Abstract
Historically, the analyses used for newborn screening (NBS) were biochemical, but increasingly, molecular genetic analyses are being introduced in the workflow. We describe the application of molecular genetic analyses in the Danish NBS programme and show that second-tier molecular genetic testing is useful to reduce the false positive rate while simultaneously providing information about the precise molecular genetic variant and thus informing therapeutic strategy and easing providing information to parents. When molecular genetic analyses are applied as second-tier testing, valuable functional data from biochemical methods are available and in our view, such targeted NGS technology should be implemented when possible in the NBS workflow. First-tier NGS technology may be a promising future possibility for disorders without a reliable biomarker and as a general approach to increase the adaptability of NBS for a broader range of genetic diseases, which is important in the current landscape of quickly evolving new therapeutic possibilities. However, studies on feasibility, sensitivity, and specificity are needed as well as more insight into what views the general population has towards using genetic analyses in NBS. This may be sensitive to some and could have potentially negative consequences for the NBS programme.
1. Introduction
Newborn screening (NBS) is a public health programme for early diagnosis of treatable, mostly genetic, diseases, such as congenital adrenal hyperplasia and selected inherited metabolic diseases (IEM) such as phenylketonuria (PKU) and medium-chain acyl-CoA dehydrogenase deficiency (MCADD). Early treatment in a latent stage of disease can, in many cases, prevent disease manifestations, which for diseases on many NBS panels includes irreversible brain and organ damage and death [1,2,3]. Internationally, NBS was started >50 years ago and in Denmark for PKU in 1975 (Figure 1). NBS for PKU became the paradigm for a well-functioning, cost-effective screening. It enabled the expansion of the disorders screened for: in Denmark, hypothyroidism was added in 1977 and expanded newborn screening (eNBS) with addition of 23 diseases tested by tandem mass spectrometry (Figure 1) was implemented in 2002. Today the Danish routine screening panel includes 18 treatable diseases (Figure 2); see also Figure 1 for a timeline of Danish NBS. In the most common set-up, the methods of screening involve the biochemical determination of analytes such as amino acids and their ratios, acylcarnitines or hormones. For confirmation of the screening positive samples, further biochemical methods are commonly used together with molecular genetic studies using new samples from the child. In recent years, molecular genetic studies have been introduced as second-tier testing before contacting the family for confirmative evaluation [1,4,5,6,7]; even the use of molecular genetic studies as first-tier analyses has been introduced when screening for spinal muscular atrophy (SMA) and severe combined immunodeficiency (SCID) [4], and further use of molecular genetics as a first-tier procedure in NBS is being discussed. In Denmark, we have recently introduced molecular genetic analyses for SCID (though, strictly speaking, not a variant analysis) as first-tier screening, and SMA has also been accepted for addition to the routine screening panel with the use of first-tier molecular genetic analyses. As a second-tier eNBS procedure, we used molecular genetic analyses on the initially obtained filter paper blood spot sample for a number of diagnoses alone or in parallel with biochemical testing. Finally, molecular genetic analyses are used as a follow-up procedure on screen positives for most diagnoses (in a new blood sample). Most diseases on many screening panels are genetic; thus, the use of molecular genetic analyses is relevant, and they have been shown to be feasible to conduct on DNA extracted from filter paper blood spot samples [8,9,10]. An important goal for using molecular genetic analyses is to decrease false positive rate [11] and to stratify infants with severe versus benign disease as has been shown for isovaleric aciduria [12]. Another goal could be to increase the adaptability of NBS making it possible to include treatable diseases without a relevant or reliable biochemical biomarker [13]. Finally, there is a general wish in many countries to increase the number of diseases on newborn screening panels and molecular genetic analyses would be able to fulfil this need because the methods are generally applicable for most inherited diseases [11].
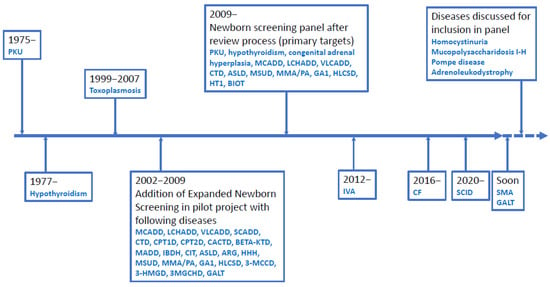
Figure 1.
Timeline for Danish Newborn Screening. Disease abbreviations are shown in the list of abbreviations.
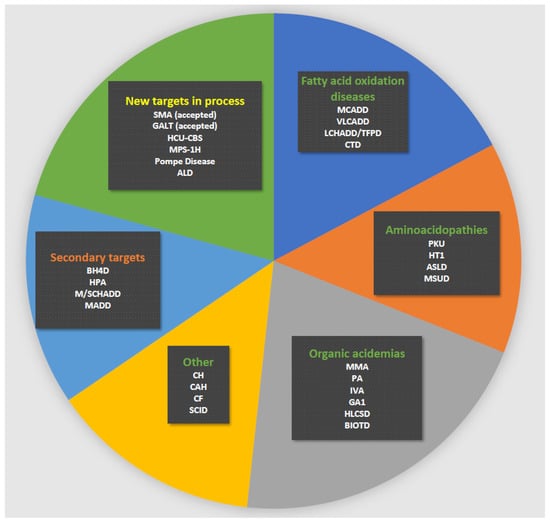
Figure 2.
Current Danish newborn screening panel. Green heading: primary targets; orange heading: secondary targets; yellow heading: new targets in process for possible inclusion. Disease abbreviations are shown in the list of abbreviations.
In this manuscript, we describe the experience with and set-up for the use of molecular genetic testing in primarily eNBS in Denmark as well as perspectives for future developments.
2. Materials and Methods
2.1. General Set-Up of Danish Newborn Screening
The general set-up for Danish newborn screening (in particular eNBS) has been described in three previous papers [2,14,15]. In this paper, focus will also be on eNBS with a short note on screening for more recently included diseases, including cystic fibrosis (CF) and SCID. The historical development is shown in Figure 1, and the actual panel of diseases screened for is shown in Figure 2. Routine newborn screening is state-run, free of charge, conducted in an informed dissent set-up and covers Denmark, the Faroe Islands and Greenland, corresponding to about 62,000 births per year (www.SSI.DK/nyfoedte (accessed on 3 May 2021)). Samples consist of capillary blood collected in the local hospital by heel prick, spotted on filter paper and dried. Samples were taken postpartum day 4–9 in the pilot period of eNBS (Figure 1) (the time of sampling used routinely until 2009) and at 48–72 h during routine NBS after 2009. Primary NBS analyses are conducted in a single centralised laboratory (Statens Serum Institute, SSI) [14,15] and results are available 2–6 days after sampling. Confirmatory testing is coordinated by and conducted in Centre for Inherited Metabolic Diseases, Copenhagen. Spare blood spots have been stored since 1982 in the Danish Newborn Screening Biobank at SSI, Copenhagen. All children with a true positive screening result for an IEM are managed in Centre for Inherited Metabolic Diseases, Copenhagen; those with CF in the two dedicated CF-centres; those with congenital hypothyroidism and congenital adrenal hyperplasia in local paediatric and paediatric endocrinology departments; and, finally, those with SCID in the paediatric transplantation centre at Copenhagen University Hospital. Review of new screening targets and application to the Danish National Board of Health for inclusion in the routine panel, as well as supervision of performance and quality assurance of NBS, is conducted by the Committee for Clinical Genetics and Screening, Danish Paediatric Society and the Danish Tandem MS working group. The systematic review process of Danish NBS, laying the ground for the current Danish NBS practice, is described in a report from the Danish National Board of Health, 2008 http://sundhedsstyrelsen.dk/~/media/2FEDEEE91A014D86B06EE1B901C66A73.ashx (accessed on 3 May 2021). Decision on the current panel was based on previous screening experiences, including results from the pilot period (Figure 1) and a review with scoring of the screening potential for selected diseases [14,15]. All diseases with a scoring >the 75th centile (and some from 50th–75th centile) were reviewed further and a final list of panel diseases was made, which was effective from February 2009 (Figure 1 and Table 1) [14,15].

Table 1.
Number of children with screen positive, true positive, false positive, false negative and not reported results during eNBS. Classic (C), mild (M), variants of uncertain significance (VUS, V) and heterozygous variants (H) represent the genotypes found during sequencing of the initial filter paper blood spot sample (see also Methods). * This is the clinical phenotype as sequencing was not performed.
