Reducing False-Positive Results in Newborn Screening Using Machine Learning
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Summary
2.2. NBS Metabolic Data Analysis Using Random Forest
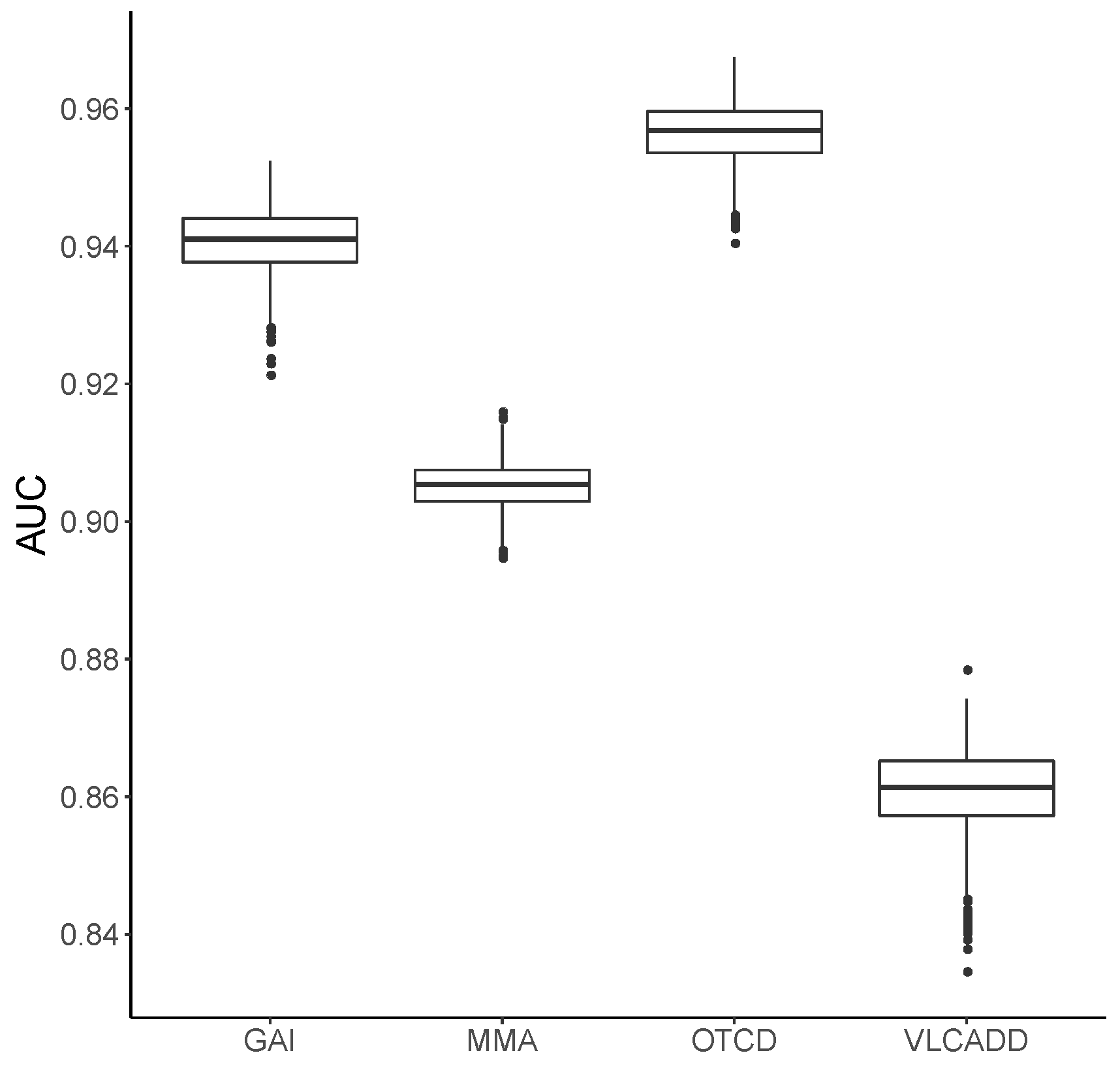
2.3. Validation of the Random Forest Model
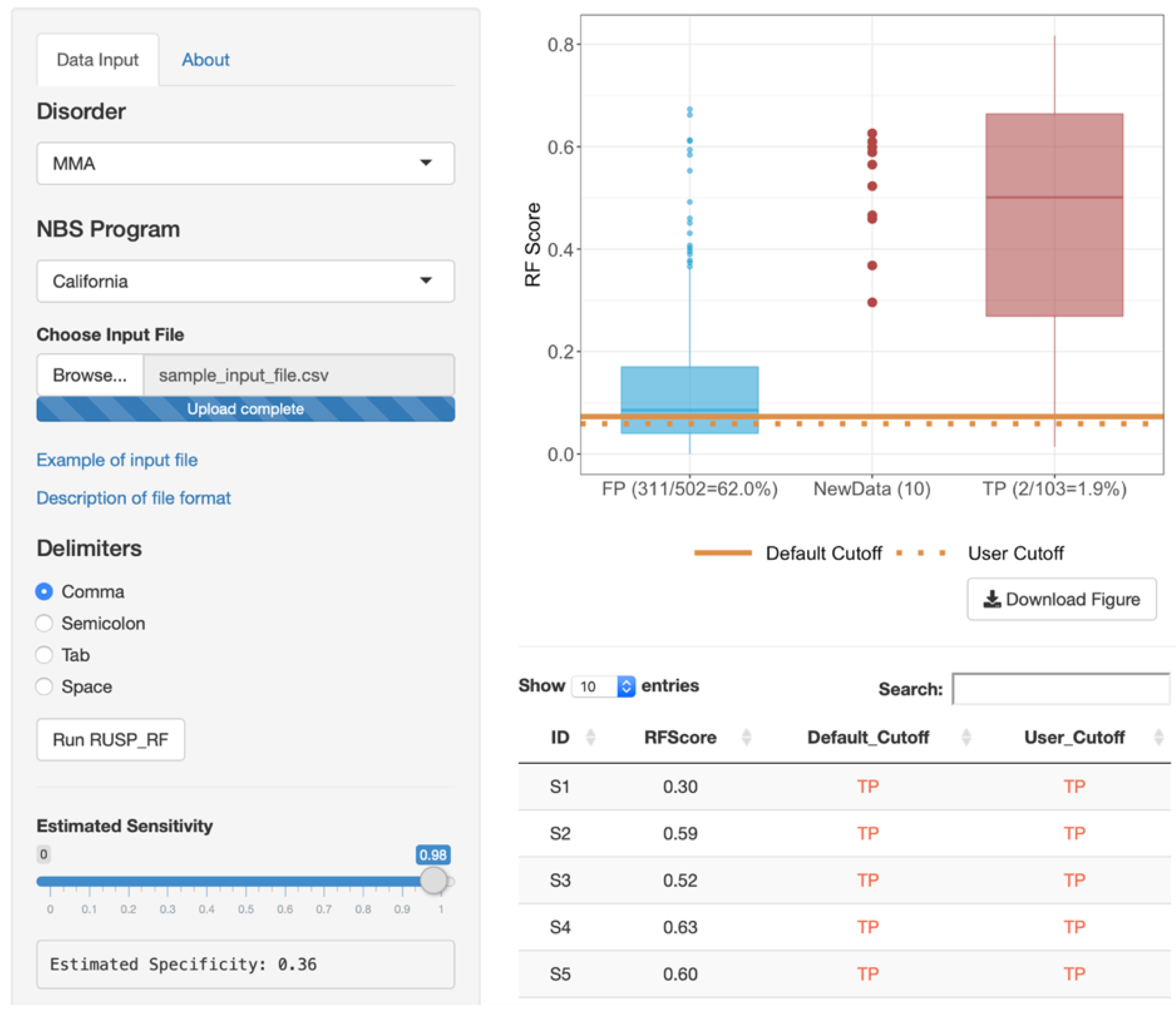
2.4. Web-Based RF Tool and Statistical Analysis
3. Results
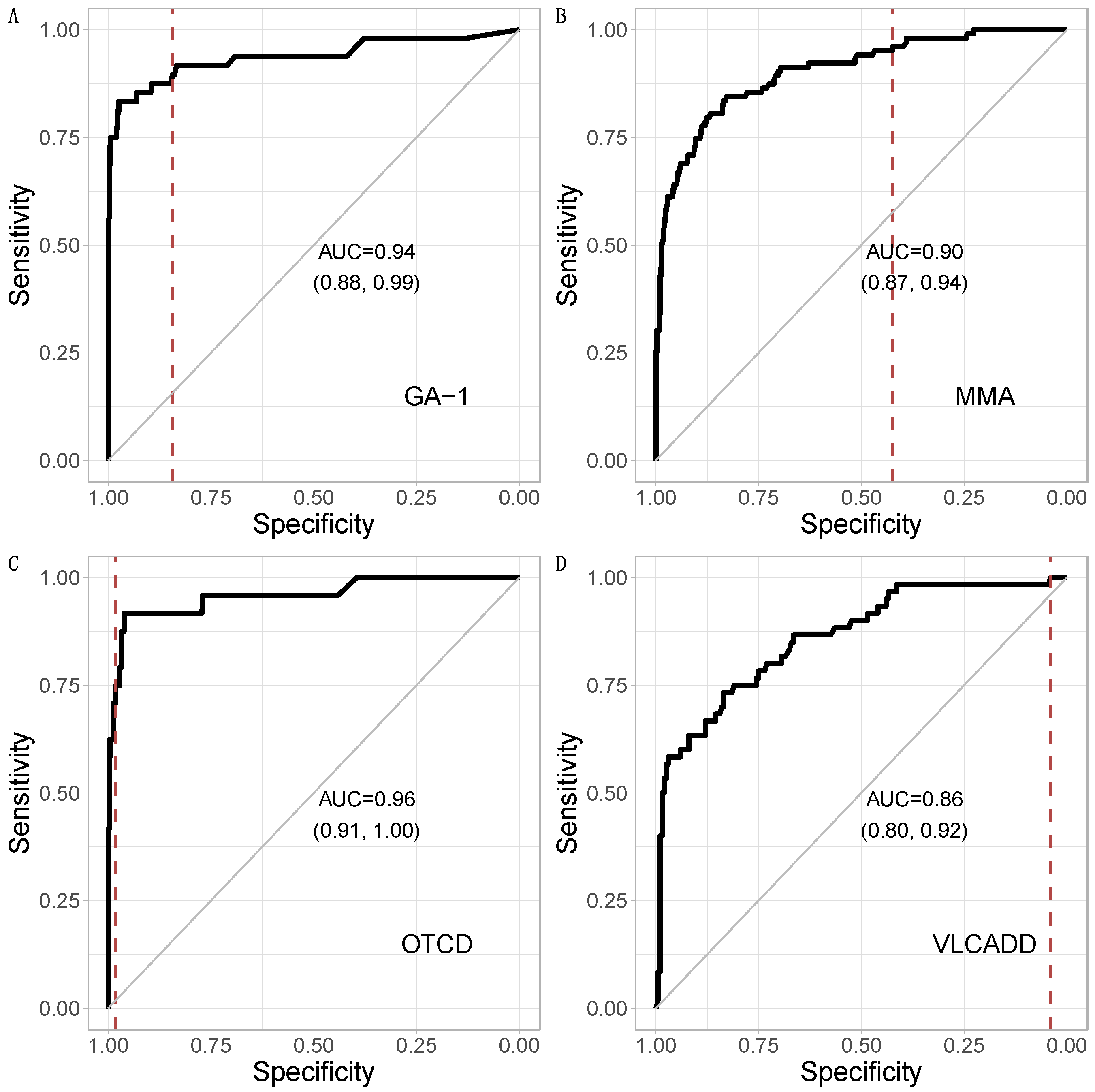
3.1. Metabolic Pattern Analysis Using Random Forest
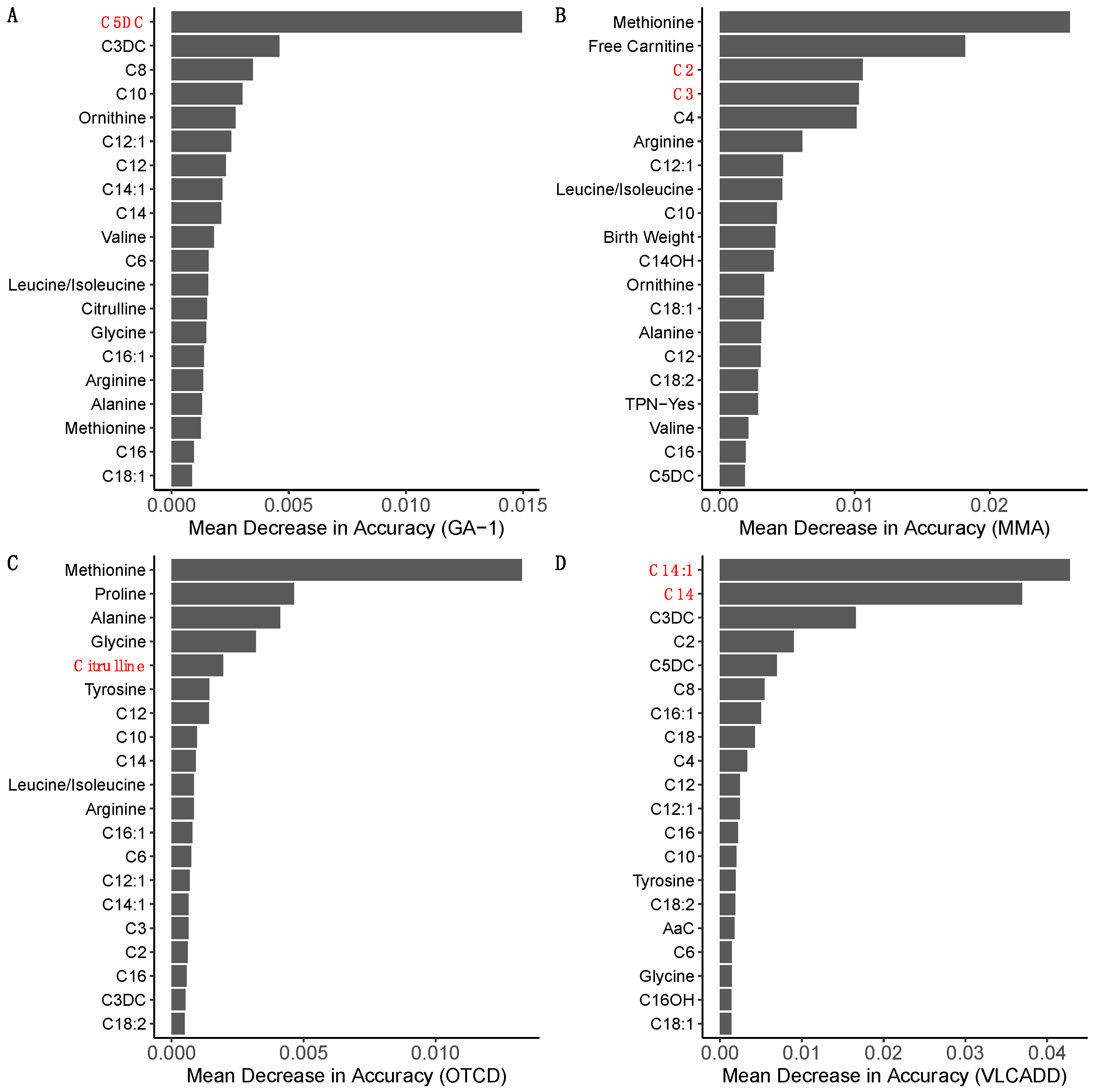
3.2. Ranking of Metabolic Analytes
3.3. Comparison of CLIR and Random Forest
3.4. Web-Based RF Tool
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kwon, C.; Farrell, P.M. The magnitude and challenge of false-positive newborn screening test results. Arch. Pediatr. Adolesc. Med. 2000, 154, 714–718. [Google Scholar] [CrossRef] [PubMed]
- Marquardt, G.; Currier, R.; McHugh, D.M.S.; Gavrilov, D.; Magera, M.J.; Matern, D.; Oglesbee, D.; Raymond, K.; Rinaldo, P.; Smith, E.H.; et al. Enhanced interpretation of newborn screening results without analyte cutoff values. Genet. Med. 2012, 14, 648–655. [Google Scholar] [CrossRef] [PubMed]
