Next-Generation Sequencing for Cystic Fibrosis: Florida Newborn Screening Experience
Abstract
1. Background and Significance
2. Methods
2.1. NBS CF Screening Algorithm
2.2. Overall Design
3. Results
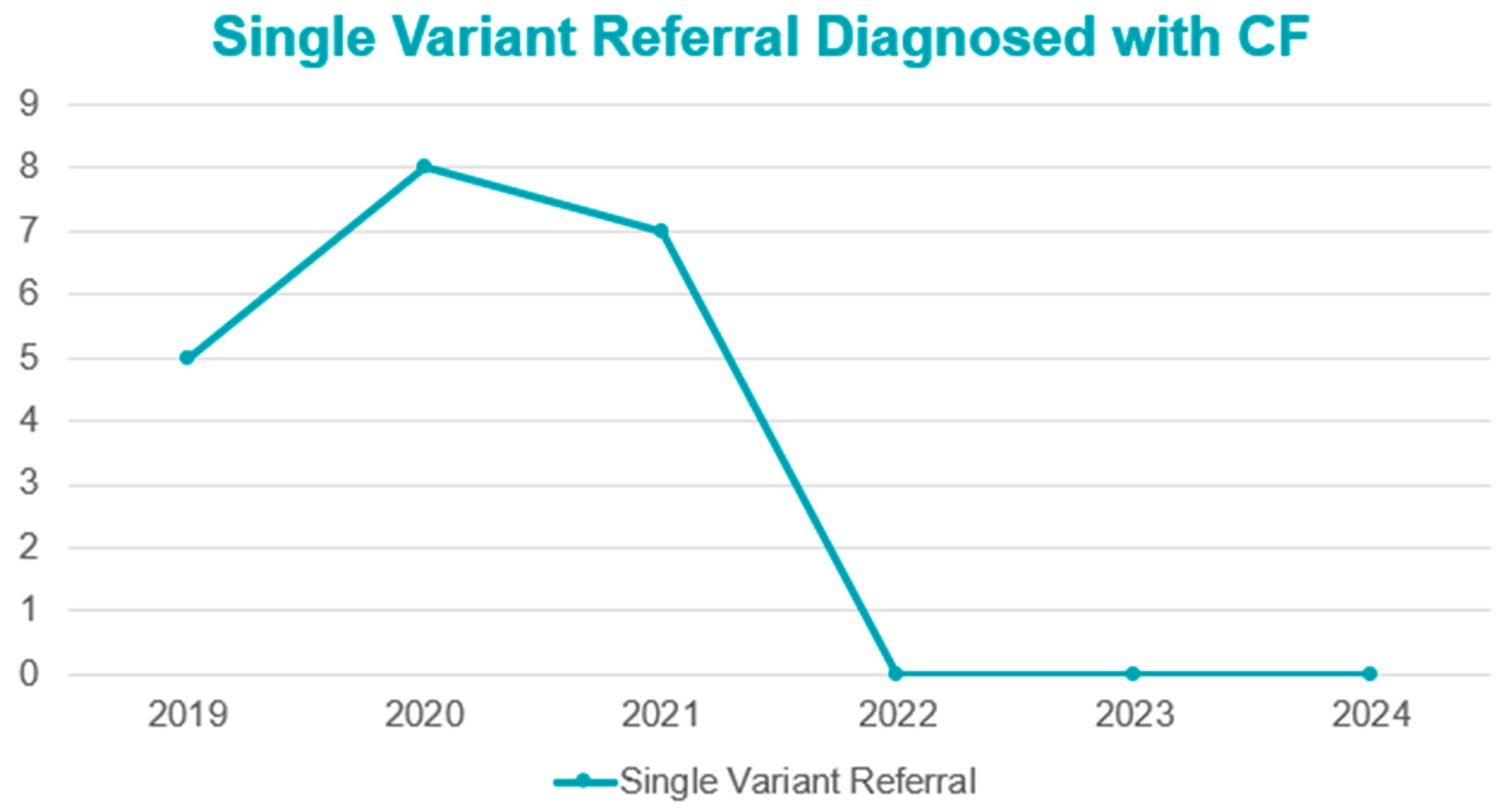
3.1. CF NBS Referrals
3.2. Diagnosis of Referrals
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Crossley, J.R.; Elliott, R.B.; Smith, P.A. Dried-blood spot screening for Cystic Fibrosis in the newborn. Lancet 1979, 1, 472–474. [Google Scholar] [CrossRef]
- Scotet, V.; L’Hostis, C.; Ferec, C. The changing epidemiology of cystic fibrosis: Incidence, survival and impact of the CFTR gene discovery. Genes 2020, 11, 589. [Google Scholar] [CrossRef]
- Farrell, P.M.; Li, Z.; Kosorok, M.R.; Laxova, A.; Green, C.G.; Collins, J.; Lai, H.-C.; Rock, M.J.; Splaingard, M.L. Bronchopulmonary disease in children with cystic fibrosis after early or delayed diagnosis. Am. J. Respir. Crit. Care Med. 2003, 168, 1100–1108. [Google Scholar] [CrossRef]
- Coffey, M.J.; Whitaker, V.; Gentin, N.; Junek, R.; Shalhoub, C.; Nightingale, S.; Hilton, J.; Wiley, V.; Wilcken, B.; Gaskin, K.J.; et al. Differences in outcomes between early and late diagnosis of cystic fibrosis in the newborn screening era. J. Pediatr. 2017, 181, 137–145. [Google Scholar] [CrossRef]
- Bender, L.M.; Cotton, S.W.; Willis, M.S. Kids in America: Newborn screening for cystic fibrosis. Lab. Med. 2011, 42, 595–601. [Google Scholar] [CrossRef][Green Version]
- Schrijver, I.; Pique, L.; Graham, S.; Pearl, M.; Cherry, A.; Kharrazi, M. The spectrum of CFTR variants in nonwhite cystic fibrosis patients: Implications for molecular diagnostic testing. J. Mol. Diagn. 2016, 18, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Sugarman, E.A.; Rohlfs, E.M.; Silverman, L.M.; Alitto, M.A. CFTR mutation distribution among US Hispanic and African American individuals; evaluation in cystic fibrosis patients and carrier screening populations. Genet. Med. 2004, 6, 392–399. [Google Scholar] [CrossRef]
