Ionic Liquid-Assisted Laser Desorption/Ionization–Mass Spectrometry: Matrices, Microextraction, and Separation
Abstract
1. Introduction
2. Matrix-Assisted Laser Desorption/Ionization–Mass Spectrometry
- Effective ionic liquid matrices (ILMs) usually should have high absorption at the same wavelength of the laser radiation.
- They should have capability to protonate (positive mode) or deprotonate (negative mode) the target analyte.
- They should effectively ionize the target analyte.
- They should effectively ionize all analytes in a mixture without or with minimal ion suppression.
- They should cause no fragmentation of the analytes.
- They should form no adduct species with the investigated analytes.
- They should be miscible with the analyte solution and co-crystalize with the investigated analytes.
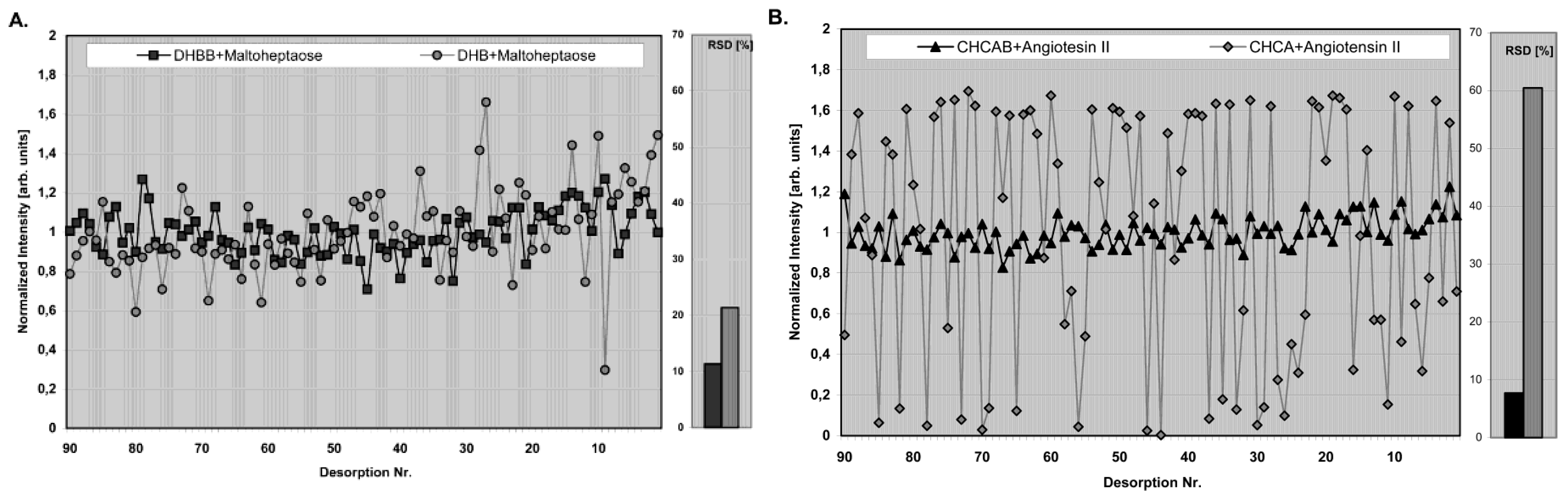
- They should ensure high reproducibility with very low relative standard deviation (RSD) from spot to spot.
- They should cause no change in the chemical structure of the investigated analyte.
- They should be cheap and nontoxic.
3. Ionic Liquids-Assisted Laser Desorption/Ionization Mass Spectrometry
3.1. Ionic Liquids-Assisted Laser Desorption/Ionization–Mass Spectrometry Applications for Proteins
3.2. Ionic Liquids-Assisted Laser Desorption/Ionization–Mass Spectrometry Applications for Peptides, Carbohydrates, Lipids, and Oligonucleotides
3.3. Ionic Liquid-Assisted Laser Desorption/Ionization–Mass Spectrometry Applications for Small Molecules
3.4. Ionic Liquid-Assisted Laser Desorption/Ionization–Mass Spectrometry Applications for Polymer and Pathogenic Bacteria
3.5. Imaging Using Ionic Liquid Matrices
3.6. Quantitative Analysis Using Ionic Liquid Matrices-Assisted Laser Desorption/Ionization-Mass Spectrometry
4. Factors Influencing the Analysis Using Ionic Liquid Matrices
4.1. Types of Ionic Liquid Matrices and Analytes
4.2. Preparation of Ionic Liquid Matrices
4.3. Sample Preparation
4.4. Solvent
4.5. Additives
4.6. Impurities
4.7. Instrumental Parameters
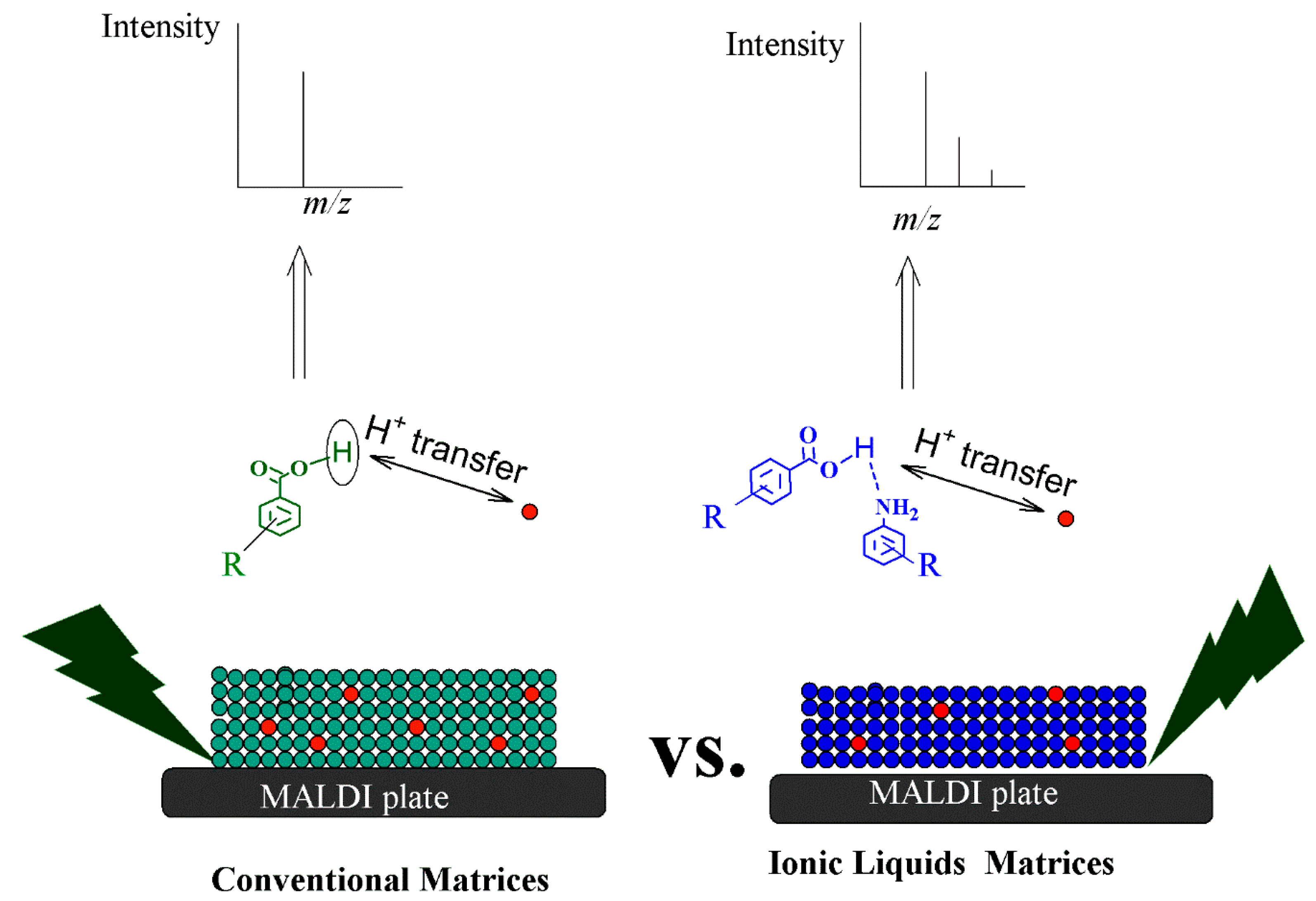
5. Principles and Mechanisms of Ionization Using Ionic Liquid Matrices
- (1)
- (2)
- Secondary ion formation, including H+ transfer, e- capture and H+ transfer, cationization, e- transfer, and ejection [138].
- (3)
- The ’’Lucky Survivor” model; this model claims that the ionization take places in the solution, and the ionized species retain their solution-state charge and exist as preformed ions within the solid state matrix [141].