Screening of the first half million newborns and first million newborns, respectively, was published earlier, including analytical details for first- and second-tier as well as follow-up procedures; readers are referred to these publications for such details [2,14,15]. The study was approved by the Ethics Committee (KF 01-152/98).
For this paper we included all children born from February 1st, 2002 until February 28th 2021 [16]. The period included the eNBS pilot period until February 2nd, 2009, with eNBS performed with written informed consent, followed by NBS performed in a routine NBS program with informed dissent (see above and Figure 1). Percentage of newborns screened in eNBS changed during the period from 65% at the start to 85% at the end of pilot period and 99.85% during routine screening from 2009. Over the whole period, 1,092,450 newborns were screened. In the pilot period, 82,930 (7.6%) were not screened using the expanded screening panel (because no consent was available), but were screened for the diseases present in the routine panel (Figure 1) [14,15].
We extracted molecular genetic data for all true positives, false positives and false negatives as well as unreported results (see below) for those diseases where molecular genetic analyses were conducted on the initial filter paper blood spot sample. Overall, we conducted first-tier molecular genetic analyses for SCID (though strictly speaking, TRECs are a surrogate genetic marker and not a variant analysis). In CF, biotinidase deficiency and holocarboxylase deficiency we performed molecular genetic analyses in the initially collected filter paper blood spot sample before contacting the family. For MCADD, VLCADD, LCHADD, and IVA molecular genetic analyses were also conducted on the initially collected filter paper blood spot sample but in parallel with biochemical testing in new samples. For the remaining diseases, molecular genetic analyses were performed later in the follow-up in new samples and these are not the focus of this manuscript. For additional disorders screened for during the pilot period from 2002 to 2009, but removed from the panel in 2009, molecular genetic analyses were conducted in a few cases as described in the results section.
For other analytical details, including biochemical first tier testing, we refer to our previous publications [14,15].
2.2. Molecular Genetic Analyses
Conventional Sanger sequencing on DNA extracted from the original filter blood spot was used to confirm a screen positive MCADD (NM000016.4, ACADM), VLCADD (NM_000018.3, ACADVL), LCHADD/TPD (NM_000182.4, HADHA; NM_000183.2, HADHB), IVA (NM_002225.3, IVD), MADD (NM_000126.3, ETFA; NM_001985.2, ETFB; NM_4453.3, ETFDH; NM_001104577.1, SLC52A1; NM_024531.4, SLC52A2; NM_033409.3, SLC52A3; NM_018339.5, RFK; NM_025207.4, FLAD1)), HLCSD (NM_000411.8, HLCS) and BTD (NM_000060.4, BTD). Exonic elements and a minimum of 15 bp of the flanking intronic regions were amplified by conventional PCR and subsequently sequenced on an Applied Biosystems 3500 DX Genetic Analyzer using BigDye® Terminator v1.1 Cycle Sequencing kit (Applied Biosystems, Waltham, MA, USA). Sequence data were analysed using the GeneSearch v.4.4.3 and Alamut Visual v.2.10 software. When available, parental DNA samples were investigated to determine the phase of variants. Only variants interpreted to be causative of the first-tier biochemical phenotype were reported. The coding and exon flanking sequence of the CFTR gene (NM_000492.4) was assessed using a commercial NGS assay (Ion AmpliSeq™ CFTR Panel) and sequenced on an Ion-S5 platform, essentially according to the manufactures description (ThermoFisher, Waltham, MA, USA). Further details can be found in [17].
Prediction of disease severity was mainly based on published or in-house data of the identified genetic variants. For novel non-published variants, clinical impact was predicted from the presumed consequence on the protein function, with loss-of-function variants (premature stop-codons, insertions/deletion creating a shift in the reading frame, etc.) categorized as severe impact. Some variants, primarily missense variants, were unknown from the literature and commonly used databases and thus did not allow for a prediction of disease severity or a specific functional consequence for the protein (variants of unknown significance). Variants were only reported if the complete genotype and the biochemical data supported a convincing conclusion. Definition of mild versus severe/classical genotypes was: mild genotypes were those where at least one of two variants from the literature was known as such or which, in unrelated individuals, have been associated with a biochemical phenotype but with no or limited clinical correlate. Classical/severe genotypes were those where both variants have a well described biochemical phenotype (published or studied in-house) and a certain associated clinical correlate with classical manifestations for the given disease.
True positive results came from newborns in whom, or in whose mothers, the suspected disease was diagnosed by confirmatory testing—see details in [2,14,15]. False positive results were from newborns in whom the suspected disease was not confirmed in the child or the mother. False negative results were from children with negative results, born in the screening period and diagnosed clinically or otherwise, e.g., by family studies, with a disease in the NBS panel.
3. Results
Findings during molecular genetic studies in the initial filter paper blood spot sample are described in the following and are summarized for diseases included in eNBS in Table 1.
3.1. Medium-Chain Acyl-CoA Dehydrogenase Deficiency (OMIM ID: 201450) (MCADD)
A total of 124 children were screened positive for MCADD, with 109 true positives and 15 false positives. Four were false negatives and were all diagnosed during family studies conducted in connection with a positive newborn screening result in a younger sibling; one of these had a classical genotype, whereas two had a mild and one had an unknown genotype. Molecular genetic analyses were conducted in all children who screened positive. Eleven of the 15 children with a false positive result were heterozygotes for a known pathogenic variant and four had no likely pathogenic variant in the ACADM gene; all 15 children with a false positive result had a normal or only slightly abnormal acylcarnitine profile (in carriers) on confirmative testing. For the present paper, genotypes have been classified into classical, mild and uncertain as described in the method section. Among the 109 children with a true screen positive result, the common classical variant c.985A>G was found in 140 of 218 alleles (64%) with 52 being homozygous and 36 compound heterozygous. Twenty-one children did not have the c.985A>G variant, and many of these children were of non-Caucasian descent (13 patients). Eighteen children (16%) had a known “mild” genotype [2,18], mostly compound heterozygosity for the c.199T>C variant (14 children) or c.127G>A (2 children); no child was homozygous for a mild mutation. Eleven children had an uncertain genotype, though all had abnormal acylcarnitines in follow-up samples. These samples came from non-Caucasian children in five cases. In one case, a mother was diagnosed with MCADD (homozygous for c.985A>G) because of low free carnitine in her newborn child [19].
3.2. Very Long-Chain Acyl-CoA Dehydrogenase Deficiency (OMIM ID: 201475) (VLCADD)
A total of 25 children were screened positive for VLCADD, with six true positives and 19 false positives. None had a false negative result. Molecular genetic analyses of ACADVL were conducted in 24 of the children. Eleven of the 19 children with a false positive result were heterozygotes for a pathogenic variant in ACADVL, while no pathogenic or likely pathogenic variant was identified in the remaining children, all of whom had normal follow-up acylcarnitines (though one had slightly raised C14:1, normalizing in a second sample taken one month after). Of the six children with true positive results, three children had a genotype of unpredictable clinical consequence and the remaining three had classical genotypes [20]. Two of the children with a genotype of unpredictable clinical consequence had pathological acylcarnitines consistent with VLCADD and have developed pronounced clinical disease consistent with early onset VLCADD disease, while the third child has clearly abnormal acylcarnitines consistent with VLCADD, but no clinical signs on standard treatment.
3.3. Long-Chain 3-Hydroxy Acyl-CoA Dehydrogenase Deficiency (OMIM ID: 609016) (LCHADD)
A total of five children were screened positive for LCHADD, all of whom were true positive. We found no false negative cases. Molecular genetic analyses were conducted in all. Three had well-known pathogenic variants in HADHA all with compound heterozygosity for the common classical variant c.1528G>C, two had a splice variant on the other allele and one had a premature stop codon variant. The remaining two children were of non-Caucasian descent and both had uncertain variants in the HADHA gene, with one being homozygous for a missense variant and one compound heterozygous for a missense and a splice variant. All five children had conclusive levels of 3-hydroxylated long-chain acylcarnitines suggestive of LCHADD and have developed clinical signs of LCHADD and are treated as such.