- Tortorelli, S.; Eckerman, J.S.; Orsini, J.J.; Stevens, C.; Hart, J.; Hall, P.L.; Alexander, J.J.; Gavrilov, D.; Oglesbee, D.; Raymond, K.; et al. Moonlighting newborn screening markers: The incidental discovery of a second-tier test for Pompe disease. Genet. Med. 2018, 20, 840–846. [Google Scholar] [CrossRef] [PubMed]
- Minter Baerg, M.M.; Stoway, S.D.; Hart, J.; Mott, L.; Peck, D.S.; Nett, S.L.; Eckerman, J.S.; Lacey, J.M.; Turgeon, C.T.; Gavrilov, D.; et al. Precision newborn screening for lysosomal disorders. Genet. Med. 2018, 20, 847–854. [Google Scholar] [CrossRef]
- Hall, P.L.; Marquardt, G.; McHugh, D.M.; Currier, R.J.; Tang, T.; Stoway, S.D.; Rinaldo, P. Postanalytical tools improve performance of newborn screening by tandem mass spectrometry. Genet. Med. 2014, 16, 889–895. [Google Scholar] [CrossRef]
- Morkrid, L.; Rowe, A.D.; Elgstoen, K.B.; Olesen, J.H.; Ruijter, G.; Hall, P.L.; Tortorelli, S.; Schulze, A.; Kyriakopoulou, L.; Wamelink, M.M.; et al. Continuous age- and sex-adjusted reference