- CFTR2.org (CFTR2 Variant List History|CFTR2): Clinical and Functional Translation of CFTR. Available online: https://cftr2.org (accessed on 23 September 2025).
- ClinVar. National Center for Biotechnology Information, National Library of Medicine, Bethesda, MD, USA. Available online: https://www.ncbi.nlm.nih.gov/clinvar/ (accessed on 23 September 2025).
- Sadeghi, H.; Kay, D.M.; Langfelder-Schwind, E.; DeCelie-Germana, J.K.; Berdella, M.; Soultan, Z.N.; Goetz, D.M.; Caggana, M.; Fortner, C.N.; Giusti, R.; et al. Characterization of 223 infants with CFTR-Related metabolic syndrome/cystic fibrosis screen positive, inconclusive diagnosis (CRMS/CFSPID) identified during the first three years of newborn screening via IRT-DNA-SEQ in New York State. J. Cyst. Fibros. 2024, 24, 404–411. [Google Scholar] [CrossRef]
- McGarry, M.E.; Sciortino, S.; Graham, S.; Bishop, T.; Gibb, E.R. Improved detection of cystic fibrosis by the California Newborn Screening Program for all races and ethnicities. Pediatr. Pulmonol. 2024, 59, 2901–2909. [Google Scholar] [CrossRef] [PubMed]
- Ren, C.L.; Borowitz, D.S.; Gonska, T.; Howenstine, M.S.; Levy, H.; Massie, J.; Milla, C.; Munck, A.; Southern, K.W. Cystic fibrosis transmembrane conductance regulator-related metabolic syndrome and cystic fibrosis screen positive, inconclusive diagnosis. J. Pediatr. 2017, 181, S45–S51.e1. [Google Scholar] [CrossRef] [PubMed]
- gnomAD. gnomAD Browser, Version v4.1.0; Broad Institute: Cambridge, MA, USA. Available online: https://gnomad.broadinstitute.org/ (accessed on 23 September 2025).
