Analytical Validation of a Genomic Newborn Screening Workflow
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Design of Validation Plates
- Eight positive newborn samples (NBPOS-1 to NBPOS-8, 3-plex of each), with pathogenic (P) or likely pathogenic (LP) variants in PAH, ACADM, MMUT, G6PD, CFTR, DDC genes, confirmed with screening and diagnostic methods.
- Eight negative newborn samples (NBNEG-1 to NBNEG-8, 3-plex of each).
- Four negative adult samples (ADNEG-1 to ADNEG-4, 4-plex of each). These are adult whole blood samples with no reported conditions.
- Same four negative adult samples spotted on DBS (DBS-ADNEG-1 to DBS-ADNEG-4, 4-plex each).
- HG002-NA24385 Genome in a Bottle (GIAB) reference DNA (https://www.nist.gov/programs-projects/genome-bottle, accessed on 22 September 2025, Coriell Institute, Camden, NJ, USA), 8-plex.
2.3. DNA Extraction
2.4. tNGS Panel Design and Sequencing
2.5. Bioinformatic Analysis
2.6. Sensitivity and Precision of Sequencing
2.7. Reproducibility of the Results
2.8. Definition and Selection of Quality Metrics
2.9. Threshold Setting
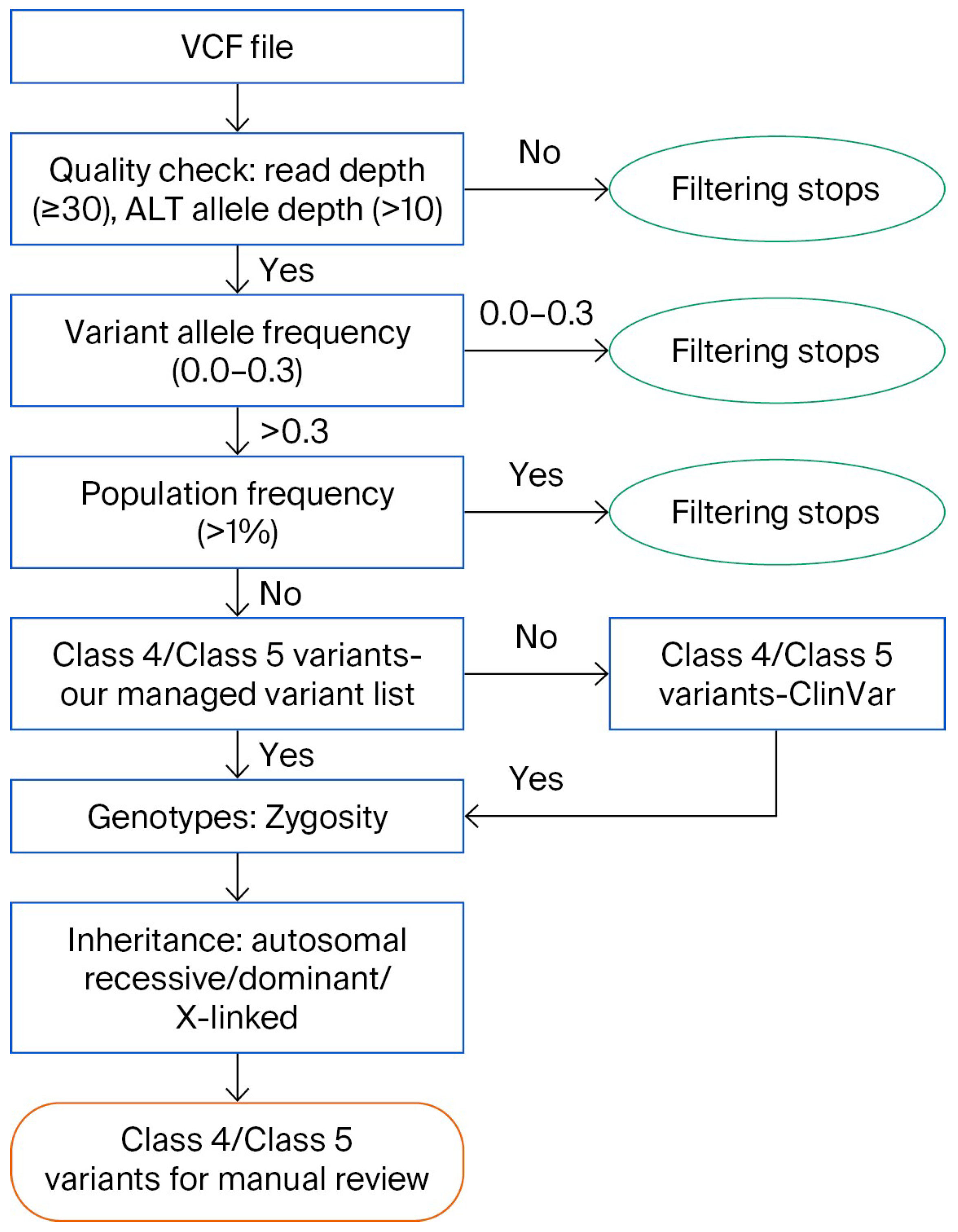
2.10. Variant Interpretation Pipeline
3. Results