intervals of urinary markers for cerebral creatine deficiency syndromes: A novel approach to the definition of reference intervals. Clin. Chem. 2015, 61, 760–768. [Google Scholar] [CrossRef]
- Baumgartner, C.; Bohm, C.; Baumgartner, D.; Marini, G.; Weinberger, K.; Olgemöller, B.; Liebl, B.; Roscher, A.A. Supervised machine learning techniques for the classification of metabolic disorders in newborns. Bioinformatics 2004, 20, 2985–2996. [Google Scholar] [CrossRef]
- Chen, W.H.; Hsieh, S.L.; Hsu, K.P.; Chen, H.P.; Su, X.Y.; Tseng, Y.J.; Chien, Y.H.; Hwu, W.L.; Lai, F. Web-based newborn screening system for metabolic diseases: Machine learning versus clinicians. J. Med. Internet Res. 2013, 15, e98. [Google Scholar] [CrossRef]
- Ho, T.K. Random decision forests. In Paper Presented at: Proceedings of the 3rd International Conference on Document Analysis and Recognition, Montreal, QC, Canada, 14–16 August 1995. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Chen, X.; Ishwaran, H. Random forests for genomic data analysis. Genomics 2012, 99, 323–329. [Google Scholar] [CrossRef]
- Wu, B.; Abbott, T.; Fishman, D.; McMurray, W.; Mor, G.; Stone, K.; Ward, D.; Williams, K.; Zhao, H. Comparison of statistical methods for classification of ovarian cancer using mass spectrometry data. Bioinformatics 2003, 19, 1636–1643. [Google Scholar] [CrossRef] [PubMed]
- Melo, C.; Navarro, L.C.; de Oliveira, D.N.; Guerreiro, T.M.; Lima, E.O.; Delafiori, J.; Dabaja, M.Z.; Ribeiro, M.D.S.; de Menezes, M.; Rodrigues, R.G.M.; et al. A Machine Learning Application Based in Random Forest for Integrating Mass Spectrometry-Based Metabolomic Data: A Simple Screening Method for Patients With Zika Virus. Front. Bioeng Biotechnol. 2018, 11, 31. [Google Scholar] [CrossRef] [PubMed]
- Kopp, B.T.; Joseloff, E.; Goetz, D.; Ingram, B.; Heltshe, S.L.; Leung, D.H.; Ramsey, B.W.; McCoy, K.; Borowitz, D. Urinary metabolomics reveals unique metabolic signatures in infants with cystic fibrosis. J. Cyst. Fibros. 2019, 18, 507–515. [Google Scholar] [CrossRef] [PubMed]