- (4)
6. Advantages of Ionic Liquid as Matrices
7. Applications of Ionic Liquids for Microextraction Using Matrix Assisted Laser Desorption/Ionization Mass Spectrometry
8. Advantages of Ionic Liquids for Microextraction
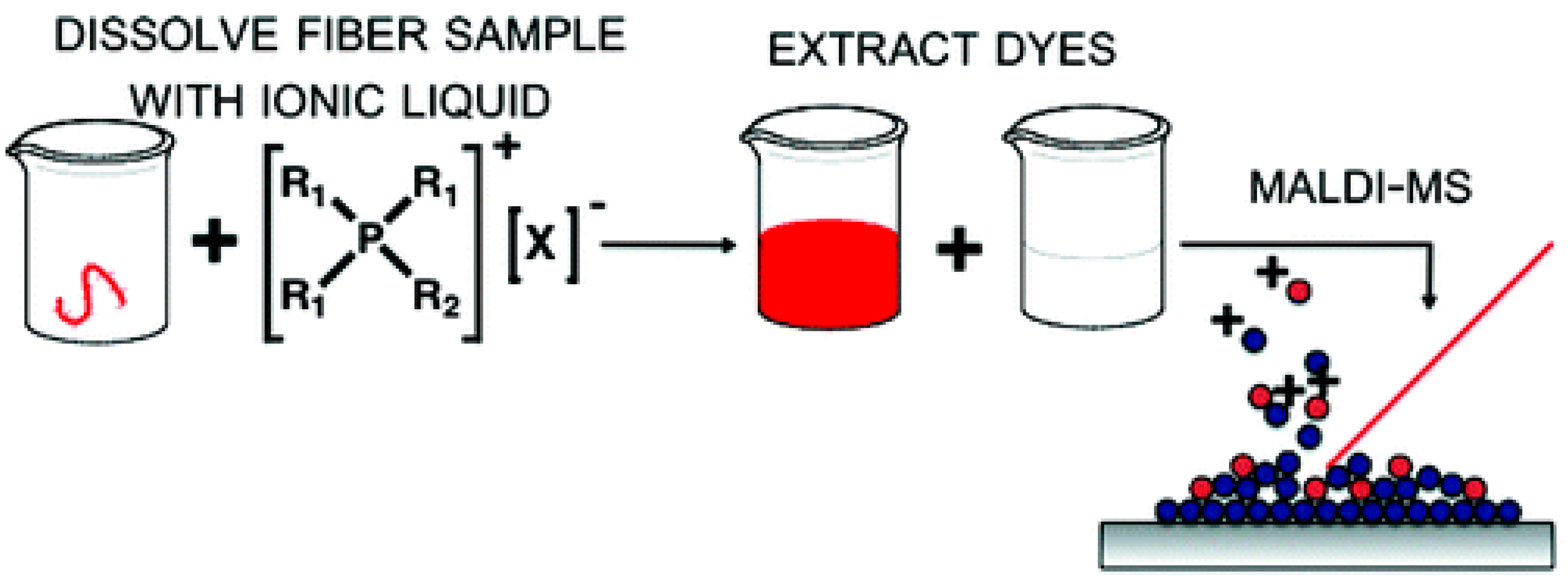
9. Applications of Ionic Liquids for Analyte Separation Using Matrix Assisted Laser Desorption/Ionization Mass Spectrometry
Advantages of Ionic Liquids for Separation
10. Challenges and Remarks
Funding
Acknowledgments
Conflicts of Interest
References
- Welton, T. Room-Temperature Ionic Liquids. Solvents for Synthesis and Catalysis. Chem. Rev. 1999, 99, 2071–2084. [Google Scholar] [CrossRef] [PubMed]
- Wasserscheid, P.; Keim, W. Ionic Liquids—New “Solutions” for Transition Metal Catalysis. Angew. Chem. 2000, 39, 3772–3789. [Google Scholar] [CrossRef]
- Wilkes, J.S.; Levisky, J.A.; Wilson, R.A.; Hussey, C.L. Dialkylimidazolium chloroaluminate melts: a new class of room-temperature ionic liquids for electrochemistry, spectroscopy and synthesis. Inorg. Chem. 1982, 21, 1263–1264. [Google Scholar] [CrossRef]
- Carmichael, A.J.; Seddon, K.R. Polarity study of some 1-alkyl-3-methylimidazolium ambient-temperature ionic liquids with the solvatochromic dye, Nile Red. J. Phys. Org. Chem. 2000, 13, 591–595. [Google Scholar] [CrossRef]
- Ding, J.; Armstrong, D.W. Chiral ionic liquids: Synthesis and applications. Chirality 2005, 17, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.L.; Ding, J.; Welton, T.; Armstrong, D.W. Characterizing ionic liquids on the basis of multiple solvation interactions. J. Am. Chem. Soc. 2002, 124, 14247–14254. [Google Scholar] [CrossRef] [PubMed]
- Hulsbosch, J.; De Vos, D.E.; Binnemans, K.; Ameloot, R. Biobased Ionic Liquids: Solvents for a Green Processing Industry? ACS Sustain. Chem. Eng. 2016, 4, 2917–2931. [Google Scholar] [CrossRef]
- Aggarwal, R.; Khullar, P.; Mandial, D.; Mahal, A.; Ahluwalia, G.K.; Bakshi, M.S. Bipyridinium and Imidazolium Ionic Liquids for Nanomaterials Synthesis: pH Effect, Phase Transfer Behavior, and Protein Extraction. ACS Sustain. Chem. Eng. 2017, 5, 7859–7870. [Google Scholar] [CrossRef]
- Dai, C.; Zhang, J.; Huang, C.; Lei, Z. Ionic Liquids in Selective Oxidation: Catalysts and Solvents. Chem. Rev. 2017, 117, 6929–6983. [Google Scholar] [CrossRef] [PubMed]
- Eftekhari, A. Supercapacitors utilising ionic liquids. Energy Storage Mater. 2017, 9, 47–69. [Google Scholar] [CrossRef]
- Wasilewski, T.; Gębicki, J.; Kamysz, W. Prospects of ionic liquids application in electronic and bioelectronic nose instruments. TrAC Trends Anal. Chem. 2017, 93, 23–36. [Google Scholar] [CrossRef]
- Tan, Z.; Liu, J.; Pang, L. Advances in analytical chemistry using the unique properties of ionic liquids. TrAC Trends Anal. Chem. 2012, 39, 218–227. [Google Scholar] [CrossRef]
- Soukup-Hein, R.J.; Warnke, M.M.; Armstrong, D.W. Ionic Liquids in Analytical Chemistry. Annu. Rev. Anal. Chem. 2009, 2, 145–168. [Google Scholar] [CrossRef] [PubMed]
- Ho, T.D.; Zhang, C.; Hantao, L.W.; Anderson, J.L. Ionic liquids in analytical chemistry: Fundamentals, advances, and perspectives. Anal. Chem. 2014, 86, 262–285. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S. Analytical applications of room-temperature ionic liquids: A review of recent efforts. Anal. Chim. Acta 2006, 556, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Jiang, G.; Liu, J.; Jönsson, J.Å. Application of ionic liquids in analytical chemistry. TrAC Trends Anal. Chem. 2005, 24, 20–27. [Google Scholar] [CrossRef]
- Shamsi, S.A.; Danielson, N.D. Utility of ionic liquids in analytical separations. J. Sep. Sci. 2007, 30, 1729–1750. [Google Scholar] [CrossRef] [PubMed]
- Berthod, A.; Ruiz-Ángel, M.J.; Carda-Broch, S. Ionic liquids in separation techniques. J. Chromatogr. A 2008, 1184, 6–18. [Google Scholar] [CrossRef] [PubMed]
- Andre, M.; Loidl, J.; Laus, G.; Schottenberger, H.; Bentivoglio, G.; Wurst, K.; Ongania, K.H. Ionic liquids as advantageous solvents for headspace gas chromatography of compounds with low vapor pressure. Anal. Chem. 2005, 77, 702–705. [Google Scholar] [CrossRef] [PubMed]
- Marszałł, M.P.; Kaliszan, R. Application of Ionic Liquids in Liquid Chromatography. Crit. Rev. Anal. Chem. 2007, 37, 127–140. [Google Scholar] [CrossRef]
- Shi, X.; Qiao, L.; Xu, G. Recent development of ionic liquid stationary phases for liquid chromatography. J. Chromatogr. A 2015, 1420, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, G.; Claudia Röwer, C.; Koy, C.; Protzel, C.; Lorenz, P.; Thiesen, H.-J.; Hakenberg, O.; Glocker, M. Ultraviolet matrix-assisted laser desorption/ionization time-of-flight mass spectrometry for phosphopeptide analysis with a solidified ionic liquid matrix. Eur. J. Mass Spectrom. 2015, 21, 65. [Google Scholar] [CrossRef] [PubMed]
- Weidmann, S.; Kemmerling, S.; Mädler, S.; Stahlberg, H.; Braun, T.; Zenobi, R. Ionic liquids as matrices in microfluidic sample deposition for high-mass matrix- assisted laser desorption/ionization mass spectrometry. Eur. J. Mass Spectrom. (Chichester, Eng.) 2012, 18, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Yao, B.; Li, C.; Shi, G. An improved Hummers method for eco-friendly synthesis of graphene oxide. Carbon 2013, 64, 225–229. [Google Scholar] [CrossRef]
- Chen, X.; Bian, Y.; Liu, F.; Teng, P.; Sun, P. Comparison of micellar extraction combined with ionic liquid based vortex-assisted liquid–liquid microextraction and modified quick, easy, cheap, effective, rugged, and safe method for the determination of difenoconazole in cowpea. J. Chromatogr. A 2017, 1518, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Wei, D.; Ivaska, A. Applications of ionic liquids in electrochemical sensors. Anal. Chim. Acta 2008, 607, 126–135. [Google Scholar] [CrossRef] [PubMed]
- Soukup-Hein, R.J.; Remsburg, J.W.; Breitbach, Z.S.; Sharma, P.S.; Payagala, T.; Wanigasekara, E.; Huang, J.; Armstrong, D.W. Evaluating the use of tricationic reagents for the detection of doubly charged anions in the positive mode by ESI-MS. Anal. Chem. 2008, 80, 2612–2616. [Google Scholar] [CrossRef] [PubMed]
- Remsburg, J.W.; Soukup-Hein, R.J.; Crank, J.A.; Breitbach, Z.S.; Payagala, T.; Armstrong, D.W. Evaluation of Dicationic Reagents for Their Use in Detection of Anions Using Positive Ion Mode ESI-MS Via Gas Phase Ion Association. J. Am. Soc. Mass Spectrom. 2008, 19, 261–269. [Google Scholar] [CrossRef] [PubMed]
- Soukup-Hein, R.J.; Remsburg, J.W.; Dasgupta, P.K.; Armstrong, D.W. A general, positive ion mode ESI-MS approach for the analysis of singly charged inorganic and organic anions using a dicationic reagent. Anal. Chem. 2007, 79, 7346–7352. [Google Scholar] [CrossRef] [PubMed]