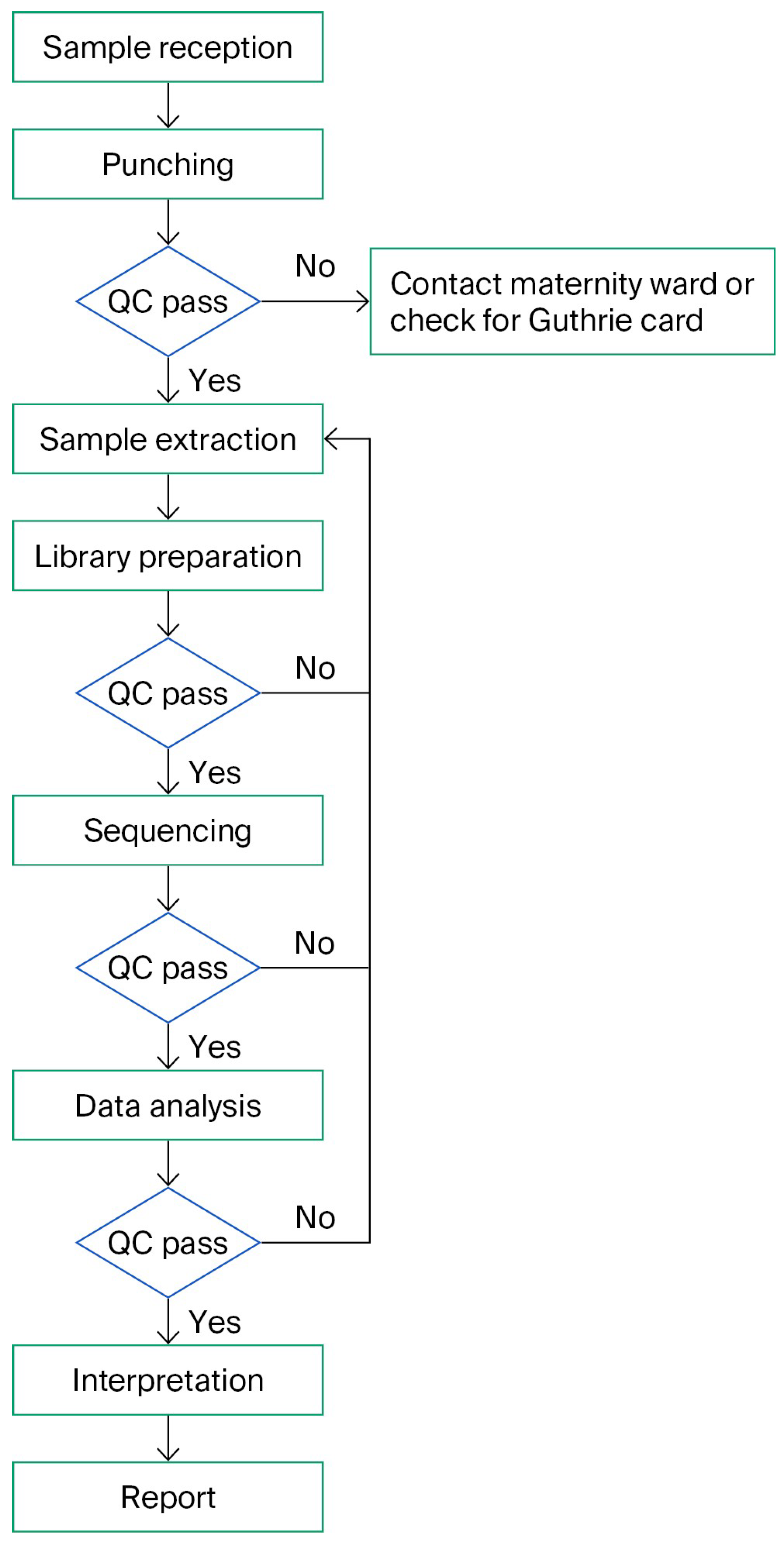
3.1. Workflow
3.2. Variant Interpretation Pipeline
3.3. Validation Samples
3.4. Performance of the Analysis
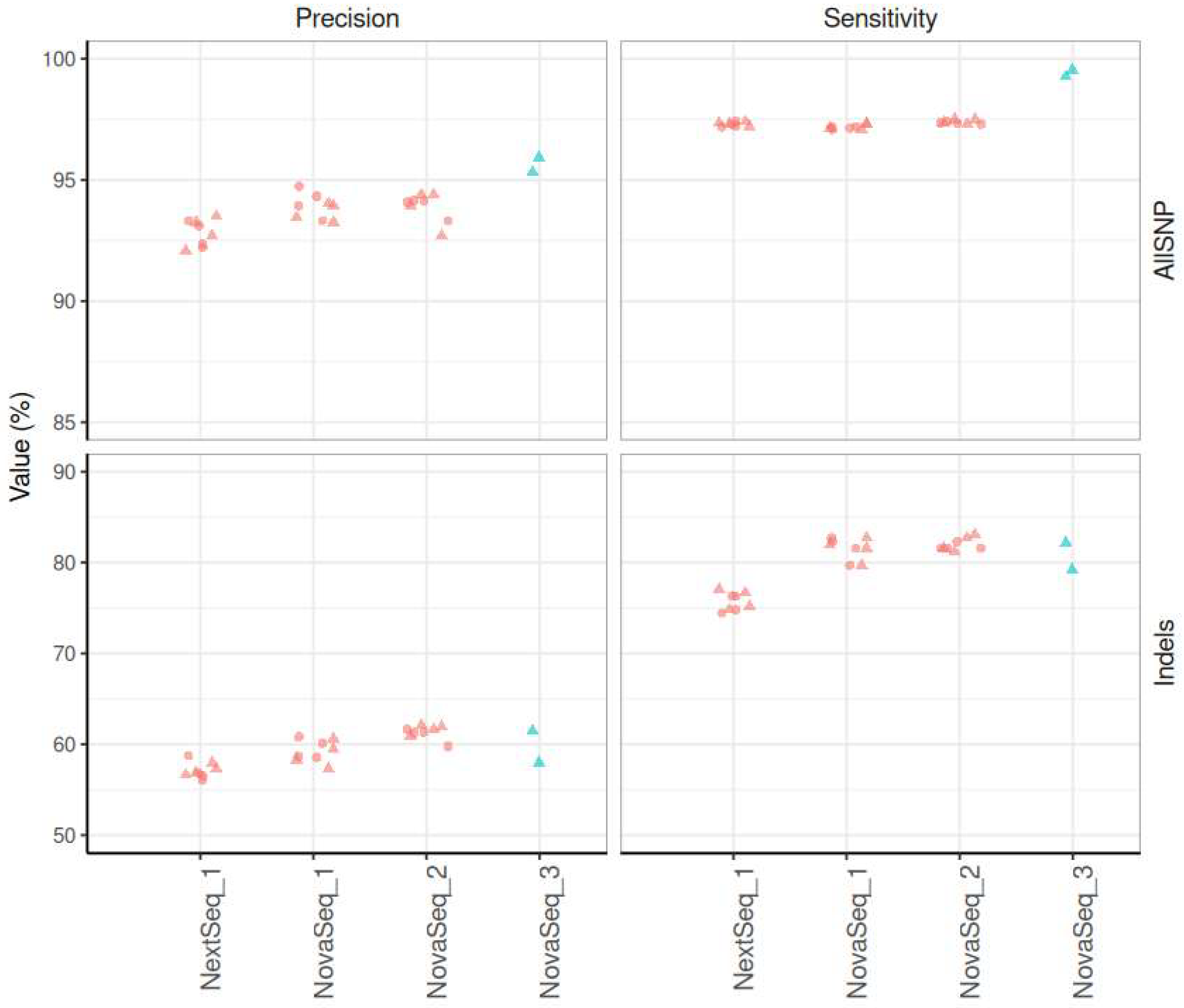
3.4.1. Sensitivity, Precision
3.4.2. Intra-Run and Inter-Run Concordance
3.5. Quality Control
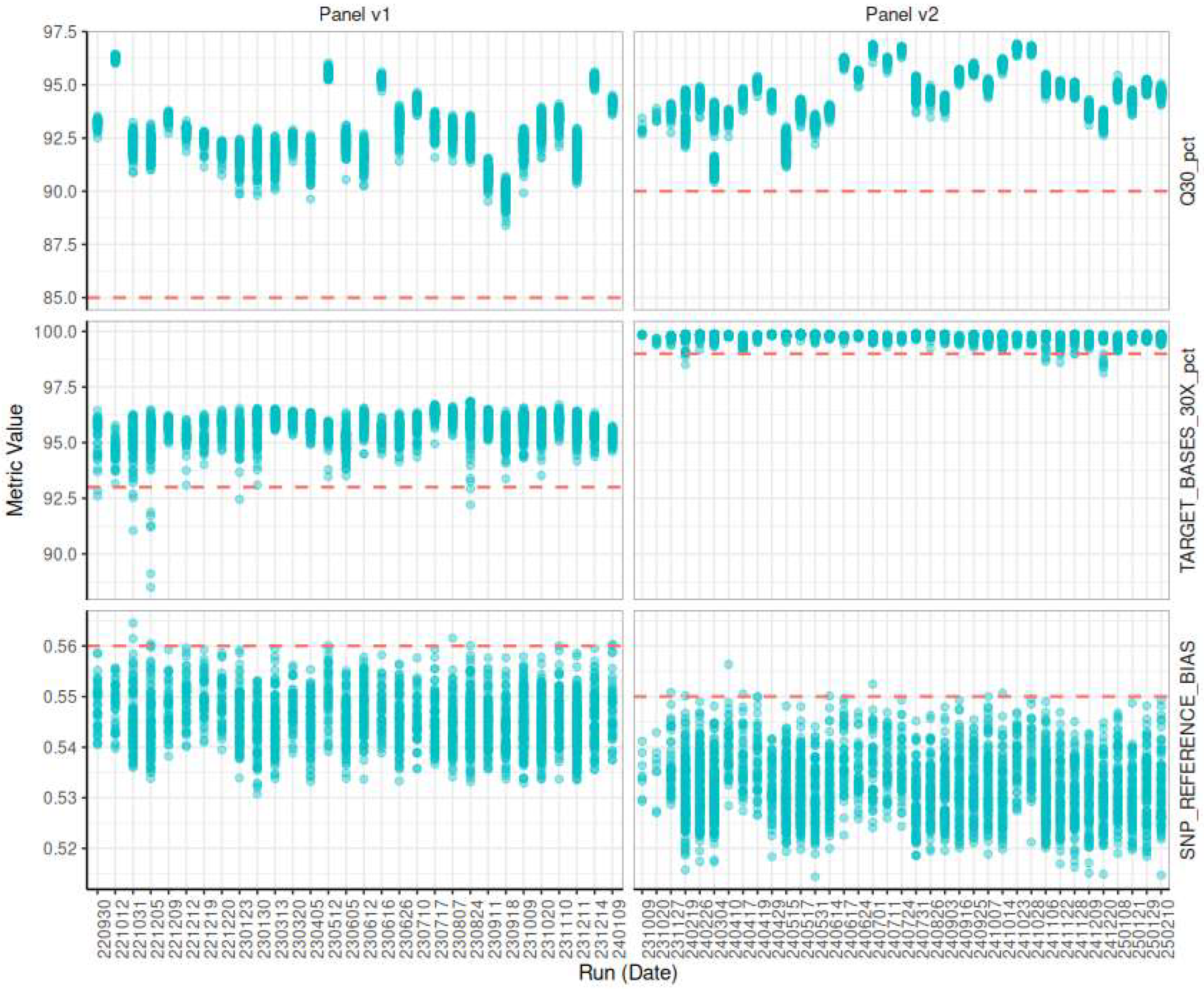
3.5.1. Evaluating Sequencing Quality
3.5.2. Evaluating Target Selection Quality
3.5.3. Evaluating Inversions and Contaminations
3.5.4. Longitudinal Monitoring
3.5.5. Decision Criteria for Sample Quality
3.6. Robustness to Workflow Variations
3.6.1. Initial DNA Quantity
3.6.2. Initial Material Variation: Whole Blood or Dried Blood Spot
3.6.3. Sequencing Instrument
3.7. Optimizations
3.7.1. DNA Extraction Automation
3.7.2. Target of Interest: Improving Performance and Clinical Impact
4. Discussion
- Immutable configuration tracking, where every analysis run is associated with a fixed pipeline version, tool versions, and parameters.
- Automated unit and integration testing for all pipeline components, ensuring that updates or infrastructure changes do not introduce regressions.
- Reference dataset benchmarking to regularly evaluate the pipeline against synthetic or known truth sets (e.g., Genome in a Bottle, synthetic mixtures), thereby safeguarding analytical performance.
- Clear separation between development and production environments, with a formal promotion workflow when pipeline changes are validated and ready for deployment.
- Data provenance mechanisms (e.g., checksums, sample lineage tracking) to ensure that outputs can be backtracked to raw data and initial parameters.
- Furthermore, harmonization with external clinical guidelines should be embedded where applicable, particularly at the variant filtration and prioritization stages.
