Scholar] [CrossRef]
- Vasquez, C.; Mohandas, P.; Louie, K.L.; Benamer, N.; Bapat, A.C.; Morley, G.E. Enhanced fibroblast-myocyte interactions in response to cardiac injury. Circ. Res. 2010, 107, 1011–1020. [Google Scholar] [CrossRef]
- Kiseleva, I.; Kamkin, A.; Pylaev, A.; Kondratjev, D.; Leiterer, K.P.; Theres, H.; Wagner, K.D.; Persson, P.B.; Gunther, J. Electrophysiological properties of mechanosensitive atrial fibroblasts from chronic infarcted rat heart. J. Mol. Cell. Cardiol. 1998, 30, 1083–1093. [Google Scholar] [CrossRef]
- Vasquez, C.; Benamer, N.; Morley, G.E. The cardiac fibroblast: Functional and electrophysiological considerations in healthy and diseased hearts. J. Cardiovasc. Pharmacol. 2011, 57, 380–388. [Google Scholar] [CrossRef]
- Zhang, Y.; Kanter, E.M.; Yamada, K.A. Remodeling of cardiac fibroblasts following myocardial infarction results in increased gap junction intercellular communication. Cardiovasc. Pathol. 2010, 19, e233–e240. [Google Scholar] [CrossRef]
- Roell, W.; Lewalter, T.; Sasse, P.; Tallini, Y.N.; Choi, B.R.; Breitbach, M.; Doran, R.; Becher, U.M.; Hwang, S.M.; Bostani, T.; et al. Engraftment of connexin 43-expressing cells prevents post-infarct arrhythmia. Nature 2007, 450, 819–824. [Google Scholar] [CrossRef]
- Quinn, T.A.; Camelliti, P.; Rog-Zielinska, E.A.; Siedlecka, U.; Poggioli, T.; O’Toole, E.T.; Knopfel, T.; Kohl, P. Electrotonic coupling of excitable and nonexcitable cells in the heart revealed by optogenetics. Proc. Natl. Acad. Sci. USA 2016, 113, 14852–14857. [Google Scholar] [CrossRef]
- Yu, J.; Entcheva, E. Inscribing Optical Excitability to Non-Excitable Cardiac Cells: Viral Delivery of Optogenetic Tools in Primary Cardiac Fibroblasts. Methods Mol. Biol. 2016, 1408, 303–317. [Google Scholar] [CrossRef]
- Wu, Y.; Li, S.S.; Jin, X.; Cui, N.; Zhang, S.; Jiang, C. Optogenetic approach for functional assays of the cardiovascular system by light activation of the vascular smooth muscle. Vascul. Pharmacol. 2015, 71, 192–200. [Google Scholar] [CrossRef][Green Version]
- Figueroa, X.F.; Duling, B.R. Gap junctions in the control of vascular function. Antioxid. Redox Signal. 2009, 11, 251–266. [Google Scholar] [CrossRef]
- Zhang, S.; Cui, N.; Wu, Y.; Zhong, W.; Johnson, C.M.; Jiang, C. Optogenetic intervention to the vascular endothelium. Vascul. Pharmacol. 2015, 74, 122–129. [Google Scholar] [CrossRef] [PubMed]
- Hulsmans, M.; Sam, F.; Nahrendorf, M. Monocyte and macrophage contributions to cardiac remodeling. J. Mol. Cell. Cardiol. 2016, 93, 149–155. [Google Scholar] [CrossRef]
- Hulsmans, M.; Clauss, S.; Xiao, L.; Aguirre, A.D.; King, K.R.; Hanley, A.; Hucker, W.J.; Wulfers, E.M.; Seemann, G.; Courties, G.; et al. Macrophages Facilitate Electrical Conduction in the Heart. Cell 2017, 169, 510–522. [Google Scholar] [CrossRef] [PubMed]
- Munshi, N.V. Resident Macrophages: Near and Dear to Your Heart. Cell 2017, 169, 376–377. [Google Scholar] [CrossRef] [PubMed]
- Hulsmans, M.; Sager, H.B.; Roh, J.D.; Valero-Munoz, M.; Houstis, N.E.; Iwamoto, Y.; Sun, Y.; Wilson, R.M.; Wojtkiewicz, G.; Tricot, B.; et al. Cardiac macrophages promote diastolic dysfunction. J. Exp. Med. 2018, 215, 423–440. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zaglia, T.; Di Bona, A.; Mongillo, M. A Light Wand to Untangle the Myocardial Cell Network. Methods Protoc. 2019, 2, 34. https://doi.org/10.3390/mps2020034
Zaglia T, Di Bona A, Mongillo M. A Light Wand to Untangle the Myocardial Cell Network. Methods and Protocols. 2019; 2(2):34. https://doi.org/10.3390/mps2020034
Chicago/Turabian StyleZaglia, Tania, Anna Di Bona, and Marco Mongillo. 2019. "A Light Wand to Untangle the Myocardial Cell Network" Methods and Protocols 2, no. 2: 34. https://doi.org/10.3390/mps2020034
APA StyleZaglia, T., Di Bona, A., & Mongillo, M. (2019). A Light Wand to Untangle the Myocardial Cell Network. Methods and Protocols, 2(2), 34. https://doi.org/10.3390/mps2020034