- Carda-Broch, S.; García-Alvarez-Coque, M.C.; Ruiz-Angel, M.J. Extent of the influence of phosphate buffer and ionic liquids on the reduction of the silanol effect in a C18 stationary phase. J. Chromatogr. A 2018, 1559, 112–117. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Angel, M.J.; Carda-Broch, S.; Berthod, A. Ionic liquids versus triethylamine as mobile phase additives in the analysis of β-blockers. J. Chromatogr. A 2006, 1119, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Turker, S.D.; Dunn, W.B.; Wilkie, J. MALDI-MS of drugs: Profiling, imaging, and steps towards quantitative analysis. Appl. Spectrosc. Rev. 2017, 52, 73–99. [Google Scholar] [CrossRef]
- Francese, S.; Bradshaw, R.; Denison, N. An update on MALDI mass spectrometry based technology for the analysis of fingermarks—Stepping into operational deployment. Analyst 2017, 142, 2518–2546. [Google Scholar] [CrossRef] [PubMed]
- Abdelhamid, H.N.; Wu, H. Soft Ionization of Metallo-Mefenamic Using Electrospray Ionization Mass Spectrometry. Mass Spectrom. Lett. 2015, 6, 43–47. [Google Scholar] [CrossRef]
- Sekar, R.; Kailasa, S.K.; Abdelhamid, H.N.; Chen, Y.-C.; Wu, H.-F. Electrospray ionization tandem mass spectrometric studies of copper and iron complexes with tobramycin. Int. J. Mass Spectrom. 2013, 338, 23–29. [Google Scholar] [CrossRef]
- Chen, Y.-C.; Abdelhamid, H.N.; Wu, H.-F. Simple and Direct Quantitative Analysis for Quinidine Drug in Fish Tissues. Mass Spectrom. Lett. 2017, 8, 8–13. [Google Scholar] [CrossRef]
- Kumaran, S.; Abdelhamid, H.N.; Wu, H.-F. Quantification analysis of protein and mycelium contents upon inhibition of melanin for: Aspergillus Niger: A study of matrix assisted laser desorption/ionization mass spectrometry (MALDI-MS). RSC Adv. 2017, 7, 30289–30294. [Google Scholar] [CrossRef]
- Abdelhamid, H.N.; Khan, M.S.; Wu, H.-F. Graphene oxide as a nanocarrier for gramicidin (GOGD) for high antibacterial performance. RSC Adv. 2014, 4, 50035–50046. [Google Scholar] [CrossRef]
- Abdelhamid, H.N. Delafossite Nanoparticle as New Functional Materials: Advances in Energy, Nanomedicine and Environmental Applications. Mater. Sci. Forum 2015, 832, 28–53. [Google Scholar] [CrossRef]
- Abdelhamid, H.N.; Wu, H.-F. A method to detect metal-drug complexes and their interactions with pathogenic bacteria via graphene nanosheet assist laser desorption/ionization mass spectrometry and biosensors. Anal. Chim. Acta 2012, 751, 94–104. [Google Scholar] [CrossRef] [PubMed]
- Abdelhamid, H.N.; Wu, H.-F. Synthesis of a highly dispersive sinapinic acid@graphene oxide (SA@GO) and its applications as a novel surface assisted laser desorption/ionization mass spectrometry for proteomics and pathogenic bacteria biosensing. Analyst 2015, 140, 1555–1565. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.-S.; Abdelhamid, H.N.; Wu, H.-F. Synthesis and antibacterial activities of graphene decorated with stannous dioxide. RSC Adv. 2014, 4, 3722. [Google Scholar] [CrossRef]
- Abdelhamid, H.N.; Wu, H.-F. Multifunctional graphene magnetic nanosheet decorated with chitosan for highly sensitive detection of pathogenic bacteria. J. Mater. Chem. B 2013, 1, 3950–3961. [Google Scholar] [CrossRef]
- Abdelhamid, H.N.; Wu, H.-F. Probing the interactions of chitosan capped CdS quantum dots with pathogenic bacteria and their biosensing application. J. Mater. Chem. B 2013, 1, 6094–6106. [Google Scholar] [CrossRef]
- Abdelhamid, H.N.; Wu, H.-F. Polymer dots for quantifying the total hydrophobic pathogenic lysates in a single drop. Colloids Surf. B. Biointerfaces 2013, 115C, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Gedda, G.; Abdelhamid, H.N.; Khan, M.S.; Wu, H.-F. ZnO nanoparticle-modified polymethyl methacrylate-assisted dispersive liquid–liquid microextraction coupled with MALDI-MS for rapid pathogenic bacteria analysis. RSC Adv. 2014, 4, 45973–45983. [Google Scholar] [CrossRef]
- Gopal, J.; Abdelhamid, H.N.; Hua, P.-Y.; Wu, H.-F. Chitosan nanomagnets for effective extraction and sensitive mass spectrometric detection of pathogenic bacterial endotoxin from human urine. J. Mater. Chem. B 2013, 1, 2463–2475. [Google Scholar] [CrossRef]
- Abdelhamid, H.N.; Wu, H.-F. Proteomics analysis of the mode of antibacterial action of nanoparticles and their interactions with proteins. TrAC Trends Anal. Chem. 2014, 65, 30–46. [Google Scholar] [CrossRef]
- Wu, H.F.; Gopal, J.; Abdelhamid, H.N.; Hasan, N. Quantum dot applications endowing novelty to analytical proteomics. Proteomics 2012, 12, 2949–2961. [Google Scholar] [CrossRef] [PubMed]
- Shastri, L.; Abdelhamid, H.N.; Nawaz, M.; Wu, H.-F. Synthesis, characterization and bifunctional applications of bidentate silver nanoparticle assisted single drop microextraction as a highly sensitive preconcentrating probe for protein analysis. RSC Adv. 2015, 5, 41595–41603. [Google Scholar] [CrossRef]
- Abdelhamid, H.N.; Kumaran, S.; Wu, H.-F. One-pot synthesis of CuFeO2 nanoparticles capped with glycerol and proteomic analysis of their nanocytotoxicity against fungi. RSC Adv. 2016, 6, 97629–97635. [Google Scholar] [CrossRef]
- Jiang, D.; Song, N.; Li, X.; Ma, J.; Jia, Q. Highly selective enrichment of phosphopeptides by on-chip indium oxide functionalized magnetic nanoparticles coupled with MALDI-TOF MS. Proteomics 2017, 1700213. [Google Scholar] [CrossRef] [PubMed]
- Abdelhamid, H.N.; Wu, H.-F. Facile synthesis of nano silver ferrite (AgFeO2) modified with chitosan applied for biothiol separation. Mater. Sci. Eng. C Mater. Biol. Appl. 2014, 45, 438–445. [Google Scholar] [CrossRef] [PubMed]
- Abdelhamid, H.N.; Lin, Y.C.; Wu, H.-F. Magnetic nanoparticle modified chitosan for surface enhanced laser desorption/ionization mass spectrometry of surfactants. RSC Adv. 2017, 7, 41585–41592. [Google Scholar] [CrossRef]
- Hua, P.-Y.; Manikandan, M.; Abdelhamid, H.N.; Wu, H.-F. Graphene nanoflakes as an efficient ionizing matrix for MALDI-MS based lipidomics of cancer cells and cancer stem cells. J. Mater. Chem. B 2014, 2, 7334–7343. [Google Scholar] [CrossRef]
- Abdelhamid, H.N.; Lin, Y.C.; Wu, H.F. Thymine chitosan nanomagnets for specific preconcentration of mercury (II) prior to analysis using SELDI-MS. Microchim. Acta 2017, 184, 1517–1527. [Google Scholar] [CrossRef]
- Abdelhamid, H.N.; Wu, H.-F. Ultrasensitive, rapid, and selective detection of mercury using graphene assisted laser desorption/ionization mass spectrometry. J. Am. Soc. Mass Spectrom. 2014, 25, 861–868. [Google Scholar] [CrossRef] [PubMed]
- Abdelhamid, H.N.; Talib, A.; Wu, H.F. One pot synthesis of gold—Carbon dots nanocomposite and its application for cytosensing of metals for cancer cells. Talanta 2017, 166, 357–363. [Google Scholar] [CrossRef] [PubMed]
- Abdelhamid, H.N. Ionic liquids for mass spectrometry: matrices, separation and microextraction. TrAC Trends Anal. Chem. 2015. [Google Scholar] [CrossRef]