3.4. Multiple Acyl-CoA Dehydrogenase Deficiency (OMIM ID: 231680) (MADD)
MADD is not a primary target in Danish newborn screening, but five children turned out to be positive for more than one fatty acid oxidation disorder (MCADD and VLCADD) and were investigated. Unfortunately, only two were molecular genetically analysed using the initial blood spot. One presenting with a classical infantile clinical picture of MADD died and was therefore not molecular genetically investigated. In two children, the acylcarnitine profiles were normalized in the confirmative samples and the children were healthy and also not molecular genetically investigated. The remaining two children had persistent classical MADD acylcarnitines in follow-up samples, and in one child homozygosity for a known pathogenic variant in ETFDH was found. In the second child, only one ETFB variant of unknown significance was found in exon 3, and further investigations, including investigations of ETFB mRNA and Whole Genome Sequencing (WGS) with variant calling filtered to show only variants relevant for IEM, so could not establish a genetic diagnosis. Moreover, the WGS data did not reveal any variants in ETFB intron 2 and 3 to explain the ETFB exon 3 skipping events that were observed in low amounts (about 10%) of ETFB mRNAs. Both children were riboflavin-responsive and are developing normally.
3.5. Carnitine Palmitoyl Transferase 1 Deficiency (OMIM ID: 600528) (CPT1D)
Screening for CPT1D was only undertaken for a short period (2002–2007). Concerning CPT1D, 48 children were screened positive and of these, 26 were homozygous for the c.1436C>T variant common in the Inuit population. The Danish newborn screening program also covers Greenland, explaining the high frequency observed in our data. The clinical consequences associated with homozygosity for the c.1436C>T variant is unclear [21], and children with this genotype were not reported. In the remaining children acylcarnitine profiles normalized and sequencing of the CPT1 gene was not conducted; one of these children later presented clinically, arguing for the use of second-tier CPT1 sequencing as part of NBS [22].
3.6. Biotinidase Deficiency (OMIM ID: 253260) (BIOTD)
A total of 79 children were screened positive, with 47 true positives, 18 false positives and 14 not reported (see below). We found no false negative children. Molecular genetic analyses were performed in all except three with a false positive result and normal biotinidase activities. Screening for biotinidase deficiency started in 2009, and enzyme testing has been the first-tier analysis in the whole of the period, but the follow-up algorithm changed: from 2009 to 2018 all children with a positive initial screening test were contacted for confirmative testing (biotinidase activity determination and molecular genetic studies); after 2018 a second-tier testing on the initial filter paper blood spot sample with sequencing of the biotinidase (BTD) gene was introduced, and only children with two pathogenic or likely pathogenic variants were reported. This removed all false positives. Enzyme testing in a new sample was performed at admission in all children reported to have two likely pathogenic/pathogenic variants. Cut-off for treating biotinidase deficiency was 30% residual activity.
Among the 18 children with false positive results, six children had at least one allele with the common c.1330G>C variant with two children being homozygous (with normal enzyme activities above 30% residual activity). No other likely pathogenic/pathogenic BTD variants were found among the remaining 12 false positive children. Among the 14 non-reported children, heterozygosity for the common c.1330G>C variant was found in a single child and no BTD variants in the remaining children. Among the 47 true positives, 37 children had at least one allele with the common c.1330G>C variant, with one being homozygous (with biotinidase below cut-off). Three true positives (according to biotinidase activity) were seemingly only heterozygous, with two of the variants being the c.1330G>C variant. The remaining true positives were either compound heterozygous for the c.1330G>C variant and another BTD variant (34 children) or homozygous/compound heterozygous for other BTD variants (10 children).
3.7. Holocarboxylase Synthase Deficiency (OMIM ID: 253270) (HLCSD)
3-Hydroxyisovalerylcarnitine (C5OH) is a marker for holocarboxylase synthase deficiency (HLCSD), common in the Faroe Islands, and a good screening target. However, C5OH is also a marker for 3-Methylcrotonyl-CoA Carboxylase deficiency (OMIM IDs: 210200, 210210) (3-MCCD), which was in the initial screening panel together with HLCSD in the pilot period [14]. After review of the screening panel, 3-MCCD was removed in 2009. From 2009, we sequenced the HLCS gene in all newborns with a raised C5OH and only reported those with two pathogenic alleles of the gene. As an exception, caused by the increased frequency of HLCSD in the Faroe Islands, 11 Faroese children were reported and put on biotin supplementation until sequencing results were ready; 1 turned out to have HLCSD deficiency.
There were a total (in the pilot period where 3-MCCD was reported and in the period after, where 3-MCCD was not reported) of 117 children with raised C5OH. There were 19 cases caused by 3-MCCD, among whom seven were children and 12 were mothers having a child who had a raised C5OH in their screening sample; nine were false positive for 3-MCCD (in the pre-2009 samples). In addition, one each had 3-hydroxy-3-methyl-glutaryl-CoA lyase deficiency (OMIM ID: 246450) (3-HMGCLD) and 3-methylglutaconyl-CoA hydratase deficiency (OMIM ID: 250950) (3-MGCHD). Five had holocarboxylase synthase deficiency. It is important to note that 82 children were not reported according to the above algorithm, with four being heterozygous for an HLCS variant. One child was false negative for holocarboxylase synthase deficiency with low C5OH (0.62 µmol/L (cut-off >1.4 µmol/L)).
3.8. Isovaleric Acidemia (OMIM ID: 243500) (IVA)
Screening for IVA was introduced in 2012 and 10 children were positive for isovaleric acidemia (with LC-MS/MS confirmed presence of the IVD C5 isomer) with six true screen positive children. Sequencing showed two pathogenetic variants in all six with five of them being compound heterozygous for the known mild c.941C>T variant in the isovaleryl-CoA dehydrogenase (IVD) gene and one being homozygous. Thus, all of them could be predicted to have a mild phenotype, confirmed by a mild clinical course in all of them. The remaining four screen-positive samples were false positive; two were heterozygous, and two had no disease-associated IVD variants. One of the false positives died neonatally and was found to be homozygous for a pathogenic ENNPP1 variant associated with infantile arterial calcification—this was probably not related to the raised 3-hydroxyisovalerylcarnitine in the filter paper blood spot sample and a follow-up sample was not available. The other three children had normal acylcarnitine profiles in follow-up samples.
3.9. Cystic Fibrosis (OMIM ID: 219700) (CF)
During the period May 2016 (start of CF screening) until February 2021, 11,654 (3.8% of screened newborns) had a raised immunoreactive trypsinogen (IRT) used for first-tier testing. In second-tier test (testing for presence of F508del), we found 50 children homozygous for F508del, 605 children were F508del heterozygous and 330 children without F508del had IRT in fail-safe range. The 330 fail-safe children together with the 605 heterozygous children were included in our third-tier testing with next generation sequencing (NGS) of the entire cystic fibrosis transmembrane conductance regulator (CFTR) gene and among them we found 21 who were compound heterozygous for two pathogenic variants. One child had two candidate variants in cis (revealed by segregation analysis) and was reported as a carrier. Finally, among the fail-safe children, we found two carriers with pathogenic variants and they were reported together with the F508del heterozygotes found during second-tier testing as carriers. Thus, for CF, the policy is to report carriers in contrast to all other diseases on the screening panel. For details of Danish CF newborn screening, see [17], but a few conclusions of importance for the molecular genetic studies are that: 16% of alleles found were rare variants classified as non-pathogenic in relation to CF; the variant p.R117H was found in 1.6% of samples, but the p.R117H-5T was not found (of which only the p.R117H-5T would be reported); five (7%) of the children receiving a genetic report turned out to have a cystic fibrosis screen positive inconclusive diagnosis (CFSPID) because of the finding of variants of uncertain significance and needed long-term follow-up; two children were false negative: one with no F508del variant who did not reach fail-safe level and one child who did not reach first-tier IRT level because of meconium ileus.
3.10. Severe Combined Immunodeficiency (OMIM ID: 300400) (SCID)
Screening for SCID has only been part of the Danish newborn screening panel since February 2020 and is the first example of molecular genetic first-tier analyses in Danish NBS, where the amount of T-cell receptor excision circles (TRECs) is tested using real-time quantitative PCR. One year after starting screening, we have not encountered any SCID cases, but have found four children with low TRECs, two of whom were premature newborns and two children had secondary diagnoses causing low TRECs.