- Peng, G.; Shen, P.; Gandotra, N.; Le, A.; Fung, E.; Jelliffe-Pawlowski, L.; Davis, R.W.; Enns, G.M.; Zhao, H.; Cowan, T.M.; et al. Combining newborn metabolic and DNA analysis for second-tier testing of methylmalonic acidemia. Genet. Med. 2019, 21, 896–903. [Google Scholar] [CrossRef]
- American College of Medical Genetics Newborn Screening Expert Group. Newborn screening: Toward a uniform screening panel and system—Executive summary. Pediatrics 2006, 117, S296–S307. [Google Scholar] [CrossRef]
- Oshiro, T.M.; Perez, P.S.; Baranauskas., J.A. How Many Trees in a Random Forest? In Machine Learning and Data Mining in Pattern Recognition; Perner, P., Ed.; Springer: Berlin/Heidelberg, Germany, 2012. [Google Scholar]
- Varma, S.; Simon, R. Bias in error estimation when using cross-validation for model selection. BMC Bioinform. 2006, 7, 91. [Google Scholar] [CrossRef]
- Nicodemus, K.K. Letter to the editor: On the stability and ranking of predictors from random forest variable importance measures. Brief. Bioinform. 2011, 12, 369–373. [Google Scholar] [CrossRef]
- Shiny: Web Application Framework for R. Available online: https://shiny.rstudio.com (accessed on 1 August 2019).