- Nicolardi, S.; van der Burgt, Y.E.M.; Codée, J.D.C.; Wuhrer, M.; Hokke, C.H.; Chiodo, F. Structural Characterization of Biofunctionalized Gold Nanoparticles by Ultrahigh-Resolution Mass Spectrometry. ACS Nano 2017, 11, 8257–8264. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Shen, Y.; Franke, D.; Sebastián, V.; Bawendi, M.G.; Jensen, K.F. Characterization of Indium Phosphide Quantum Dot Growth Intermediates Using MALDI-TOF Mass Spectrometry. J. Am. Chem. Soc. 2016, 138, 13469–13472. [Google Scholar] [CrossRef] [PubMed]
- Abdelhamid, H.N.; Chen, Z.-Y.; Wu, H.-F. Surface tuning laser desorption/ionization mass spectrometry (STLDI-MS) for the analysis of small molecules using quantum dots. Anal. Bioanal. Chem. 2017, 409, 4943–4950. [Google Scholar] [CrossRef] [PubMed]
- Abdelhamid, H.N. Organic matrices, ionic liquids, and organic matrices@nanoparticles assisted laser desorption/ionization mass spectrometry. TrAC Trends Anal. Chem. 2017, 89, 68–98. [Google Scholar] [CrossRef]
- Abdelhamid, H.N. Applications of Nanomaterials and Organic Semiconductors for Bacteria &Biomolecules Analysis/Biosensing Using Laser Analytical Spectroscopy. Master’s Thesis, National Sun-Yat Sen University, Kaohsiung, Taiwan, 2013. [Google Scholar]
- Abdelhamid, H.N. Ionic Liquids Matrices for Laser Assisted Desorption/Ionization Mass Spectrometry. Mass Spectrom. Purif. Tech. 2015, 1, 109–119. [Google Scholar] [CrossRef]
- Abdelhamid, H.N. Physicochemical Properties of Proteomic Ionic Liquids Matrices for MALDI-MS. J. Data Min. Genom. Proteom. 2016, 7, 189. [Google Scholar] [CrossRef]
- Chiang, C.-K.; Chen, W.-T.; Chang, H.-T. Nanoparticle-based mass spectrometry for the analysis of biomolecules. Chem. Soc. Rev. 2011, 40, 1269–1281. [Google Scholar] [CrossRef] [PubMed]
- Nasser Abdelhamid, H.; Wu, H.F. Furoic and mefenamic acids as new matrices for matrix assisted laser desorption/ionization-(MALDI)-mass spectrometry. Talanta 2013, 115, 442–450. [Google Scholar] [CrossRef] [PubMed]
- Shahnawaz Khan, M.; Abdelhamid, H.N.; Wu, H.-F. Near infrared (NIR) laser mediated surface activation of graphene oxide nanoflakes for efficient antibacterial, antifungal and wound healing treatment. Colloids Surf. B Biointerfaces 2015, 127C, 281–291. [Google Scholar] [CrossRef] [PubMed]
- Abdelhamid, H.N.; Wu, B.-S.; Wu, H.-F. Graphene coated silica applied for high ionization matrix assisted laser desorption/ionization mass spectrometry: A novel approach for environmental and biomolecule analysis. Talanta 2014, 126, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Abdelhamid, H.N.; Bhaisare, M.L.; Wu, H.-F. Ceria nanocubic-ultrasonication assisted dispersive liquid-liquid microextraction coupled with matrix assisted laser desorption/ionization mass spectrometry for pathogenic bacteria analysis. Talanta 2014, 120, 208–217. [Google Scholar] [CrossRef] [PubMed]
- Abdelhamid, H.N.; Wu, H.-F. Gold nanoparticles assisted laser desorption/ionization mass spectrometry and applications: from simple molecules to intact cells. Anal. Bioanal. Chem. 2016, 408, 4485–4502. [Google Scholar] [CrossRef] [PubMed]
- Abdelhamid, H.N.; Talib, A.; Wu, H.-F. Correction: Facile synthesis of water soluble silver ferrite (AgFeO2) nanoparticles and their biological application as antibacterial agents. RSC Adv. 2015, 5, 39952–39953. [Google Scholar] [CrossRef]
- Abdelhamid, H.N.; Talib, A.; Wu, H.-F. Facile synthesis of water soluble silver ferrite (AgFeO2) nanoparticles and their biological application as antibacterial agents. RSC Adv. 2015, 5, 34594–34602. [Google Scholar] [CrossRef]
- Abdelhamid, H.N.; Wu, H.-F. Monitoring metallofulfenamic–bovine serum albumin interactions: A novel method for metallodrug analysis. RSC Adv. 2014, 4, 53768–53776. [Google Scholar] [CrossRef]
- Chen, Z.-Y.; Abdelhamid, H.N.; Wu, H.-F. Effect of surface capping of quantum dots (CdTe) on proteomics. Rapid Commun. Mass Spectrom. 2016, 30, 1403–1412. [Google Scholar] [CrossRef] [PubMed]
- Abdelhamid, H.N. Ionic liquids for mass spectrometry: Matrices, separation and microextraction. TrAC Trends Anal. Chem. 2016, 77, 122–138. [Google Scholar] [CrossRef]
- Armstrong, D.W.; Zhang, L.-K.; He, L.; Gross, M.L. Ionic Liquids as Matrixes for Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry. Anal. Chem. 2001, 73, 3679–3686. [Google Scholar] [CrossRef] [PubMed]
- Tholey, A.; Heinzle, E. Ionic (liquid) matrices for matrix-assisted laser desorption/ionization mass spectrometry—Applications and perspectives. Anal. Bioanal. Chem. 2006, 386, 24–37. [Google Scholar] [CrossRef] [PubMed]
- Fukuyama, Y.; Funakoshi, N.; Takeyama, K.; Hioki, Y.; Nishikaze, T.; Kaneshiro, K.; Kawabata, S.; Iwamoto, S.; Tanaka, K. 3-Aminoquinoline/p-Coumaric Acid as a MALDI Matrix for Glycopeptides, Carbohydrates, and Phosphopeptides. Anal. Chem. 2014, 86, 1937–1942. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Terasawa, K.; Kaneshiro, K.; Uchimura, H.; Yamamoto, R.; Fukuyama, Y.; Shimizu, K.; Sato, T.-A.; Tanaka, K. Improvement of mass spectrometry analysis of glycoproteins by MALDI-MS using 3-aminoquinoline/α-cyano-4-hydroxycinnamic acid. Anal. Bioanal. Chem. 2013, 405, 4289–4293. [Google Scholar] [CrossRef] [PubMed]
- Fukuyama, Y.; Nakaya, S.; Yamazaki, Y.; Tanaka, K. Ionic Liquid Matrixes Optimized for MALDI-MS of Sulfated/Sialylated/Neutral Oligosaccharides and Glycopeptides. Anal. Chem. 2008, 80, 2171–2179. [Google Scholar] [CrossRef] [PubMed]
- Towers, M.W.; Mckendrick, J.E.; Cramer, R. Introduction of 4-Chloro-α-cyanocinnamic Acid Liquid Matrices for High Sensitivity UV-MALDI MS. J. Proteome Res. 2010, 9, 1931–1940. [Google Scholar] [CrossRef] [PubMed]
- Calvano, C.D.; Carulli, S.; Palmisano, F. Aniline/α-cyano-4-hydroxycinnamic acid is a highly versatile ionic liquid for matrix-assisted laser desorption/ ionization mass spectrometry. Rapid Commun. Mass Spectrom. 2009, 23, 1659–1668. [Google Scholar] [CrossRef] [PubMed]
- Snovida, S.I.; Chen, V.C.; Perreault, H. Use of a 2,5-Dihydroxybenzoic Acid/Aniline MALDI Matrix for Improved Detection and On-Target Derivatization of Glycans: A Preliminary Report. Anal. Chem. 2006, 78, 8561–8568. [Google Scholar] [CrossRef] [PubMed]
- Mank, M.; Stahl, B.; Boehm, G. 2,5-Dihydroxybenzoic Acid Butylamine and Other Ionic Liquid Matrixes for Enhanced MALDI-MS Analysis of Biomolecules. Anal. Chem. 2004, 76, 2938–2950. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.L.; Gross, M.L. Ionic-liquid matrices for quantitative analysis by MALDI-TOF mass spectrometry. J. Am. Soc. Mass Spectrom. 2004, 15, 1833–1837. [Google Scholar] [CrossRef] [PubMed]
- Palmblad, M.; Cramer, R. Liquid Matrix Deposition on Conductive Hydrophobic Surfaces for Tuning and Quantitation in UV-MALDI Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2007, 18, 693–697. [Google Scholar] [CrossRef] [PubMed]
- Park, K.M.; Hyeon, T.; Kim, M.S.; Moon, J.H. In Situ Quantification and Profiling of Phosphatidylcholine in Mouse Brain Tissue by Matrix-assisted Laser Desorption Ionization with a Liquid Matrix. Bull. Korean Chem. Soc. 2017, 38, 636–641. [Google Scholar] [CrossRef]