- Rock, M.J.; Baker, M.; Antos, N.; Farrell, P.M. Refinement of newborn screening for cystic fibrosis with next generation sequencing. Pediatr. Pulmonol. 2023, 58, 778–787. [Google Scholar] [CrossRef]
- LeGrys, V.A.; Yankaskas, J.R.; Quittell, L.M.; Marshall, B.C.; Mogayzel, P.J. Diagnostic sweat testing: The Cystic Fibrosis Foundation guidelines. J. Pediatr. 2007, 151, 85–89. [Google Scholar] [CrossRef]
- Shenoy, A.; Spyropoulos, D.; Peeke, K.; Smith, D.; Cellucci, M.; Chidekel, A. Newborn Screening for Cystic Fibrosis: Infant and Laboratory Factors Affecting Successful Sweat Test Completion. Int. J. Neonatal Screen. 2020, 7, 1. [Google Scholar] [CrossRef]
- Tluczek, A.; Koscik, R.L.; Farrell, P.M.; Rock, M.J. Psychosocial risk associated with newborn screening for cystic fibrosis: Parents’ experience while awaiting the sweat-test appointment. Pediatrics 2005, 115, 1692–1703. [Google Scholar] [CrossRef] [PubMed]
- Foil, K.; Christon, L.; Kerrigan, C.; Flume, P.A.; Drinkwater, J.; Szentpetery, S. Experiences of cystic fibrosis newborn screening and genetic counseling. J. Community Genet. 2023, 14, 621–626. [Google Scholar] [CrossRef] [PubMed]
- Brown, R.F.; Close, C.T.; Mailes, M.G.; Gonzalez, L.J.; Goetz, D.M.; Filigno, S.S.; Preslar, R.; Tran, Q.T.; Hempstead, S.E.; Lomas, P.; et al. Cystic fibrosis foundation position paper: Redefining the cystic fibrosis care team. J. Cyst. Fibros. 2024, 23, 1045–1054. [Google Scholar] [CrossRef] [PubMed]
- Green, D.M.; Lahiri, T.; Raraigh, K.S.; Ruiz, F.; Spano, J.; Antos, N.; Bonitz, L.; Christon, L.; Gregoire-Bottex, M.; Hale, J.E.; et al. Cystic Fibrosis Foundation Evidence-Based Guideline for the Management of CRMS/CFSPID. Pediatrics 2024, 153, e2023064657. [Google Scholar] [CrossRef]
- Wolfe, A.G.; Gilley, S.P.; Waldrop, S.W.; Olson, C.; Harding, E.; Widmer, K.; Gumer, L.B.; Haemer, M.; Hoppe, J.E. Case Report: Cystic fibrosis with kwashiorkor: A rare presentation in the era of universal newborn screening. Front. Pediatr. 2023, 10, 1083155. [Google Scholar] [CrossRef]
- Qussous, K.; Abdulhamid, I.; Kleyn, M.; Schuen, J.; Nasr, S.Z. Five cases of missed cystic fibrosis heterozygous mutations identified after a positive newborn screen on a sibling. Respir. Med. Case Rep. 2022, 36, 101572. [Google Scholar] [CrossRef]
- Dunn, C.T.; Skrypek, M.M.; Powers, A.L.; Laguna, T.A. The need for vigilance: The case of a false-negative newborn screen for cystic fibrosis. Pediatrics 2011, 128, e446–e449. [Google Scholar] [CrossRef]
- McGarry, M.E.; Raraigh, K.S.; Farrell, P.; Shropshire, F.; Padding, K.; White, C.; Dorley, M.C.; Hicks, S.; Ren, C.L.; Tullis, K.; et al. Cystic fibrosis newborn screening: A systematic review-driven consensus guideline from the United States cystic fibrosis foundation. Int. J. Neonatal Screen. 2025, 11, 24. [Google Scholar] [CrossRef] [PubMed]


| Year | CF | CRMS/CFSPID | Carriers |
|---|---|---|---|
| 2019 | 40 | 10 | 410 |
| 2020 | 38 | 12 | 335 |