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Andermann, A. Revisting wilson and Jungner in the genomic age: A review of screening criteria over the past 40 years. Bull. World Health Organ. 2008, 86, 317–319. [Google Scholar] [CrossRef] [PubMed]
- Fernhoff, P.M. Newborn Screening for Genetic Disorder. Pediatr. Clin. N. Am. 2009, 56, 505–513. [Google Scholar] [CrossRef] [PubMed]
- Engel, A.G. Congenital Myasthenic Syndromes. In Rosenberg’s Molecular and Genetic Basis of Neurological and Psychiatric Disease, 5th ed.; Academic Press: Cambridge, MA, USA, 2015; pp. 1191–1208. [Google Scholar] [CrossRef]
- Brennenstuhl, H.; Jung-Klawitter, S.; Assmann, B.; Opladen, T. Inherited Disorders of Neurotransmitters: Classification and Practical Approaches for Diagnosis and Treatment. Neuropediatrics 2019, 50, 2–14. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Prasad, A. Inborn Errors of Metabolism and Epilepsy: Current Understanding, Diagnosis, and Treatment Approaches. Int. J. Mol. Sci. 2017, 18, 1384. [Google Scholar] [CrossRef]
- Amir, F.; Atzinger, C.; Massey, K.; Greinwald, J.; Hunter, L.L.; Ulm, E.; Kettler, M. The Clinical Journey of Patients with Riboflavin Transporter Deficiency Type 2. J. Child Neurol. 2020, 35, 283–290. [Google Scholar] [CrossRef]
- Roberts, E.A.; Schilsky, M.L. Current and Emerging Issues in Wilson’s Disease. N. Engl. J. Med. 2023, 389, 922–938. [Google Scholar] [CrossRef]
- Van Der Burg, M.; Mahlaoui, N.; Gaspar, H.B.; Pai, S.-Y. Universal Newborn Screening for Severe Combined Immunodeficiency (SCID). Front. Pediatr. 2019, 7, 373. [Google Scholar] [CrossRef]
- Boemer, F.; Caberg, J.-H.; Beckers, P.; Dideberg, V.; Di Fiore, S.; Bours, V.; Marie, S.; Dewulf, J.; Marcelis, L.; Deconinck, N.; et al. Three years pilot of spinal muscular atrophy newborn screening turned into official program in Southern Belgium. Sci. Rep. 2021, 11, 19922. [Google Scholar] [CrossRef]
- Müller-Felber, W.; Blaschek, A.; Schwartz, O.; Gläser, D.; Nennstiel, U.; Brockow, I.; Wirth, B.; Burggraf, S.; Röschinger, W.; Becker, M.; et al. Newbornscreening SMA—From Pilot Project to Nationwide Screening in Germany. J. Neuromuscul. Dis. 2023, 10, 55–65. [Google Scholar] [CrossRef]
- Servais, L.; Dangouloff, T.; Muntoni, F.; Scoto, M.; Baranello, G. Spinal muscular atrophy in the UK: The human toll of slow decisions. Lancet 2025, 405, 619–620. [Google Scholar] [CrossRef]
- Ziegler, A.; Koval-Burt, C.; Kay, D.M.; Suchy, S.F.; Begtrup, A.; Langley, K.G.; Hernan, R.; Amendola, L.M.; Boyd, B.M.; Bradley, J.; et al. Expanded Newborn Screening Using Genome Sequencing for Early Actionable Conditions. JAMA 2025, 333, 232. [Google Scholar] [CrossRef]
- Chen, T.; Fan, C.; Huang, Y.; Feng, J.; Zhang, Y.; Miao, J.; Wang, X.; Li, Y.; Huang, C.; Jin, W.; et al. Genomic Sequencing as a First-Tier Screening Test and Outcomes of Newborn Screening. JAMA Netw. Open 2023, 6, e2331162. [Google Scholar] [CrossRef]
- Boemer, F.; Hovhannesyan, K.; Piazzon, F.; Minner, F.; Mni, M.; Jacquemin, V.; Mashhadizadeh, D.; Benmhammed, N.; Bours, V.; Jacquinet, A.; et al. Population-based, first-tier genomic newborn screening in the maternity ward. Nat. Med. 2025, 31, 1339–1350. [Google Scholar] [CrossRef]