- Jones, J.J.; Batoy, S.M.A.B.; Wilkins, C.L.; Liyanage, R.; Lay, J.O. Ionic liquid matrix-induced metastable decay of peptides and oligonucleotides and stabilization of phospholipids in MALDI FTMS analyses. J. Am. Soc. Mass Spectrom. 2005, 16, 2000–2008. [Google Scholar] [CrossRef] [PubMed]
- Bronzel, J.L.; Milagre, C.D.F.; Milagre, H.M.S. Analysis of low molecular weight compounds using MALDI- and LDI-TOF-MS: Direct detection of active pharmaceutical ingredients in different formulations. J. Mass Spectrom. 2017, 52, 752–758. [Google Scholar] [CrossRef] [PubMed]
- Leipert, J.; Treitz, C.; Leippe, M.; Tholey, A. Identification and Quantification of N-Acyl Homoserine Lactones Involved in Bacterial Communication by Small-Scale Synthesis of Internal Standards and Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2017. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.L.; Gross, M.L.; Hsu, F.F. Ionic-liquid matrices for improved analysis of phospholipids by MALDI-TOF mass spectrometry. J. Am. Soc. Mass Spectrom. 2005, 16, 679–682. [Google Scholar] [CrossRef] [PubMed]
- Spencer, S.E.; Kim, S.Y.; Kim, S.B.; Schug, K.A. Matrix-assisted laser desorption/ionization–time of flight-mass spectrometry profiling of trace constituents of condom lubricants in the presence of biological fluids. Forensic Sci. Int. 2011, 207, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.H.; Park, K.M.; Ahn, S.H.; Lee, S.H.; Kim, M.S. Investigations of Some Liquid Matrixes for Analyte Quantification by MALDI. J. Am. Soc. Mass Spectrom. 2015, 26, 1657–1664. [Google Scholar] [CrossRef] [PubMed]
- Crank, J.A.; Armstrong, D.W. Towards a Second Generation of Ionic Liquid Matrices (ILMs) for MALDI-MS of Peptides, Proteins, and Carbohydrates. J. Am. Soc. Mass Spectrom. 2009, 20, 1790–1800. [Google Scholar] [CrossRef] [PubMed]
- Kaneshiro, K.; Fukuyama, Y.; Iwamoto, S.; Sekiya, S.; Tanaka, K. Highly Sensitive MALDI Analyses of Glycans by a New Aminoquinoline-Labeling Method Using 3-Aminoquinoline/α-Cyano-4-hydroxycinnamic Acid Liquid Matrix. Anal. Chem. 2011, 83, 3663–3667. [Google Scholar] [CrossRef] [PubMed]
- Tissot, B.; Gasiunas, N.; Powell, A.K.; Ahmed, Y.; Zhi, Z.; Haslam, S.M.; Morris, H.R.; Turnbull, J.E.; Gallagher, J.T.; Dell, A. Towards GAG glycomics: Analysis of highly sulfated heparins by MALDI-TOF mass spectrometry. Glycobiology 2007, 17, 972–982. [Google Scholar] [CrossRef] [PubMed]
- Ramos Catharino, R.; de Azevedo Marques, L.; Silva Santos, L.; Baptista, A.S.; Glória, E.M.; Calori-Domingues, M.A.; Facco, E.M.P.; Eberlin, M.N. Aflatoxin Screening by MALDI-TOF Mass Spectrometry. Anal. Chem. 2005, 77, 8155–8157. [Google Scholar] [CrossRef] [PubMed]
- Snovida, S.I.; Rak-Banville, J.M.; Perreault, H. On the Use of DHB/Aniline and DHB/N,N-Dimethylaniline Matrices for Improved Detection of Carbohydrates: Automated Identification of Oligosaccharides and Quantitative Analysis of Sialylated Glycans by MALDI-TOF Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2008, 19, 1138–1146. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Shen, S.; Wu, D.; Cai, P.; Pan, Y. Novel ionic liquid matrices for qualitative and quantitative detection of carbohydrates by matrix assisted laser desorption/ionization mass spectrometry. Anal. Chim. Acta 2017, 985, 114–120. [Google Scholar] [CrossRef] [PubMed]
- Snovida, S.I.; Perreault, H. A 2,5-dihydroxybenzoic acid/N,N-dimethylaniline matrix for the analysis of oligosaccharides by matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun. Mass Spectrom. 2007, 21, 3711–3715. [Google Scholar] [CrossRef] [PubMed]
- Schnöll-Bitai, I.; Ullmer, R.; Hrebicek, T.; Rizzi, A.; Lacik, I. Characterization of the molecular mass distribution of pullulans by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry using 2,5-dihydroxybenzoic acid butylamine (DHBB) as liquid matrix. Rapid Commun. Mass Spectrom. 2008, 22, 2961–2970. [Google Scholar] [CrossRef] [PubMed]
- PEI, X.-L.; HUANG, Y.-Y.; GONG, C.; XU, X. Matrix-assisted Laser Desorption/Ionization-Mass Spectrometry Imaging of Oligosaccharides in Soybean and Bean Leaf with Ionic Liquid as Matrix. Chinese J. Anal. Chem. 2017, 45, 1155–1163. [Google Scholar] [CrossRef]
- Laremore, T.N.; Murugesan, S.; Park, T.J.; Avci, F.Y.; Zagorevski, D.V.; Linhardt, R.J. Matrix-assisted laser desorption/ionization mass spectrometric analysis of uncomplexed highly sulfated oligosaccharides using ionic liquid matrices. Anal. Chem. 2006, 78, 1774–1779. [Google Scholar] [CrossRef] [PubMed]
- Abdelhamid, H.N.; Khan, M.S.; Wu, H.-F. Design, characterization and applications of new ionic liquid matrices for multifunctional analysis of biomolecules: A novel strategy for pathogenic bacteria biosensing. Anal. Chim. Acta 2014, 823, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Nishikaze, T.; Fukuyama, Y.; Kawabata, S.I.; Tanaka, K. Sensitive analyses of neutral N -glycans using anion-doped liquid matrix G3CA by negative-ion matrix-assisted laser desorption/ionization mass spectrometry. Anal. Chem. 2012, 84, 6097–6103. [Google Scholar] [CrossRef] [PubMed]
- Ling, L.; Xiao, C.; Jiang, L.; Wang, S.; Li, Y.; Chen, X.; Guo, X. A cool and high salt-tolerant ionic liquid matrix for preferential ionization of phosphopeptides by negative ion MALDI-MS. New J. Chem. 2017, 41, 12241–12249. [Google Scholar] [CrossRef]
- Zhang, S.; Yao, Z.-P. Improved detection of phosphopeptides by negative ion matrix-assisted laser desorption/ionization mass spectrometry using a proton sponge co-matrix. Anal. Chim. Acta 2012, 711, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Przybylski, C.; Gonnet, F.; Bonnaffé, D.; Hersant, Y.; Lortat-Jacob, H.; Daniel, R. HABA-based ionic liquid matrices for UV-MALDI-MS analysis of heparin and heparan sulfate oligosaccharides. Glycobiology 2009, 20, 224–234. [Google Scholar] [CrossRef] [PubMed]
- Zabet-Moghaddam, M.; Heinzle, E.; Tholey, A. Qualitative and quantitative analysis of low molecular weight compounds by ultraviolet matrix-assisted laser desorption/ionization mass spectrometry using ionic liquid matrices. Rapid Commun. Mass Spectrom. 2004, 18, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Bonnel, D.; Franck, J.; Mériaux, C.; Salzet, M.; Fournier, I. Ionic matrices pre-spotted matrix-assisted laser desorption/ionization plates for patient maker following in course of treatment, drug titration, and MALDI mass spectrometry imaging. Anal. Biochem. 2013, 434, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Lemaire, R.; Tabet, J.C.; Ducoroy, P.; Hendra, J.B.; Salzet, M.; Fournier, I. Solid Ionic Matrixes for Direct Tissue Analysis and MALDI Imaging. Anal. Chem. 2006, 78, 809–819. [Google Scholar] [CrossRef] [PubMed]