- R: A Language and Environment for Statistical Computing. Available online: https://www.r-project.org (accessed on 1 August 2019).
- Liaw, A.; Wiener, M. Classification and regression by randomForest. R. News. 2002, 2, 18–22. [Google Scholar]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.-C.; Müller, M. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef]
- Kuhn, M. Building predictive models in R using the caret package. J. Stat. Softw. 2008, 28, 1–26. [Google Scholar] [CrossRef]
- Zytkovicz, T.H.; Fitzgerald, E.F.; Marsden, D.; Larson, C.A.; Shih, V.E.; Johnson, D.M.; Strauss, A.W.; Comeau, A.M.; Eaton, R.B.; Grady, G.F. Tandem mass spectrometric analysis for amino, organic, and fatty acid disorders in newborn dried blood spots: A two-year summary from the New England Newborn Screening Program. Clin. Chem. 2001, 47, 1945–1955. [Google Scholar] [CrossRef] [PubMed]
- Weisfeld-Adams, J.D.; Morrissey, M.A.; Kirmse, B.M.; Salveson, B.R.; Wasserstein, M.P.; McGuire, P.J.; Sunny, S.; Cohen-Pfeffer, J.L.; Yu, C.; Caggana, M.; et al. Newborn screening and early biochemical follow-up in combined methylmalonic aciduria and homocystinuria, cblC type, and utility of methionine as a secondary screening analyte. Mol. Genet. Metab. 2010, 99, 116–123. [Google Scholar] [CrossRef] [PubMed]
- Rinaldo, P.; Whitley, R.J.; Rhead, W.J.; Hannon, W.H. Evidence-Based Rationale for Expanded Newborn Screening. N. Engl. J. Med. 2009, 348, 2304–2312. [Google Scholar]
- McClead, R.E.J.; Rozen, R.; Fox, J.; Rosenberg, L.; Menke, J.; Bickers, R.; Morrow, G., 3rd. Clinical application of DNA analysis in a family with OTC deficiency. Am. J. Med. Genet. 1986, 25, 513–518. [Google Scholar] [CrossRef]
- McHugh, D.; Cameron, C.A.; Abdenur, J.E.; Abdulrahman, M.; Adair, O.; Al Nuaimi, S.A.; Ahlman, H.; Allen, J.J.; Antonozzi, I.; Archer, S.; et al. Clinical validation of cutoff target ranges in newborn screening of metabolic disorders by tandem mass spectrometry: A worldwide collaborative project. Genet. Med. 2011, 13, 230–254. [Google Scholar] [CrossRef]
- Di Donato, S.; Rimoldi, M.; Garavaglia, B.; Uziel, G. Propionylcarnitine excretion in propionic and methylmalonic acidurias: A cause of carnitine deficiency. Clin. Chim. Acta 1984, 139, 13–21. [Google Scholar] [CrossRef]
- Bisanzi, S.; Morrone, A.; Donati, M.A.; Pasquini, E.; Spada, M.; Strisciuglio, P.; Parenti, G.; Parini, R.; Papadia, F.; Zammarchi, E. Genetic analysis in nine unrelated Italian patients affected by OTC deficiency: Detection of novel mutations in the OTC gene. Mol. Genet. Metab. 2002, 76, 137–144. [Google Scholar] [CrossRef]
- De Sain-van der Velden, M.G.; Rinaldo, P.; Elvers, B.; Henderson, M.; Walter, J.H.; Prinsen, B.H.; Verhoeven-Duif, N.M.; de Koning, T.J.; van Hasselt, P. The Proline/Citrulline Ratio as a Biomarker for OAT Deficiency in Early Infancy. JIMD Rep. 2012, 6, 95–99. [Google Scholar]