3.11. q-Spinal Muscular Atrophy (OMIM ID: 253300) (SMA)
We have prepared and validated a screening program for spinal muscular atrophy based on molecular genetic assessment for the common exon 7 deletion, and are currently waiting for political approval.
3.12. Other Diagnoses
For the remaining diagnoses in the Danish Newborn Screening Panel, molecular genetic studies are not a part of the screening procedures on the initial filter paper blood spot sample and are only performed as part of confirmatory testing. These diseases include methylmalonic acidemia (many OMIM IDs, but the most relevant here are: 251000, 251100, 251110); propionic acidemia (OMIM ID: 606054); glutaric aciduria type 1 (OMIM ID: 231670); phenylketonuria (OMIM ID: 261660); maple syrup urine disease (MSUD) (OMIM ID: 248600); argininosuccinate lyase deficiency (OMIM ID: 207900); hepatorenal tyrosinemia (OMIM ID: 276700); carnitine transporter deficiency (OMIM ID: 600528); and congenital adrenal hyperplasia (OMIM ID: 201910).
Here, we only want to comment on MSUD: there was a total of 57 children screen positive for MSUD and of these, three had classical MSUD. Two children presented with intermittent MSUD in the reported period, and both had normal leucine and normal leucine/phenylalanine and leucine/alanine ratios in their newborn filter paper blood spot samples. We subsequently sequenced the genes associated with MSUD (DLD, DBT, BCKDHA, BCKDHB and PPM1K) in 30 of the above 54 false positive samples. No children with two pathogenic variants were found. One was a carrier.
4. Discussion
In this study we showed that use of molecular genetic studies in the initial filter paper blood spot sample may decrease the false positive rate, make possible the molecular genetic filtering of diseases and stratify the severity of the disease in question in the true positive newborn. The results argue for an extension of such use of molecular genetic studies as a second-tier procedure and gives some insight as to what can be expected if used as a first-tier procedure.
4.1. Molecular Genetic Studies May Decrease the False Positive Rate
Our focus when introducing molecular genetic investigations in NBS has been to minimise the number of children receiving a false positive result. The Danish NBS programme performs relatively well and for the eNBS programme, the false positive rates during 2002 to 2021 were at 0.034%, corresponding to one false positive for every 2900 screened newborns. While this is relatively low, it implies that since the start of eNBS in 2002, 380 families have been investigated for a disease in the eNBS panel, that the newborn did not have. Investigations may include both expensive and invasive additional testing and a discussion of the benefit–harm ratio is relevant. While some studies cannot show any consequence of this to the families, others have shown an increased number of contacts with the health care system as well as admissions, thus suggesting a vulnerable child syndrome [11,23,24,25,26,27]. Both concerning the confirmatory investigations in the newborn period and the possible further investigations and admissions later on, the price may be considerable also from an economical point of view. Thus, there is a clear incentive to reduce the number of false positives. Some examples of success in our programme include the screening for biotinidase and HLCS deficiency. For biotinidase deficiency, we had two false positives/year before and none after introduction of second-tier genetic testing. The same was true for HLCSD screening effectively reducing the false positive rate. For 11 of 15 MCADD false positives and 11 of 19 VLCADD false positives we found heterozygosity for a pathogenic variant in ACADM and ACADVL, respectively, and the remaining had no likely pathogenic/pathogenic variants. These results strongly indicated their status as false positives even without the results of confirmative acylcarnitine profiles, which were normal or slightly abnormal (in case of true carriers). Though this did not prevent the reporting of the children because molecular genetic analyses in our set-up were conducted in parallel with biochemical confirmative testing for MCADD and VLCADD, it shows the potential of second-tier molecular genetic analyses giving sufficient data for not reporting these children; and also giving the reason for the abnormal first-tier acylcarnitines in carrier children. Data from the Norwegian NBS program also give support to these conclusions [5].
4.2. Molecular Genetic Studies May Filter Diseases and Disease Subtypes
As first-tier analyses are mostly biochemical and not molecular genetic, it may be difficult to separate diseases which share a common biomarker. For some of these diseases, we may wish to filter away those diseases that do not fulfil requirements for being neonatally screened for. In the view of the authors and the Danish Health Authorities, 3-MCCD is one such example, because in most individuals 3-MCCD is a biochemical finding, rather than a clinical problem [28]. Thus, we wanted to filter away and not report 3-MCCD and only report HLCS deficiency. In our cohort, one non-reported patient presented clinically in a metabolic crisis with 3-MCCD for which the child was successfully treated. This is unfortunate, but it should be seen in the following context: 81 children that were not reported and thus avoided being referred for additional testing; and 19 individuals, who were found to have 3-MCCD before we started the molecular genetic testing and who are all clinically normal today—this may be viewed as an unnecessary medicalisation. This algorithm represents an indirect filtering as we do not know whether the children have 3-MCCD—only that they do not have HLCSD. A similar algorithm using more direct filtering could be used in other settings; children with the c.199T>C variant in the ACADM gene have a mild phenotype, and it has been discussed—also in Denmark—whether this should be reported or at least not reported in children homozygous for the variant [29]. In the screening for MCADD, 16% had a genotype predicting a mild phenotype, which may be one reason for the three times higher number of children diagnosed with MCADD in Denmark during screening than before [2,14] and a concern in relation to possible unnecessary medicalisation. The same may be true for the common mild variant (c.941C>T) in the IVD gene [30]. In the Danish screening algorithm for CF the low penetrant p.R117H variant is only reported if it is found on a 5T background, also representing a filtering of the screening cohort [17]. The above are examples of situations where an entire disease or a subgroup of children with a certain disease type may be molecular genetically filtered away and not reported, possibly reducing harm. However, for such strategy we need to have an in-depth knowledge about how the molecular pathology is related to clinical manifestations or the lack of these [4]. Such knowledge is not always available [7], and we need to collect more molecular genetic data into generally available variant databases together with clinical data. Second-tier molecular genetic testing may also provide the clinician with important prognostic information, where examples may be similar to those described for the mild variants in ACADM and IVD above. Additional examples could be predictions of vitamin B12-responsive cases with MMA, Kuvan-responsive cases with PKU or riboflavin-responsive cases with MADD—all very important and prognostic information to give parents as well as to direct therapy [31].
Screening for some diseases is made difficult by the existence of enzymatic pseudodeficiency. One such disease is mucopolysaccharidosis type 1 (MPS1 deficiency, Hurler syndrome), which we consider including in the NBS panel. Pseudodeficiency alleles may be disclosed by second-tier molecular genetic testing, considerably reducing the number of children reported for confirmative testing [32] (and this number may be further reduced with parallel second-tier testing of glycosaminoglycans [33]).
4.3. Use of Molecular Genetic Studies: Possibilities and Drawbacks
Next-generation sequencing as part of NBS is being introduced in other countries [6]. Similar to Denmark, NGS is used in Norway as part of CF NBS and as second-tier testing for metabolic diseases [5,34]. The UK has started analysing the use of NGS in NBS [10], but otherwise experience is still limited [6,7]. There are a number of disease candidates for NGS-based diagnostics, such as MCADD, VLCADD, IVA and CF mentioned above. MADD is another candidate, which, however, may also present and exemplify controversies. At present, seven genes are known to cause characteristic MADD acylcarnitines and potential riboflavin-responsive and treatable diseases, which could be tested for with NGS as a second-tier test [35]. However, more recently, genetic deficiencies of respiratory chain complexes have also been associated with MADD-like acylcarnitines [36,37]; this challenges and possibly leads to the extension of the genetic evaluation, increasing the risk of identifying variants with unknown clinical relevance or associated with late-onset and untreatable diseases, which are not a focus for NBS [7].
4.3.1. Reporting of Carriers
The reporting of carriers after third-tier NGS testing for CF is performed in the Danish NBS programme [17], but not for other diseases and—in the view of the authors—the reporting of carriers is not an aim of NBS. It is clear that second-tier molecular testing will identify carriers as they are probably enriched in false positive first-tier samples, as we observed in both CF [17] and fatty oxidation defects. In our view the finding of carriers during NBS may be used as an aetiology for being false positive and a way to reduce the false positive rate, but should not be reported. In contrast to this, reporting of carrier status may be performed relevantly in the context of preconception testing (or cascade screening early in pregnancy) as is presently being discussed as a second set-up for screening for rare, untreatable diseases in Denmark [38].