- Ropartz, D.; Bodet, P.-E.; Przybylski, C.; Gonnet, F.; Daniel, R.; Fer, M.; Helbert, W.; Bertrand, D.; Rogniaux, H. Performance evaluation on a wide set of matrix-assisted laser desorption ionization matrices for the detection of oligosaccharides in a high-throughput mass spectrometric screening of carbohydrate depolymerizing enzymes. Rapid Commun. Mass Spectrom. 2011, 25, 2059–2070. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.; Lanthier, P.; Liu, X.; Sandhu, J.K.; Stanimirovic, D.; Li, J. MALDI mass spectrometry imaging of gangliosides in mouse brain using ionic liquid matrix. Anal. Chim. Acta 2009, 639, 57–61. [Google Scholar] [CrossRef] [PubMed]
- Byliński, H.; Gębicki, J.; Dymerski, T.; Namieśnik, J. Direct Analysis of Samples of Various Origin and Composition Using Specific Types of Mass Spectrometry. Crit. Rev. Anal. Chem. 2017, 47, 340–358. [Google Scholar] [CrossRef] [PubMed]
- Carda-Broch, S.; Berthod, A.; Armstrong, D.W. Ionic matrices for matrix-assisted laser desorption/ionization time-of-flight detection of DNA oligomers. Rapid Commun. Mass Spectrom. 2003, 17, 553–560. [Google Scholar] [CrossRef] [PubMed]
- Ham, B.M.; Jacob, J.T.; Cole, R.B. MALDI-TOF MS of phosphorylated lipids in biological fluids using immobilized metal affinity chromatography and a solid ionic crystal matrix. Anal. Chem. 2005, 77, 4439–4447. [Google Scholar] [CrossRef] [PubMed]
- Meriaux, C.; Franck, J.; Wisztorski, M.; Salzet, M.; Fournier, I. Liquid ionic matrixes for MALDI mass spectrometry imaging of lipids. J. Proteom. 2010, 73, 1204–1218. [Google Scholar] [CrossRef] [PubMed]
- Shrivas, K.; Hayasaka, T.; Goto-Inoue, N.; Sugiura, Y.; Zaima, N.; Setou, M. Ionic matrix for enhanced MALDI imaging mass spectrometry for identification of phospholipids in mouse liver and cerebellum tissue sections. Anal. Chem. 2010, 82, 8800–8806. [Google Scholar] [CrossRef] [PubMed]
- Hurtado, P.; Hortal, A.R.; Martínez-Haya, B. Matrix-assisted laser desorption/ionization detection of carbonaceous compounds in ionic liquid matrices. Rapid Commun. Mass Spectrom. 2007, 21, 3161–3164. [Google Scholar] [CrossRef] [PubMed]
- Gabler, C.; Pittenauer, E.; Dörr, N.; Allmaier, G. Imaging of a Tribolayer Formed from Ionic Liquids by Laser Desorption/Ionization-Reflectron Time-of-Flight Mass Spectrometry. Anal. Chem. 2012, 84, 10708–10714. [Google Scholar] [CrossRef] [PubMed]
- Shariatgorji, M.; Nilsson, A.; Källback, P.; Karlsson, O.; Zhang, X.; Svenningsson, P.; Andren, P.E. Pyrylium salts as reactive matrices for MALDI-MS imaging of biologically active primary amines. J. Am. Soc. Mass Spectrom. 2015, 26, 934–939. [Google Scholar] [CrossRef] [PubMed]
- Berthod, A.; Crank, J.A.; Rundlett, K.L.; Armstrong, D.W. A second-generation ionic liquid matrix-assisted laser desorption/ ionization matrix for effective mass spectrometric analysis of biodegradable polymers. Rapid Commun. Mass Spectrom. 2009, 23, 3409–3422. [Google Scholar] [CrossRef] [PubMed]
- Serrano, C.A.; Zhang, Y.; Yang, J.; Schug, K.A. Matrix-assisted laser desorption/ionization mass spectrometric analysis of aliphatic biodegradable photoluminescent polymers using new ionic liquid matrices. Rapid Commun. Mass Spectrom. 2011, 25, 1152–1158. [Google Scholar] [CrossRef] [PubMed]
- Kooijman, P.C.; Kok, S.; Honing, M. Independent assessment of matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) sample preparation quality: Effect of sample preparation on MALDI-MS of synthetic polymers. Rapid Commun. Mass Spectrom. 2017, 31, 362–370. [Google Scholar] [CrossRef] [PubMed]
- Gabriel, S.J.; Pfeifer, D.; Schwarzinger, C.; Panne, U.; Weidner, S.M. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometric imaging of synthetic polymer sample spots prepared using ionic liquid matrices. Rapid Commun. Mass Spectrom. 2014, 28, 489–498. [Google Scholar] [CrossRef] [PubMed]
- Abdelhamid, H.N.; Gopal, J.; Wu, H.-F. Synthesis and application of ionic liquid matrices (ILMs) for effective pathogenic bacteria analysis in matrix assisted laser desorption/ionization (MALDI-MS). Anal. Chim. Acta 2013, 767, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.H.; Gopal, J.; Wu, H.F. Ionic solution and nanoparticle assisted MALDI-MS as bacterial biosensors for rapid analysis of yogurt. Biosens. Bioelectron. 2012, 31, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Djidja, M.-C.; Claude, E.; Scriven, P.; Allen, D.W.; Carolan, V.A.; Clench, M.R. Antigen retrieval prior to on-tissue digestion of formalin-fixed paraffin-embedded tumour tissue sections yields oxidation of proline residues. Biochim. Biophys. Acta Proteins Proteom. 2017, 1865, 901–906. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Giese, R.W. Recommendations for quantitative analysis of small molecules by matrix-assisted laser desorption ionization mass spectrometry. J. Chromatogr. A 2017, 1486, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Tholey, A.; Zabet-Moghaddam, M.; Heinzle, E. Quantification of peptides for the monitoring of protease-catalyzed reactions by matrix-assisted laser desorption/ionization mass spectrometry using ionic liquid matrixes. Anal. Chem. 2006, 78, 291–297. [Google Scholar] [CrossRef] [PubMed]
- Giménez, E.; Benavente, F.; Barbosa, J.; Sanz-Nebot, V. Ionic liquid matrices for MALDI-TOF-MS analysis of intact glycoproteins. Anal. Bioanal. Chem. 2010, 398, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Weidner, S.; Knappe, P.; Panne, U. MALDI-TOF imaging mass spectrometry of artifacts in “dried droplet” polymer samples. Anal. Bioanal. Chem. 2011, 401, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Tholey, A. Ionic liquid matrices with phosphoric acid as matrix additive for the facilitated analysis of phosphopeptides by matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun. Mass Spectrom. 2006, 20, 1761–1768. [Google Scholar] [CrossRef] [PubMed]
- Fukuyama, Y.; Takeyama, K.; Kawabata, S.; Iwamoto, S.; Tanaka, K. An optimized matrix-assisted laser desorption/ionization sample preparation using a liquid matrix, 3-aminoquinoline/α -cyano-4-hydroxycinnamic acid, for phosphopeptides. Rapid Commun. Mass Spectrom. 2012, 26, 2454–2460. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, S.; Martinez-Chapa, S.O. Principles and Mechanism of MALDI-ToF-MS Analysis. In Fundamentals of MALDI-ToF-MS Analysis; Springer: Singapore, 2017; pp. 1–19. [Google Scholar]
- Zenobi, R.; Knochenmuss, R. Ion formation in MALDI mass spectrometry. Mass Spectrom. Rev. 1998, 17, 337–366. [Google Scholar] [CrossRef]
- Lu, I.-C.; Lee, C.; Lee, Y.-T.; Ni, C.-K. Ionization Mechanism of Matrix-Assisted Laser Desorption/Ionization. Annu. Rev. Anal. Chem. 2015, 8, 21–39. [Google Scholar] [CrossRef] [PubMed]
- Chu, K.Y.; Lee, S.; Tsai, M.-T.; Lu, I.-C.; Dyakov, Y.A.; Lai, Y.H.; Lee, Y.-T.; Ni, C.-K. Thermal Proton Transfer Reactions in Ultraviolet Matrix-Assisted Laser Desorption/Ionization. J. Am. Soc. Mass Spectrom. 2014, 25, 310–318. [Google Scholar] [CrossRef] [PubMed]
- Karas, M.; Glückmann, M.; Schäfer, J. Ionization in matrix-assisted laser desorption/ionization: Singly charged molecular ions are the lucky survivors. J. Mass Spectrom. 2000, 35, 1–12. [Google Scholar] [CrossRef]