- Hennermann, J.B.; Roloff, S.; Gellermann, J.; Gruters, A.; Klein, J. False-positive newborn screening mimicking glutaric aciduria type I in infants with renal insufficiency. J. Inherited Metab. Dis. 2009, 32, S355–S359. [Google Scholar] [CrossRef]
- Diekman, E.; de Sain-van der Velden, M.; Waterham, H.; Kluijtmans, L.; Schielen, P.; van Veen, E.B.; Ferdinandusse, S.; Wijburg, F.; Visser, G. The Newborn Screening Paradox: Sensitivity vs. Overdiagnosis in VLCAD Deficiency. JIMD Rep. 2016, 27, 101–106. [Google Scholar]
- Kolker, S.; Boy, S.P.; Heringer, J.; Müller, E.; Maier, E.M.; Ensenauer, R.; Mühlhausen, C.; Schlune, A.; Greenberg, C.R.; Koeller, D.M.; et al. Complementary dietary treatment using lysine-free, arginine-fortified amino acid supplements in glutaric aciduria type I—A decade of experience. Mol. Genet. Metab. 2012, 107, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Atkins, A.E.; Tarini, B.A.; Phillips, E.K.; Calhoun, A. Misclassification of VLCAD carriers due to variable confirmatory testing after a positive NBS result. J. Community Genet. 2019, 10, 447–451. [Google Scholar] [CrossRef] [PubMed]
- Merritt, J.L., 2nd; Vedal, S.; Abdenur, J.E.; Au, S.M.; Barshop, B.A.; Feuchtbaum, L.; Harding, C.O.; Hermerath, C.; Lorey, F.; Sesser, D.E.; et al. Infants suspected to have very-long chain acyl-CoA dehydrogenase deficiency from newborn screening. Mol. Genet. Metab. 2014, 111, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Vernooij-van Langen, A.M.; Loeber, J.G.; Elvers, B.; Triepels, R.H.; Roefs, J.; Gille, J.J.; Reijntjens, S.; Dompeling, E.; Dankert-Roelse, J.E. The influence of sex, gestational age, birth weight, blood transfusion, and timing of the heel prick on the pancreatitis-associated protein concentration in newborn screening for cystic fibrosis. J. Inherited Metab. Dis. 2013, 36, 147–154. [Google Scholar] [CrossRef]
| Disorder | Confirmed Positive | First-Tier NBS | Second-Tier Analysis (RF, This Study) | ||
|---|---|---|---|---|---|
| False Positives | PPV | False Positives * | PPV | ||
| GA-1 | 48 | 1344 | 3.10% | 150 | 22.30% |
| MMA | 103 | 502 | 16.40% | 276 | 26.40% |
| OTCD | 24 | 496 | 3.50% | 11 | 62.10% |
| VLCADD | 60 | 200 | 23.10% | 196 | 23.40% |
| GA-1 | MMA | OTCD | VLCADD | Control * | |
|---|---|---|---|---|---|
| Gestational Age, week | |||||
| <37 | 340 (24.4%) | 175 (28.9%) | 181 (34.8%) | 42 (16.2%) | 5490 (5.5%) |
| 37–41 | 1005 (72.2%) | 412 (68.1%) | 325 (62.5%) | 206 (79.2%) | 93,603 (94.0%) |
| >41 | 47 (3.4%) | 18 (3.0%) | 14 (2.7%) | 12 (4.6%) | 444 (0.4%) |
| Birth Weight, g | |||||
| <2500 | 279 (20.0%) | 173 (28.6%) | 130 (25.0%) | 26 (10.0%) | 4045 (4.1%) |
| 2500–4000 | 1025 (73.6%) | 381 (63.0%) | 354 (68.1%) | 223 (85.8%) | 87,268 (87.7%) |