4.3.2. Use as First-Tier Test
NGS in the context of a first-tier setting is being discussed and may have some advantages. Most diseases on many newborn screening panels are caused by pathogenic variants in well-known genes and diagnosis using molecular genetic analyses may seem relevant and are feasible to conduct on filter paper blood spot samples [8,9,10]. As alluded to earlier, a main goal for NBS quality development is to decrease the false positive rate and with first-tier NGS, it could theoretically be possible to obtain the same reduction in false positive rates as we observed for second-tier use of molecular genetic analyses [11]. It may also lead to stratification of infants with severe versus benign disease as for isovaleric aciduria [12]. Another goal for the first-tier use of molecular genetic methods would be to increase the adaptability of NBS making it possible to diagnose treatable diseases without a reliable biochemical biomarker [13]; thus, as shown here for intermittent MSUD, non-classical variants may not be diagnosed by routine biochemical screening [39,40,41]. Additionally, there is a general wish in many countries to increase the number of diseases on NBS panels, and molecular genetic analyses would be able to fulfil this need because the methods are generally applicable for most inherited diseases [7,11]. New therapies are being developed quickly for a number of inherited diseases with many therapies being dependent on early therapeutic initiation shortly after birth; one example is the quickly advancing therapeutic advances in spinal muscular atrophy which have been answered by a quick introduction of a molecularly based newborn screening for SMA in some countries, e.g., in Belgium [42] and very soon in Denmark.
First-tier NGS could be a means to decrease false negative rate. We found 18 children with false negative results. Two (of whom one died) had intermittent MSUD, which cannot be identified by biochemical screening [40,41]; we sequenced children with leucin above cut-off but found none. Thus, the use of second-tier molecular genetic testing would not have helped. Other children with false negative results had CTD, HLCSD and MCADD. All of these false screen negative children had pathogenic variants in the relevant genes and may have been diagnosed via first-tier NGS technology, thus potentially decreasing the false negative rate. However, in a recent paper with post hoc application of targeted whole-exome sequencing (WES) as a first-tier screening procedure, sensitivity for the included 48 inborn errors was only 88% and thus not as high as tandem mass spectrometry, which, in the same study, had a sensitivity of 99% [43]. We encountered children with low biotinidase activity who seemingly were only heterozygotes for a BTD variant as well as one child with a MADD biochemical profile heterozygous for an ETFB variant and in a set-up with first-tier targeted NGS, these children would not have been reported, increasing false negative rate. This argues for a parallel use of targeted NGS technology and biochemical methods [44], though data on sensitivity and specificity in other settings are scarce. Concerning WES specificity in an untargeted set-up, the above study [43] disclosed findings in genes unrelated to the inborn error found for the child in question, decreasing specificity to 98%—much lower than with tandem mass spectrometry, arguing for a targeted set-up if used for NBS.
We would not report carriers in a possible first-tier NGS approach, removing the ability to diagnose mothers affected with diseases such as MCADD, GA1, and CTD because of an abnormal biochemical profile in their newborn. The diagnosis of mothers is not an aim of NBS, but a number of affected mothers have been reported in the Danish NBS program with reference to the treatability of the diseases, and we have seen this as an extra benefit of NBS.
4.3.3. Secondary/Incidental/VUS Findings
Use of NGS in NBS has a risk of identifying secondary/incidental findings as well as variants of uncertain significance (VUS). Risks associated with such findings are: use of increased time because of difficulties in variant interpretation postponing reporting; reporting of benign variants or variants for late-onset diseases, leading to unnecessary medicalising of the child, giving unnecessary treatment and creating patients-in-waiting among other risks. These risks are particularly high in a non-targeted set-up, while it should be minimised when the analysis is targeted exclusively to genes in the NBS panel [10,44], most of which are well-studied genes and well-represented in variant databases used for scoring the variants. However, we found a number of VUS in such known genes, including ACADM, ACADVL and CFTR, and this has also been shown previously for other disease genes included in NBS panels, including PAH gene for phenylketonuria and GALT for galactosemia [45]. The increasing ethnical variation in Western populations will increase the number of VUS even further, which will challenge correct interpretation in an even highly targeted NGS approach as experienced during Danish CF NBS [17]. For all 11 MCADD VUS, and for those reported for VLCADD, it would have proven difficult to report the findings if no biochemical data were available, thus increasing the false negative rate in a first-tier NGS approach. Although variant databases are available with data on specific variants, variants may still be scored very differently by different laboratories with low concordance [5,34,46,47]. All of these problems are well-known when investigating patients with a phenotype; however, during NBS, no clinical phenotype is available and a biochemical phenotype is only available if biochemical testing is conducted alongside NGS testing—with the present knowledge, this would probably be valuable to continue with [48].
4.3.4. Turnaround Time
Turnaround time is a critical concern for NBS. We know from diagnostic WGS for critically ill infants that time to a result varies from 2 weeks to less than 24 h [49,50]; experiences from this are based on sequencing performed mostly untargeted, and a targeted approach for exclusively the diseases in the NBS panel could probably make analysis and evaluation more efficient [43]. If performed in parallel with tandem mass spectrometry, algorithms could be made to report immediately clearly abnormal biochemical findings such as raised succinylacetone for tyrosinemia, thus not increasing turnaround time. Using umbilical cord blood could potentially decrease the time to reporting of a positive newborn, but biochemical testing may not have sufficient sensitivity [51]. If, however, NGS technology becomes a reliable first-tier procedure, this analysis could be used for testing umbilical cord blood thus potentially reducing diagnostic age.
5. Conclusions and Perspectives
The use of molecular genetic technologies as part of second-tier testing is a useful tool to reduce the false positive rate while at the same time giving information about the precise molecular genetic defect to the clinician and thus informing therapeutic strategy and easing giving information about the disease and its prognosis to parents. It also gives the possibility to filter away “unwanted” diagnoses sharing a biomarker with a “wanted” diagnosis. When used in such a set-up, the valuable functional data obtained via tandem mass spectrometry or other biochemical methods will still be available and usability could be extended further via the inclusion of metabolomics or the Collaborative Laboratory Integrated Reports (CLIR) tool to further define phenotype [5,52,53,54,55]. In our view, targeted NGS technology as a second-tier technology should be implemented when possible in the NBS workflow.
First-tier NGS technology may be a promising future possibility, especially for disorders without a reliable biomarker. Pilot studies focusing on feasibility, sensitivity, and specificity should be implemented. Additionally, studies to show ways to integrate NGS technology with classical methods, metabolomics, the CLIR software or transcriptomics should be performed. Finally, we need to explore the views on the use of NGS technology as part of NBS for treatable diseases carried by the general population and in particular by those of reproductive age [27]. To conduct genetic analyses in NSB may be sensitive to some people, and it is of great importance to address all concerns, including parental fears of the inappropriate use of genetic data, in order to prevent potentially negative consequences for the NBS programme as a whole, including maintaining the high degree of trust and the more than 99.9% participation rate in the Danish NBS programme.
Author Contributions
Writing—original draft, A.M.L.; Writing—review & editing, F.W., K.S., M.B.-H., N.G., B.S.A., D.M.H., M.D. and R.K.J.O. All authors have read and agreed to the published version of the manuscript.
Funding
The work was supported by grants from the Ronald McDonald Børnefond, Danmarks Sundhedsfond, Direktør Ib Henriksens Fond, Ragnhild Ibsens Legat til Medicinsk Forskning, Gerda og Aage Haenschs Fond, Dronning Louises Børnehospitals Forskningsfond, Læge Sofus Carl Emil Friis og Hustru Olga Doris Friis’s Legat, Aase og Ejnar Danielsens Fond, Oda og Hans Svenningsens Fond, Fonden af 1870, Vanførefonden, Fonden til Lægevidenskabens Fremme, Aarhus County Research Initiative and the Danish Medical Research Council.
Institutional Review Board Statement
The study was approved by the Ethics Committee (KF 01-152/98).
Informed Consent Statement
An informed dissent procedure was applied for routine NBS, while written informed consent was obtained during the pilot study (see also Methods Section).