- Niu, S.; Zhang, W.; Chait, B.T. Direct comparison of infrared and ultraviolet wavelength matrix-assisted laser desorption/ionization mass spectrometry of proteins. J. Am. Soc. Mass Spectrom. 1998, 9, 1–7. [Google Scholar] [CrossRef]
- Knochenmuss, R. MALDI mechanisms: wavelength and matrix dependence of the coupled photophysical and chemical dynamics model. Analyst 2014, 139, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Cramer, R.; Karas, M.; Jaskolla, T.W. Enhanced MALDI MS sensitivity by weak base additives and glycerol sample coating. Anal. Chem. 2014, 86, 744–751. [Google Scholar] [CrossRef] [PubMed]
- Zabet-Moghaddam, M.; Heinzle, E.; Lasaosa, M.; Tholey, A. Pyridinium-based ionic liquid matrices can improve the identification of proteins by peptide mass-fingerprint analysis with matrix-assisted laser desorption/ionization mass spectrometry. Anal. Bioanal. Chem. 2006, 384, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Laremore, T.N.; Zhang, F.; Linhardt, R.J. Ionic liquid matrix for direct UV-MALDI-TOF-MS analysis of dermatan sulfate and chondroitin sulfate oligosaccharides. Anal. Chem. 2007, 79, 1604–1610. [Google Scholar] [CrossRef] [PubMed]
- Ullmer, R.; Rizzi, A.M. Use of a novel ionic liquid matrix for MALDI-MS analysis of glycopeptides and glycans out of total tryptic digests. J. Mass Spectrom. 2009, 44, 1596–1603. [Google Scholar] [CrossRef] [PubMed]
- Zabet-Moghaddam, M.; Krüger, R.; Heinzle, E.; Tholey, A. Matrix-assisted laser desorption/ionization mass spectrometry for the characterization of ionic liquids and the analysis of amino acids, peptides and proteins in ionic liquids. J. Mass Spectrom. 2004, 39, 1494–1505. [Google Scholar] [CrossRef] [PubMed]
- Fukuyama, Y.; Funakoshi, N.; Iwamoto, S.; Tanaka, K. Adding methanol to α -cyano-4-hydroxycinnamic acid butylamine salt as a liquid matrix to form a homogeneous spot on a focusing plate for highly sensitive and reproducible analyses in matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun. Mass Spectrom. 2014, 28, 662–664. [Google Scholar] [CrossRef] [PubMed]
- Bungert, D.; Bastian, S.; Heckmann-Pohl, D.M.; Giffhorn, F.; Heinzle, E.; Tholey, A. Screening of sugar converting enzymes using quantitative MALDI-ToF mass spectrometry. Biotechnol. Lett. 2004, 26, 1025–1030. [Google Scholar] [CrossRef] [PubMed]
- Park, K.M.; Moon, J.H.; Lee, S.H.; Kim, M.S. Quantitative transfer of polar analytes on a solid surface to a liquid matrix in MALDI profiling. J. Mass Spectrom. 2016, 51, 1152–1156. [Google Scholar] [CrossRef] [PubMed]
- Clark, K.D.; Emaus, M.N.; Varona, M.; Bowers, A.N.; Anderson, J.L. Ionic liquids: solvents and sorbents in sample preparation. J. Sep. Sci. 2018, 41, 209–235. [Google Scholar] [CrossRef] [PubMed]
- Shrivas, K.; Tapadia, K. Ionic liquid matrix-based dispersive liquid–liquid microextraction for enhanced MALDI–MS analysis of phospholipids in soybean. J. Chromatogr. B 2015, 1001, 124–130. [Google Scholar] [CrossRef] [PubMed]
- Murali, M.S.; Bonville, N.; Choppin, G.R. Uranyl Ion Extraction into Room Temperature Ionic Liquids: Species Determination by ESI and MALDI-MS. Solvent Extr. Ion Exch. 2010, 28, 495–509. [Google Scholar] [CrossRef]
- Luo, H.; Dai, S.; Bonnesen, P.V. Solvent Extraction of Sr2+ and Cs+ Based on Room-Temperature Ionic Liquids Containing Monoaza-Substituted Crown Ethers. Anal. Chem. 2004, 76, 2773–2779. [Google Scholar] [CrossRef] [PubMed]
- Lovejoy, K.S.; Purdy, G.M.; Iyer, S.; Sanchez, T.C.; Robertson, A.; Koppisch, A.T.; Del Sesto, R.E. Tetraalkylphosphonium-Based Ionic Liquids for a Single-Step Dye Extraction/MALDI MS Analysis Platform. Anal. Chem. 2011, 83, 2921–2930. [Google Scholar] [CrossRef] [PubMed]
- Lovejoy, K.S.; Lou, A.J.; Davis, L.E.; Sanchez, T.C.; Iyer, S.; Corley, C.A.; Wilkes, J.S.; Feller, R.K.; Fox, D.T.; Koppisch, A.T.; Del Sesto, R.E. Single-pot extraction-analysis of dyed wool fibers with ionic liquids. Anal. Chem. 2012, 84, 9169–9175. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, F.; Wu, H.-F. Characterization of pathogenic bacteria using ionic liquid via single drop microextraction combined with MALDI-TOF MS. Analyst 2011, 136, 4020–4027. [Google Scholar] [CrossRef] [PubMed]
- Sekiya, S.; Taniguchi, K.; Tanaka, K. On-target separation of analyte with 3-aminoquinoline/α-cyano-4-hydroxycinnamic acid liquid matrix for matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun. Mass Spectrom. 2012, 26, 693–700. [Google Scholar] [CrossRef] [PubMed]
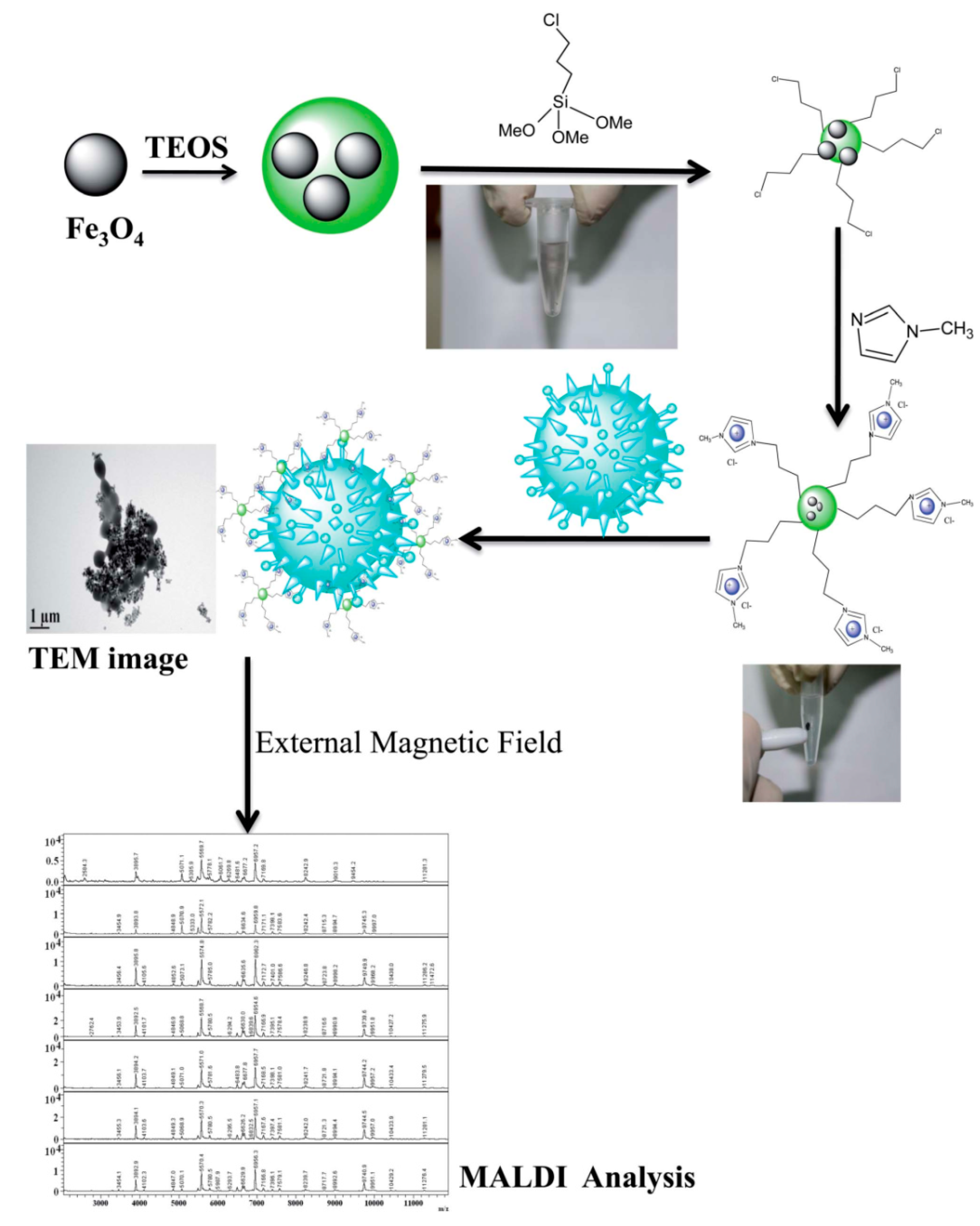
- Bhaisare, M.L.; Abdelhamid, H.N.; Wu, B.-S.; Wu, H.-F. Rapid and direct MALDI-MS identification of pathogenic bacteria from blood using ionic liquid-modified magnetic nanoparticles (Fe3O4@SiO2). J. Mater. Chem. B 2014, 2, 4671–4683. [Google Scholar] [CrossRef]
- Santos, L.S.; Haddad, R.; Höehr, N.F.; Pilli, R.A.; Eberlin, M.N. Fast Screening of Low Molecular Weight Compounds by Thin-Layer Chromatography and “On-Spot” MALDI-TOF Mass Spectrometry. Anal. Chem. 2004, 76, 2144–2147. [Google Scholar] [CrossRef] [PubMed]
- Earle, M.J.; Esperança, J.M.S.S.; Gilea, M.A.; Canongia Lopes, J.N.; Rebelo, L.P.N.; Magee, J.W.; Seddon, K.R.; Widegren, J.A. The distillation and volatility of ionic liquids. Nature 2006, 439, 831–834. [Google Scholar] [CrossRef] [PubMed]
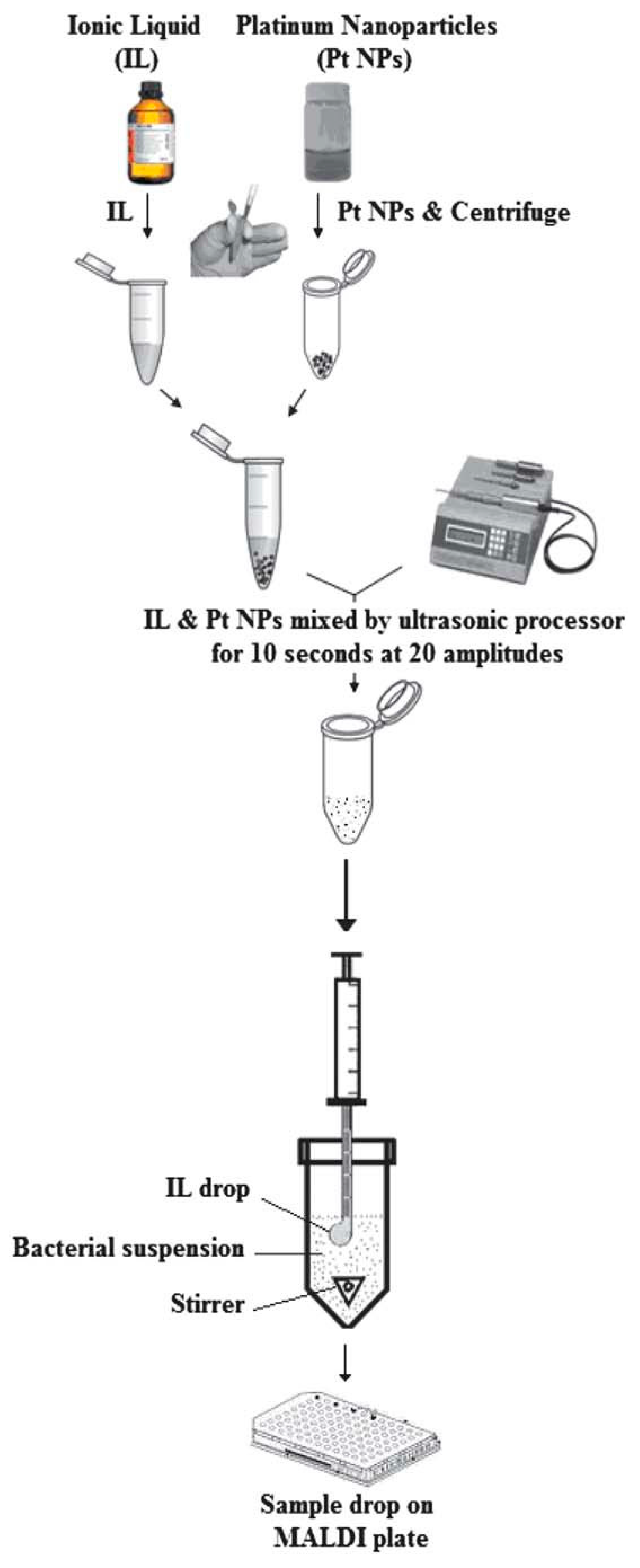
| Acid | Base | Analyte | Conditions | Low Limit of Detection (LOD, pmol) | Linear Range (pmol) | Ref. |
|---|---|---|---|---|---|---|
| CHCA | 1-methylimidazole, aniline, pyridine, N,N-diethylamine, triethylamine, tripropylamine, tributylamine | ODNs, proteins 5′-d(CTTTCCTC) and 5′-d(TCTTCCCTT), bradykinin, Tyr-bradykinin, substance P, melittin, and bovine insulin |
| 2 μM to 50 μM | [87] | |
| 3-aminoquinoline | Tetrapeptide RFDS, bradykinin fragment 1-7, angiotensin I, substance P, Glu-fibrinopeptide, ANP 104-123, ACTH 18-39, Somatostatin, and ACTH 7-38 |
| 1 | 0.001–2 | [88] | |
| Phosphatidylcholine (PC) in mouse brain tissue |
| 30 | 1–100 | [89] | ||
| n-butylamine, N,N-diethylaniline, aniline, N,N-diethylaniline | Bradykinin, substance-P, melittin, allatostatin IV oligonucleotide 5′-GGATTC-3′ phosphatidylcholine, L-α-phosphatidylcholine-β-palmitoyl-oleoyl, ([PC 16:0, 18:1]), and phosphatidylethanolamine, 1-2,dioleoyl-sn-glycerol-3-phospho-ethanolamine, ([PE 18:1, 18:1]) |
| 5000 | [90] | ||
| Triethylamine, diisopropylammine | Drugs |
| [91] | |||
| 2-aminopentane (AP) | N-acyl homoserine lactones (AHL) |
| 0.125–5 | [92] | ||
| 1-methylimidazole, aniline, pyridine, tripropylamine, tributylamine | Phosphatidylcholine (PC), phosphatidic acid (PA), phophatidylethanolamine (PE), serine (PS), glycerol (PG), and inositol (PI) |
| 127 × 103 | [93] | ||
| N,N-diisopropylethylammonium | Polymers and additives found in lubricant residues |
| 0.5% and 0.003% lubricant in biological fluid | [94] | ||