| 2021 | 40 | 12 | 570 |
| 2022 | 25 | 54 | 890 |
| 2023 | 23 | 80 | 838 |
| 2024 * | 32 | 106 | 852 |
| 2022 | 2023 | 2024 | |
|---|---|---|---|
| F508del | 320 | 235 | 390 |
| PolyT-T5-TG12 | 298 | 365 | 367 |
| PolyT-T5-TG13 | 39 | 21 | 31 |
| R117H | 65 | 31 | 51 |
| F508C | 35 | 32 | 57 |
| 3120 + 1G > A | 29 | 22 | 33 |
| R1162L | 23 | 14 | 33 |
| R75Q | 16 | 30 | 64 |
| T966T | 16 | 25 | 34 |
| I506V | 18 | 12 | 33 |
| Number of Variants | 2022 CRMS Cases | 2022 Sweat Cl Classification | 2023 CRMS Cases | 2023 Sweat Cl Classification | 2024 CRMS Cases | 2024 Sweat Cl Classification |
|---|---|---|---|---|---|---|
| 6 variants | 2 | 1 intermediate, 1 normal | 1 | 1 normal | 0 | - |
| 5 variants | 0 | - | 1 | 1 normal | 1 | 1 intermediate |
| 4 variants | 5 | 5 normal | 7 | 2 intermediate, 5 normal | 4 | 4 normal |
| 3 variants | 20 | 7 intermediate, 13 normal | 28 | 7 intermediate, 21 normal | 43 | 5 intermediate, 35 normal, 3 QNS |
| 2 variants | 20 | 2 intermediate, 18 normal | 35 | 6 intermediate, 26 normal, 3 not performed | 48 | 7 intermediate, 38 normal, 1 positive, 1 QNS |
| 1 variant | 7 | 5 intermediate, 1 normal, 1 QNS | 8 | 2 intermediate, 3 normal, 2 not performed | 10 | 6 intermediate, 4 normal |
| Total CRMS cases | 54 (15 intermediate, 36 normal, 1 QNS, 2 not performed) | 80 (17 intermediate, 55 normal, 1 QNS, 5 not performed) | 106 (19 intermediate, 82 normal, 4 QNS, 1 positive) | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Published by MDPI on behalf of the International Society for Neonatal Screening. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Green, D.M.; Polasky, J.; Weatherly, M.; Stalker, H.; Blanchard, C.; Kushner, C.; Couluris, M.; Ryland, P.; Sunitha, I.; Fong, J.; et al. Next-Generation Sequencing for Cystic Fibrosis: Florida Newborn Screening Experience. Int. J. Neonatal Screen. 2025, 11, 94. https://doi.org/10.3390/ijns11040094
Green DM, Polasky J, Weatherly M, Stalker H, Blanchard C, Kushner C, Couluris M, Ryland P, Sunitha I, Fong J, et al. Next-Generation Sequencing for Cystic Fibrosis: Florida Newborn Screening Experience. International Journal of Neonatal Screening. 2025; 11(4):94. https://doi.org/10.3390/ijns11040094
Chicago/Turabian StyleGreen, Deanna M., Jean Polasky, Mark Weatherly, Heather Stalker, Colleen Blanchard, Cheryl Kushner, Marisa Couluris, Patricia Ryland, Iruvanti Sunitha, Joseph Fong, and et al. 2025. "Next-Generation Sequencing for Cystic Fibrosis: Florida Newborn Screening Experience" International Journal of Neonatal Screening 11, no. 4: 94. https://doi.org/10.3390/ijns11040094
APA StyleGreen, D. M., Polasky, J., Weatherly, M., Stalker, H., Blanchard, C., Kushner, C., Couluris, M., Ryland, P., Sunitha, I., Fong, J., Crump, S., Reeves, E., & Barnette, K. (2025). Next-Generation Sequencing for Cystic Fibrosis: Florida Newborn Screening Experience. International Journal of Neonatal Screening, 11(4), 94. https://doi.org/10.3390/ijns11040094