Acknowledgments
We are indebted to the staff of the obstetric and paediatric departments for participating in the recruitment and diagnostic workup. The dedicated work by technicians in CIMD, Copenhagen University Hospital, and the Research Unit for Molecular Medicine, Aarhus University Hospital, and in particular Helle Highland Nygaard is sincerely acknowledged. The Committee for Clinical Genetics and Screening, Danish Paediatric Society and The Danish National Board of Health are thanked for their excellent cooperation. Several authors of this publication are members of the European Reference Network for Rare Hereditary Metabolic Disorders (MetabERN)—Project ID No 739543.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| 3-HMGD | 3-hydroxy-3-methyl-glutaryl-CoA lyase deficiency (OMIM ID: 246450) |
| 3-MCCD | 3-methylcrotonyl-CoA-carboxylase deficiency (OMIM IDs: 210200, 210210) |
| 3-MGCHD | 3-methylglutaconyl-CoA hydratase deficiency (OMIM ID: 250950) |
| ALD | adrenoleucodystrophy (OMIM ID: 300100) |
| ARG | hyperargininemia due to arginase deficiency (OMIM ID: 207800) |
| ASLD | argininosuccinate lyase deficiency (OMIM ID: 207900) |
| BETA-KTD | beta-ketothiolase deficiency (OMIM ID: 203750) |
| BH4D | biopterin cofactor deficiencies (OMIM IDs 233910, 261530, 261640) |
| BIOTD | biotinidase deficiency (OMIM ID: 253260) |
| CACTD | carnitine/acylcarnitine translocase deficiency (OMIM ID: 212138) |
| CAH | congenital adrenal hyperplasia (OMIM ID: 201910 |
| CF | cystic fibrosis (OMIM ID: 219700) |
| CFTR | cystic fibrosis transmembrane conductance regulator |
| CH | congenital hypothyroidism |
| CIT | citrullinemia type 1 due to arginosuccinate synthase deficiency (OMIM ID: 215700) |
| CPT1D | carnitine palmitoyl transferase 1 deficiency (OMIM ID: 600528) |
| CPT2D | carnitine palmitoyl transferase 2 deficiency (OMIM IDs: 255110, 600649, 608836) |
| CTD | carnitine transporter deficiency (OMIM ID: 600528) |
| eNBS | expanded newborn screening |
| FAOD | fatty acid oxidation disease |
| GA1 | glutaric aciduria type 1 due to glutaryl-CoA dehydrogenase deficiency (OMIM ID: 231670) |
| GALT | galactosemia due to galactose 1-phosphate uridyl transferase deficiency (OMIM ID: 230400) |
| IEM | inborn error of metabolism |
| HCU-CBS | homocystinuria due to cystathionine-beta-synthase deficiency (OMIM ID: 236200) |
| HHH | hyperornithinemia, hyperammonemia, homocitrullinuria due to ornithine translocase deficiency (OMIM ID: 238970) |
| HLCSD | holocarboxylase synthase deficiency (OMIM ID: 253270) |
| HPA | non-PKU mild hyperphenylalaninemia (OMIM ID: 261660) |
| HT1 | hepatorenal tyrosinemia (type 1) due to fumarylacetoacetase deficiency (OMIM ID: 276700) |
| IVA | isovaleric acidemia due to isovaleryl-CoA dehydrogenase deficiency (OMIM ID: 243500) |
| LCHADD | long-chain 3-hydroxy acyl-CoA dehydrogenase deficiency (OMIM ID 609016) |
| MADD | multiple acyl-CoA dehydrogenase deficiency (OMIM ID: 231680) |
| MCADD | medium-chain acyl-CoA dehydrogenase deficiency (OMIM ID: 201450) |
| MMA | methylmalonic aciduria (many OMIM IDs, most relevant here are: 251000, 251100, 251110); |
| MPS-1H | mucopolysaccharidosis type 1H (OMIM ID: 607014) |
| MS/MS | tandem mass spectrometry |
| M/SCHADD | medium/-short-chain L-3-hydroxy acyl-CoA dehydrogenase deficiency (OMIM ID: 231530) |
| MSUD | classic maple syrup urine disease due to branched-chain alfa-keto acid dehydrogenase deficiency (OMIM ID: 248600) |
| NBS | newborn screening |
| NGS | next generation sequencing |
| PA | propionic acidemia due to propionyl-CoA carboxylase deficiency (OMIM ID: 606054) |
| PKU | classical phenylketonuria due to phenylalanine hydroxylase deficiency (OMIM ID: 261660) |
| PPV | positive predictive value |
| SCADD | short-chain acyl-CoA dehydrogenase deficiency (OMIM ID: 201470) |
| SCID | severe combined immunodeficiency |
| SSI | Statens Serum Institute |
| TPD | trifunctional protein deficiency (OMIM ID: 609015); |
| VLCADD | very long-chain acyl-CoA dehydrogenase deficiency (OMIM ID: 201475) |
| WGS | whole genome sequencing |
References
- Almannai, M.; Marom, R.; Sutton, V.R. Newborn screening: A review of history, recent advancements, and future perspectives in the era of next generation sequencing. Curr. Opin. Pediatr. 2016, 28, 694–699. [Google Scholar] [CrossRef] [PubMed]
- Andresen, B.S.; Lund, A.; Hougaard, D.M.; Christensen, E.; Gahrn, B.; Christensen, M.; Bross, P.; Vested, A.; Simonsen, H.; Skogstrand, K.; et al. MCAD deficiency in Denmark. Mol. Genet. Metab. 2012, 106, 175–188. [Google Scholar] [CrossRef]
- Boy, N.; Mengler, K.; Thimm, E.; Schiergens, K.A.; Marquardt, T.; Weinhold, N.; Marquardt, I.; Das, A.M.; Freisinger, P.; Grünert, S.C.; et al. Newborn screening: A disease-changing intervention for glutaric aciduria type 1. Ann. Neurol. 2018, 83, 970–979. [Google Scholar] [CrossRef]
- Furnier, S.; Durkin, M.; Baker, M. Translating Molecular Technologies into Routine Newborn Screening Practice. Int. J. Neonatal Screen. 2020, 6, 80. [Google Scholar] [CrossRef]
- Tangeraas, T.; Sæves, I.; Klingenberg, C.; Jørgensen, J.; Kristensen, E.; Gunnarsdottir, G.; Hansen, E.V.; Strand, J.; Lundman, E.; Ferdinandusse, S.; et al. Performance of Expanded Newborn Screening in Norway Supported by Post-Analytical Bioinformatics Tools and Rapid Second-Tier DNA Analyses. Int. J. Neonatal Screen. 2020, 6, 51. [Google Scholar] [CrossRef] [PubMed]
- Loeber, J.; Platis, D.; Zetterström, R.; Almashanu, S.; Boemer, F.; Bonham, J.; Borde, P.; Brincat, I.; Cheillan, D.; Dekkers, E.; et al. Neonatal Screening in Europe Revisited: An ISNS Perspective on the Current State and Developments Since 2010. Int. J. Neonatal Screen. 2021, 7, 15. [Google Scholar] [CrossRef] [PubMed]
- Remec, Z.I.; Podkrajsek, K.T.; Lampret, B.R.; Kovac, J.; Groselj, U.; Tesovnik, T.; Battelino, T.; Debeljak, M. Next-Generation Sequencing in Newborn Screening: A Review of Current State. Front. Genet. 2021, 12, 662254. [Google Scholar] [CrossRef]
- Bhattacharjee, A.; Sokolsky, T.; Wyman, S.K.; Reese, M.G.; Puffenberger, E.; Strauss, K.; Morton, H.; Parad, R.B.; Naylor, E.W. Development of DNA Confirmatory and High-Risk Diagnostic Testing for Newborns Using Targeted Next-Generation DNA Sequencing. Genet. Med. 2015, 17, 337–347. [Google Scholar] [CrossRef]
- Poulsen, J.B.; Lescai, F.; Grove, J.; Bækvad-Hansen, M.; Christiansen, M.; Hagen, C.M.; Maller, J.; Stevens, C.; Li, S.; Li, Q.; et al. High-Quality Exome Sequencing of Whole-Genome Amplified Neonatal Dried Blood Spot DNA. PLoS ONE 2016, 11, e0153253. [Google Scholar] [CrossRef]
- Van Campen, J.C.; Sollars, E.S.A.; Thomas, R.C.; Bartlett, C.M.; Milano, A.; Parker, M.D.; Dawe, J.; Winship, P.R.; Peck, G.; Grafham, D.; et al. Next Generation Sequencing in Newborn Screening in the United Kingdom National Health Service. Int. J. Neonatal Screen. 2019, 5, 40. [Google Scholar] [CrossRef]
- Malvagia, S.; Forni, G.; Ombrone, D.; La Marca, G. Development of Strategies to Decrease False Positive Results in Newborn Screening. Int. J. Neonatal Screen. 2020, 6, 84. [Google Scholar] [CrossRef]
- Mütze, U.; Henze, L.; Gleich, F.; Lindner, M.; Grünert, S.C.; Spiekerkoetter, U.; Santer, R.; Blessing, H.; Thimm, E.; Ensenauer, R.; et al. Newborn screening and disease variants predict neurological outcome in isovaleric aciduria. J. Inherit. Metab. Dis. 2021. [Google Scholar] [CrossRef]
- Vasquez-Loarte, T.; Thompson, J.D.; Merritt, J.L. Considering Proximal Urea Cycle Disorders in Expanded Newborn Screening. Int. J. Neonatal Screen. 2020, 6, 77. [Google Scholar] [CrossRef]
- Lund, A.M.; Hougaard, D.M.; Simonsen, H.; Andresen, B.S.; Christensen, M.; Dunø, M.; Skogstrand, K.; Olsen, R.; Jensen, U.G.; Cohen, A.; et al. Biochemical screening of 504,049 newborns in Denmark, the Faroe Islands and Greenland—Experience and development of a routine program for expanded newborn screening. Mol. Genet. Metab. 2012, 107, 281–293. [Google Scholar] [CrossRef] [PubMed]
- Lund, A.; Wibrand, F.; Skogstrand, K.; Cohen, A.; Christensen, M.; Jäpelt, R.B.; Dunø, M.; Skovby, F.; Nørgaard-Pedersen, B.; Gregersen, N.; et al. Danish expanded newborn screening is a successful preventive public health programme. Dan. Med. J. 2020, 67, 1–8. [Google Scholar]
- Statistics Denmark. 2019. Available online: https://www.dst.dk/en (accessed on 3 May 2021).