| >4000 | 88 (6.3%) | 51 (8.4%) | 36 (6.9%) | 11 (4.2%) | 8224 (8.3%) |
| Sex | |||||
| Male | 845 (60.7%) | 321 (53.1%) | 325 (62.5%) | 165 (63.5%) | 51,352 (51.6%) |
| Female | 542 (38.9%) | 281 (46.4%) | 194 (37.3%) | 93 (35.8%) | 47,882 (48.1%) |
| Unknown | 5 (0.4%) | 3 (0.5%) | 1 (0.2%) | 2 (0.8%) | 303 (0.3%) |
| Race/Ethnicity | |||||
| Asian | 136 (9.8%) | 63 (10.4%) | 33 (6.3%) | 40 (15.4%) | 14275 (14.3%) |
| Black | 212 (15.2%) | 25 (4.1%) | 50 (9.6%) | 15 (5.8%) | 6630 (6.7%) |
| Hispanic | 444 (31.9%) | 407 (67.3%) | 224 (43.1%) | 94 (36.2%) | 49,400 (49.6%) |
| White | 554 (39.8%) | 92 (15.2%) | 197 (37.9%) | 102 (39.2%) | 26341 (26.5%) |
| Other/Unknown | 46 (3.3%) | 18 (3.0%) | 16 (3.1%) | 9 (3.5%) | 2891 (2.9%) |
| Age at Blood Collection, hour | |||||
| <12 | 246 (17.7%) | 142 (23.5%) | 45 (8.7%) | 47 (18.1%) | 21,564 (21.7%) |
| 12–24 | 877 (63.0%) | 319 (52.7%) | 259 (49.8%) | 183 (70.4%) | 71,396 (71.7%) |
| >24 | 269 (19.3%) | 144 (23.8%) | 216 (41.5%) | 30 (11.5%) | 6577 (6.6%) |
| Total Parenteral Nutrition | |||||
| No | 1178 (84.6%) | 393 (65.0%) | 453 (87.1%) | 248 (95.4%) | 97,269 (97.7%) |
| Yes | 146 (10.5%) | 187 (30.9%) | 57 (11.0%) | 3 (1.2%) | 998 (1.0%) |
| Unknown | 68 (4.9%) | 25 (4.1%) | 10 (1.9%) | 9 (3.5%) | 1270 (1.3%) |
| MDA Ranking | GA-1 | MMA | OTCD | VLCADD |
|---|---|---|---|---|
| 1 | C5DC a | Methionine b [28] | Methionine b [29,30] | C14:1 a |
| 2 | C3DC | Free Carnitine [31] | Proline b [29,32,33] | C14 a |
| 3 | C8 b [28] | C2 a | Alanine [29] | C3DC |
| 4 | C10 [34] | C3 a | Glycine [29] | C2 b [35] |
| 5 | Ornithine [36] | C4 | Citrulline a | C5DC |
| Predicted by Algorithm | NBS Results (Truth) | ||
|---|---|---|---|
| TP | FP | ||
| CLIR | TP | 44 | 67 |
| FP | 4 | 911 | |
| Random Forest | TP | 43 | 53 |
| FP | 5 | 925 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, G.; Tang, Y.; Cowan, T.M.; Enns, G.M.; Zhao, H.; Scharfe, C. Reducing False-Positive Results in Newborn Screening Using Machine Learning. Int. J. Neonatal Screen. 2020, 6, 16. https://doi.org/10.3390/ijns6010016
Peng G, Tang Y, Cowan TM, Enns GM, Zhao H, Scharfe C. Reducing False-Positive Results in Newborn Screening Using Machine Learning. International Journal of Neonatal Screening. 2020; 6(1):16. https://doi.org/10.3390/ijns6010016
Chicago/Turabian StylePeng, Gang, Yishuo Tang, Tina M. Cowan, Gregory M. Enns, Hongyu Zhao, and Curt Scharfe. 2020. "Reducing False-Positive Results in Newborn Screening Using Machine Learning" International Journal of Neonatal Screening 6, no. 1: 16. https://doi.org/10.3390/ijns6010016
APA StylePeng, G., Tang, Y., Cowan, T. M., Enns, G. M., Zhao, H., & Scharfe, C. (2020). Reducing False-Positive Results in Newborn Screening Using Machine Learning. International Journal of Neonatal Screening, 6(1), 16. https://doi.org/10.3390/ijns6010016