| 3-aminoquinoline, N,N-diethylaniline | Peptides Y5R, Y6, and substance P arginine, imipramine, and serotonin |
| 10−2 | 10−2–103 | [95] | |
| N,N-iisopropylethylammonium, N-isopropyl-N-methyl-t-butylammonium, N-isopropyl-N-methyl-N-tert-butylammonium, N,N-diisopropylethylammonium | Bradykinin, polyethylene glycol 4600, insulin, cytochrome c, bovine serum albumin (BSA), catalase, urease, dextran enzymatic synthesis, Saccharomyces cerevisiae. |
| 50–100 | [96] | ||
| 3-aminoquinoline (3-AQ) | Glycan |
| 1 × 10−3 | [97] | ||
| 1-methylimidazolium | Glycosaminoglycan (GAG) polysaccharides |
| [98] | |||
| CHCA | Triethylamine | Aflatoxins B1, B2, G1, and G2 |
| 0.05 | [99] | |
| 2,5-dihydroxybenzoic acid (DHB), CHCA, Sinapic acid | Butylamine, Triethylamine | Glycoconjugates, peptides, and proteins oligosaccharides, polymers desialylation of sialylactose, sialidase from Clostridium perfringens |
| 0.3–2.5 | [86] | |
| CHCA and ferulic acid | N,N-iisopropylethylammonium, N,N-diisopropylethylammonium, N-isopropyl-N-methyl-N-tert-butylammonium, di(2-aminopentane) | Mannan, β-Cyclodextran dextran, polyethylene glycol 4600 |
| 103 | [96] | |
| DHB | Aniline, N,N-dimethylaniline (DMA) | Sialylated Glycans |
| 30 | [100] | |
| N-methylaniline (N-MA), N-ethylaniline (N-EA) | Maltohexaose, maltoheptaose, dextran 2000 (D2000) and dextran 4000 (D4000), 1-Kestose (GF2), nystose (GF3) and 1,1,1-kestopentaose (GF4) |
| 0.01 | 10–80 | [101] | |
| N,N-dimethylaniline (DMA) | N-linked oligosaccharides Ovalbumin (chicken egg white albumin), maltohexaose, maltoheptaose, dextran standard 1000 |
| 7–22.4 | 0.7–22.4 | [102] | |
| Butylamine | Pullulans Pul-5900 5.9 Pul-11,800 11.8 Pul-22,800 22.8 Pul-47,300 47.3 Pul-112,000 112.0 |
| 0.8–4.4 | [103] | ||
| Oligosaccharides sucrose (disaccharide), raffinose (trisaccharide), stachyose (tetrasaccharide), ß-cyclodextrin, L-proline, D,L-pyroglutamic acid, L-arginine hydrochloride, D,L-tyrosine, angiotensin II, reduced glutathione and sunflower oil |
| 38 | 340–555 | [104] | ||
| CHCA p-coumaric | 1,1,3,3-tetramethylguanidium (TMG) | Sulfated/sialylated/neutral oligosaccharides |
| 0.001 | [82] | |
| CHCA and DHB | 1-methylimidazolium | Sucrose octasulfate, and an octasulfatedpentasaccharide, Arixtra |
| 8–40 | [105] | |
| Mefenamic acid | Aniline (ANI), Pyridine (Pyr), Dimethyl aniline (DMANI), 2-methyl picoline (2-P)) | Drugs, carbohydrate, and amino acids. |
| 1–20 | [106] | |
| p-coumaric acid | 1,1,3,3-tetramethylguanidium (TMG) | Anion adducted N-glycans |
| 0.001 for NO3–, 0.001 for BF4– | [107] | |
| THAP | Phosphopeptides |
| [108] | |||
| ATT | DMAN |
| 5 × 10−4 | 0–100 | [109] | |
| HABA | 1,1,3,3-tetramethylguanidine Spermine | Polysulfated carbohydrates such as heparin (HP) and heparan sulfate (HS) |
| 67 | [110] | |
| DHB CHCA SA | Tributylamine (TBA), Pyridine (Py), 1-methylimidazole(MI) | Arabinose, biotin, thiamine, NAD, ascorbic acid, a-ketoglutarate, ATP |
| 0.01 | 0.25–2.5 | [111] |
| ILs | Extraction/Separation Technique | Analytes | Instrumental Parameters | LOD | Conditions | Ref. |
|---|---|---|---|---|---|---|
| CHCAB | DLLME | Phospholipids from soybean |
| 5 and 18 fmol (LOQ) | 5 min extraction time in the presence of 30 mg/mL CHCAB and 1.2% NaCl, using chloroform as an extracting solvent and methanol as a dispersing solvent | [153] |
| 1-alkyl-3-methylimidazolium PF6 (Cnmim, n = 4 and 8) CHCA | LLME | Uranyl nitrate |
| 0.014–0.098 M | 0.1–0.5 M using NaNO3 in 1.0 M HNO3, TBP (tributyl phosphate) concentration of 1.0 M in the RTILS or in dodecane | [154] |
| 3-methylimidazolium bis[(trifluoromethyl)sulfonyl]amide and 1-butyl-3-methylimidazolium bis[(trifluoromethyl)sulfonyl]amide, 1-hexyl-3-methylimidazolium bis[(trifluoromethyl)sulfonyl] amide and 1-octyl-3-methylimidazolium bis[(trifluoromethyl)sulfonyl]amide | Sr2+ and Cs+ |
| 1.5 mM | 1 mL of IL, extracted with 10 mL of cation-containing aqueous solution (1.5 mM) for 60 min in a vibrating mixer. | [155] | |
| PR4+ cations and ferulate (FA), CHCA, and DHB anions | single-step extraction | Dyes from textiles, malachite green, nile blue nile red, bromothymol blue, fluorescein, kiton red |
| 0–98% | Samples were centrifuged at 2000 rpm for 30 min, pH 7.5–10, 50–90 °C | [156] |
| Tetrabutylphosponium chloride IL [Bu4P][Cl] | Single-Pot Extraction | dyes associated with structurally robust wool fibers |
| 0.005 mg of dye per mg of dyed wool into the IL | A cloudy red solution was produced after 24 h. The solution was filtered through a 0.45 µM syringe filter and spotted on the MALDI–MS plate in 1 µL aliquots, either neat or diluted 10,000-fold in methanol | [157] |
| Platinum nanoparticles mixed 1-butyl-3-methylimidazolium hexafluorophosphate | SDME | Escherichia coli and Serratia marcescens |
| 106cfu mL−1 | A glass vial was filled with 1 mL of sample solution, spiked with the bacteria; the sample solution was agitated on a magnetic stirrer at room temperature,a 2.0 mL portion of platinum nanoparticles prepared in IL was drawn into a 10 mL microsyringe | [158] |
| 3-Aminoquinoline/CHCA (3AQ/CHCA) | On-target separation | peptides and oligosaccharides |
| 5 pmol | Vaporization of water derived from analyte solvent | [159] |
| Cationic ionic liquid-modified Fe3O4@SiO2 magnetic nanoparticles (CILMS) | Magnetic field | E. coli, Pseudomonas aeruginosa, and Staphylococcus aureus, |
| 3.4 × 103, 3.2 × 103, and 4.2 × 103 cfu mL−1 | <5 min, RT, and use of external magnetic field | [160] |
| Triethylamine/CHCA | TLC | three arborescidine alkaloids, the anesthesics levobupivacaine and mepivacaine, and the antibiotic tetracycline |
| 5–10 ng | Elution with CHCl3/MeOH 9:1 | [161] |
| 1-butyl-3methylimidazolium hexafluorophosphate | on-target separation | Bifidobacterium lactis (Bb12), Lactobacillus acidophilus (La5), Streptococcus thermophilus and Lactobacillus bulgaricus from AB yogurt |
| 107–109cfu/mL | 10 μL of yogurt was added to 100 μL of IL (containing 0.35 mg of AgNPs) and incubated for 10 min before spotting on the MALDI plate. | [129] |
© 2018 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdelhamid, H.N. Ionic Liquid-Assisted Laser Desorption/Ionization–Mass Spectrometry: Matrices, Microextraction, and Separation. Methods Protoc. 2018, 1, 23. https://doi.org/10.3390/mps1020023
Abdelhamid HN. Ionic Liquid-Assisted Laser Desorption/Ionization–Mass Spectrometry: Matrices, Microextraction, and Separation. Methods and Protocols. 2018; 1(2):23. https://doi.org/10.3390/mps1020023
Chicago/Turabian StyleAbdelhamid, Hani Nasser. 2018. "Ionic Liquid-Assisted Laser Desorption/Ionization–Mass Spectrometry: Matrices, Microextraction, and Separation" Methods and Protocols 1, no. 2: 23. https://doi.org/10.3390/mps1020023
APA StyleAbdelhamid, H. N. (2018). Ionic Liquid-Assisted Laser Desorption/Ionization–Mass Spectrometry: Matrices, Microextraction, and Separation. Methods and Protocols, 1(2), 23. https://doi.org/10.3390/mps1020023