- Skov, M.; Baekvad-Hansen, M.; Hougaard, D.M.; Skogstrand, K.; Lund, A.; Pressler, T.; Olesen, H.V.; Duno, M. Cystic fibrosis newborn screening in Denmark: Experience from the first 2 years. Pediatr. Pulmonol. 2019, 55, 549–555. [Google Scholar] [CrossRef]
- Andresen, B.S.; Dobrowolski, S.F.; O’Reilly, L.; Muenzer, J.; McCandless, S.E.; Frazier, D.M.; Udvari, S.; Bross, P.; Knudsen, I.; Banas, R.; et al. Medium-chain acyl-CoA dehydrogenase (MCAD) mutations identified by MS/MS-based prospective screening of newborns differ from those observed in patients with clinical symptoms: Identification and characterization of a new, prevalent mutation that results in mild MCAD deficiency. Am. J. Hum. Genet. 2001, 68, 1408–1418. [Google Scholar]
- Aksglæde, L.; Christensen, M.; Olesen, J.H.; Dunø, M.; Olsen, R.K.; Andresen, B.S.; Hougaard, D.M.; Lund, A.M. Abnormal Newborn Screening in a Healthy Infant of a Mother with Undiagnosed Medium-Chain Acyl-CoA Dehydrogenase Deficiency. J. Inherit Metab. Dis. Rep. 2015, 23, 67–70. [Google Scholar]
- Andresen, B.S.; Olpin, S.; Poorthuis, B.J.; Scholte, H.R.; Vianey-Saban, C.; Wanders, R.; Ijlst, L.; Morris, A.; Pourfarzam, M.; Bartlett, K.; et al. Clear correlation of genotype with disease phenotype in very-long-chain acyl-CoA dehydrogenase deficiency. Am. J. Hum. Genet. 1999, 64, 479–494. [Google Scholar] [CrossRef]
- Rajakumar, C.; Ban, M.R.; Cao, H.; Young, T.K.; Bjerregaard, P.; Hegele, R.A. Carnitine palmitoyltransferase IA polymorphism P479L is common in Greenland Inuit and is associated with elevated plasma apolipoprotein A-I. J. Lipid Res. 2009, 50, 1223–1228. [Google Scholar] [CrossRef] [PubMed]
- Borch, L.; Lund, A.M.; Wibrand, F.; Christensen, E.; Søndergaard, C.; Gahrn, B.; Hougaard, D.M.; Andresen, B.S.; Gregersen, N.; Olsen, R.K.J. Normal Levels of Plasma Free Carnitine and Acylcarnitines in Follow-Up Samples from a Presymptomatic Case of Carnitine Palmitoyl Transferase 1 (CPT1) Deficiency Detected Through Newborn Screening in Denmark. JIMD Rep. 2011, 3, 11–15. [Google Scholar] [CrossRef]
- Karaceper, M.D.; Chakraborty, P.; Coyle, D.; Wilson, K.; Kronick, J.B.; Hawken, S.; Davies, C.; Brownell, M.; Dodds, L.; Feigenbaum, A.; et al. Canadian Inherited Metabolic Diseases Research, N. The health system impact of false positive newborn screening results for medium-chain acyl-CoA dehydrogenase deficiency: A cohort study. Orphanet. J. Rare Dis. 2016, 11, 12. [Google Scholar] [CrossRef] [PubMed]
- Gurian, E.A.; Kinnamon, D.; Henry, J.J.; Waisbren, S.E. Expanded Newborn Screening for Biochemical Disorders: The Effect of a False-Positive Result. Pediatrics 2006, 117, 1915–1921. [Google Scholar] [CrossRef] [PubMed]
- Tu, W.-J.; He, J.; Chen, H.; Shi, X.-D.; Li, Y. Psychological Effects of False-Positive Results in Expanded Newborn Screening in China. PLoS ONE 2012, 7, e36235. [Google Scholar] [CrossRef]
- Waisbren, S.E.; Albers, S.; Amato, S.; Ampola, M.; Brewster, T.G.; Demmer, L.; Eaton, R.B.; Greenstein, R.; Korson, M.; Larson, C.; et al. Effect of Expanded Newborn Screening for Biochemical Genetic Disorders on Child Outcomes and Parental Stress. JAMA 2003, 290, 2564–2572. [Google Scholar] [CrossRef] [PubMed]
- Parens, E.; Appelbaum, P.S. On What We Have Learned and Still Need to Learn about the Psychosocial Impacts of Genetic Testing. Häst. Cent. Rep. 2019, 49, S2–S9. [Google Scholar] [CrossRef]
- Stadler, S.C.; Polanetz, R.; Maier, E.M.; Heidenreich, S.C.; Niederer, B.; Mayerhofer, P.U.; Lagler, F.; Koch, H.G.; Santer, R.; Fletcher, J.M.; et al. Newborn screening for 3-methylcrotonyl-CoA carboxylase deficiency: Population heterogeneity of MCCA and MCCB mutations and impact on risk assessment. Hum. Mutat. 2006, 27, 748–759. [Google Scholar] [CrossRef] [PubMed]
- Waddell, L.; Wiley, V.; Carpenter, K.; Bennetts, B.; Angel, L.; Andresen, B.S.; Wilcken, B. Medium-chain acyl-CoA dehydrogenase deficiency: Genotype-biochemical phenotype correlations. Mol. Genet. Metab. 2006, 87, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Mütze, U.; Walter, M.; Keller, M.; Gramer, G.; Garbade, S.F.; Gleich, F.; Haas, D.; Posset, R.; Grünert, S.C.; Hennermann, J.B.; et al. Health Outcomes of Infants with Vitamin B12 Deficiency Identified by Newborn Screening and Early Treated. J. Pediatr. 2021. [Google Scholar] [CrossRef]
- Marsden, D.; Bedrosian, C.L.; Vockley, J. Impact of newborn screening on the reported incidence and clinical outcomes associated with medium- and long-chain fatty acid oxidation disorders. Genet. Med. 2021, 23, 816–829. [Google Scholar] [CrossRef]
- Clarke, L.A.; Giugliani, R.; Guffon, N.; Jones, S.A.; Keenan, H.A.; Munoz-Rojas, M.V.; Okuyama, T.; Viskochil, D.; Whitley, C.B.; Wijburg, F.A.; et al. Genotype-phenotype relationships in mucopolysaccharidosis type I (MPS I): Insights from the International MPS I Registry. Clin. Genet. 2019, 96, 281–289. [Google Scholar] [CrossRef] [PubMed]
- Peck, D.S.; Lacey, J.M.; White, A.L.; Pino, G.; Studinski, A.L.; Fisher, R.; Ahmad, A.; Spencer, L.; Viall, S.; Shallow, N.; et al. Incorporation of Second-Tier Biomarker Testing Improves the Specificity of Newborn Screening for Mucopolysaccharidosis Type I. Int. J. Neonatal Screen. 2020, 6, 10. [Google Scholar] [CrossRef]
- Lundman, E.; Gaup, H.J.; Bakkeheim, E.; Olafsdottir, E.J.; Rootwelt, T.; Storrøsten, O.T.; Pettersen, R.D. Implementation of newborn screening for cystic fibrosis in Norway. Results from the first three years. J. Cyst. Fibros. 2016, 15, 318–324. [Google Scholar] [CrossRef]
- Mosegaard, S.; DiPace, G.; Bross, P.; Carlsen, J.; Gregersen, N.; Olsen, R.K.J. Riboflavin Deficiency—Implications for General Human Health and Inborn Errors of Metabolism. Int. J. Mol. Sci. 2020, 21, 3847. [Google Scholar] [CrossRef]
- Vissing, C.R.; Duno, M.; Olesen, J.H.; Rafiq, J.; Risom, L.; Christensen, E.; Wibrand, F.; Vissing, C. Recurrent myoglobinuria and deranged acylcarnitines due to a mutation in the mtDNA MT-CO2 gene. Neurology 2013, 80, 1908–1910. [Google Scholar] [CrossRef]
- Roos, S.; Sofou, K.; Hedberg-Oldfors, C.; Kollberg, G.; Lindgren, U.; Thomsen, C.; Tulinius, M.; Oldfors, A. Mitochondrial complex IV deficiency caused by a novel frameshift variant in MT-CO2 associated with myopathy and perturbed acylcarnitine profile. Eur. J. Hum. Genet. 2019, 27, 331–335. [Google Scholar] [CrossRef] [PubMed]
- Smed, V.M.; Petersen, O.B.B.; Gerdes, A.-M.A.; Diness, B.R.; Roos, L.S. Genetic screening of prospective parents. Ugeskr Laeger 2021, 183, 13. [Google Scholar]
- Strauss, K.A.; Puffenberger, E.G.; Carson, V.J. Maple Syrup Urine Disease. In Genereviews(R); Adam, M.P., Ardinger, H.H., Eds.; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Estrella, J.; Wilcken, B.; Carpenter, K.; Bhattacharya, K.; Tchan, M.; Wiley, V. Expanded newborn screening in New South Wales: Missed cases. J. Inherit. Metab. Dis. 2014, 37, 881–887. [Google Scholar] [CrossRef] [PubMed]
- Puckett, R.; Lorey, F.; Rinaldo, P.; Lipson, M.; Matern, D.; Sowa, M.; Levine, S.; Chang, R.; Wang, R.; Abdenur, J. Maple syrup urine disease: Further evidence that newborn screening may fail to identify variant forms. Mol. Genet. Metab. 2010, 100, 136–142. [Google Scholar] [CrossRef]
- Boemer, F.; Caberg, J.-H.; Dideberg, V.; Dardenne, D.; Bours, V.; Hiligsmann, M.; Dangouloff, T.; Servais, L. Newborn screening for SMA in Southern Belgium. Neuromuscul. Disord. 2019, 29, 343–349. [Google Scholar] [CrossRef]
- Adhikari, A.N.; Gallagher, R.C.; Wang, Y.; Currier, R.J.; Amatuni, G.; Bassaganyas, L.; Chen, F.; Kundu, K.; Kvale, M.; Mooney, S.D.; et al. The role of exome sequencing in newborn screening for inborn errors of metabolism. Nat. Med. 2020, 26, 1–6. [Google Scholar] [CrossRef]
- Johnston, J.; Lantos, J.D.; Goldenberg, A.; Chen, F.; Parens, E.; Koenig, B.A.; Members of the NSIGHT Ethics and Policy Advisory Board. Sequencing Newborns:A Call for Nuanced Use of Genomic Technologies. Häst. Cent. Rep. 2018, 48, S2–S6. [Google Scholar] [CrossRef]
- Narravula, A.; Garber, K.B.; Askree, S.H.; Hegde, M.; Hall, P.L. Variants of uncertain significance in newborn screening disorders: Implications for large-scale genomic sequencing. Genet. Med. 2017, 19, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Amendola, L.M.; Dorschner, M.O.; Robertson, P.D.; Salama, J.S.; Hart, R.; Shirts, B.H.; Murray, M.L.; Tokita, M.J.; Gallego, C.J.; Kim, D.S.; et al. Actionable exomic incidental findings in 6503 participants: Challenges of variant classification. Genome Res. 2015, 25, 305–315. [Google Scholar] [CrossRef]
- Amendola, L.M.; Muenzen, K.; Biesecker, L.G.; Bowling, K.M.; Cooper, G.M.; Dorschner, M.O.; Driscoll, C.; Foreman, A.K.M.; Golden-Grant, K.; Greally, J.M.; et al. Variant Classification Concordance using the ACMG-AMP Variant Interpretation Guidelines across Nine Genomic Implementation Research Studies. Am. J. Hum. Genet. 2020, 107, 932–941. [Google Scholar] [CrossRef]
- Levy, H. Ethical and Psychosocial Implications of Genomic Newborn Screening. Int. J. Neonatal Screen. 2021, 7, 2. [Google Scholar] [CrossRef] [PubMed]
- Clark, M.M.; Hildreth, A.; Batalov, S.; Ding, Y.; Chowdhury, S.; Watkins, K.; Ellsworth, K.; Camp, B.; Kint, C.I.; Yacoubian, C.; et al. Diagnosis of genetic diseases in seriously ill children by rapid whole-genome sequencing and automated phenotyping and interpretation. Sci. Transl. Med. 2019, 11, eaat6177. [Google Scholar] [CrossRef]
- Kingsmore, S.F.; Cakici, J.A.; Clark, M.M.; Gaughran, M.; Feddock, M.; Batalov, S.; Bainbridge, M.N.; Carroll, J.; Caylor, S.A.; Clarke, C.; et al. A Randomized, Controlled Trial of the Analytic and Diagnostic Performance of Singleton and Trio, Rapid Genome and Exome Sequencing in Ill Infants. Am. J. Hum. Genet. 2019, 105, 719–733. [Google Scholar] [CrossRef] [PubMed]
- Walter, J.H.; Patterson, A.; Till, J.; Besley, G.T.N.; Fleming, G.; Henderson, M.J. Bloodspot acylcarnitine and amino acid analysis in cord blood samples: Efficacy and reference data from a large cohort study. J. Inherit. Metab. Dis. 2009, 32, 95–101. [Google Scholar] [CrossRef]
- Yoon, H.R. Screening newborns for metabolic disorders based on targeted metabolomics using tandem mass spectrometry. Ann. Pediatr. Endocrinol. Metab. 2015, 20, 119–124. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gavrilov, D.K.; Piazza, A.L.; Pino, G.; Turgeon, C.; Matern, D.; Oglesbee, D.; Raymond, K.; Tortorelli, S.; Rinaldo, P. The Combined Impact of CLIR Post-Analytical Tools and Second Tier Testing on the Performance of Newborn Screening for Disorders of Propionate, Methionine, and Cobalamin Metabolism. Int. J. Neonatal Screen. 2020, 6, 33. [Google Scholar] [CrossRef] [PubMed]
- Sörensen, L.; Von Döbeln, U.; Åhlman, H.; Ohlsson, A.; Engvall, M.; Naess, K.; Backman-Johansson, C.; Nordqvist, Y.; Wedell, A.; Zetterström, R.H. Expanded Screening of One Million Swedish Babies with R4S and CLIR for Post-Analytical Evaluation of Data. Int. J. Neonatal Screen. 2020, 6, 42. [Google Scholar] [CrossRef] [PubMed]
- Hall, P.; Marquardt, G.; McHugh, D.M.; Currier, R.; Tang, H.; Stoway, S.D.; Rinaldo, P. Postanalytical tools improve performance of newborn screening by tandem mass spectrometry. Genet. Med. 2014, 16, 889–895. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).