- The BabySeq Project Team; Holm, I.A.; Agrawal, P.B.; Ceyhan-Birsoy, O.; Christensen, K.D.; Fayer, S.; Frankel, L.A.; Genetti, C.A.; Krier, J.B.; LaMay, R.C.; et al. The BabySeq project: Implementing genomic sequencing in newborns. BMC Pediatr. 2018, 18, 225. [Google Scholar]
- Kingsmore, S.F.; Wright, M.; Olsen, L.; Schultz, B.; Protopsaltis, L.; Averbuj, D.; Blincow, E.; Carroll, J.; Caylor, S.; Defay, T.; et al. Genome-based newborn screening for severe childhood genetic diseases has high positive predictive value and sensitivity in a NICU pilot trial. Am. J. Hum. Genet. 2024, 111, 2643–2667. [Google Scholar] [CrossRef]
- Jansen, M.E.; Klein, A.W.; Buitenhuis, E.C.; Rodenburg, W.; Cornel, M.C. Expanded Neonatal Bloodspot Screening Programmes: An Evaluation Framework to Discuss New Conditions With Stakeholders. Front. Pediatr. 2021, 9, 635353. [Google Scholar] [CrossRef] [PubMed]
- Minten, T.; Bick, S.; Adelson, S.; Gehlenborg, N.; Amendola, L.M.; Boemer, F.; Coffey, A.J.; Encina, N.; Ferlini, A.; Kirschner, J.; et al. Data-driven consideration of genetic disorders for global genomic newborn screening programs. Genet. Med. 2025, 27, 101443. [Google Scholar] [CrossRef]
- Dangouloff, T.; Hovhannesyan, K.; Mashhadizadeh, D.; Minner, F.; Mni, M.; Helou, L.; Piazzon, F.; Palmeira, L.; Boemer, F.; Servais, L. Feasibility and Acceptability of a Newborn Screening Program Using Targeted Next-Generation Sequencing in One Maternity Hospital in Southern Belgium. Children 2024, 11, 926. [Google Scholar] [CrossRef]
- Charloteaux, B.; Helou, L.; Karssen, L.; Kooijman, M.; Palmeira, L.; Paquet, L.; Russo, C.; Sonet, I. Humanomics 2024.10.15; GitLab: San Francisco, CA, USA, 2024. [Google Scholar] [CrossRef]
- Wagner, J.; Olson, N.D.; Harris, L.; Khan, Z.; Farek, J.; Mahmoud, M.; Stankovic, A.; Kovacevic, V.; Yoo, B.; Miller, N.; et al. Benchmarking challenging small variants with linked and long reads. Cell Genomics 2022, 2, 100128. [Google Scholar] [CrossRef]
- Broad Institute. Picard Toolkit. Broad Inst. GitHub Repos. 2019. Available online: https://broadinstitute.github.io/picard/ (accessed on 2 June 2025).
- Tawari, N.R.; Seow, J.J.W.; Perumal, D.; Ow, J.L.; Ang, S.; Devasia, A.G.; Ng, P.C. ChronQC: A quality control monitoring system for clinical next generation sequencing. Bioinformatics 2018, 34, 1799–1800. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. Off. J. Am. Coll. Med. Genet. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- Kopanos, C.; Tsiolkas, V.; Kouris, A.; Chapple, C.E.; Albarca Aguilera, M.; Meyer, R.; Massouras, A. VarSome: The human genomic variant search engine. Bioinformatics 2019, 35, 1978–1980. [Google Scholar] [CrossRef]
- NBN EN ISO 15189:2022. Medical laboratories—Requirements for quality and competence. Brussels: Bureau for Standardization, Belgium, 2022.
- Ding, Y.; Owen, M.; Le, J.; Batalov, S.; Chau, K.; Kwon, Y.H.; Van Der Kraan, L.; Bezares-Orin, Z.; Zhu, Z.; Veeraraghavan, N.; et al. Scalable, high quality, whole genome sequencing from archived, newborn, dried blood spots. npj Genomic Med. 2023, 8, 5. [Google Scholar] [CrossRef] [PubMed]
- Turner, D.J.; Miretti, M.; Rajan, D.; Fiegler, H.; Carter, N.P.; Blayney, M.L.; Beck, S.; Hurles, M.E. Germline rates of de novo meiotic deletions and duplications causing several genomic disorders. Nat. Genet. 2008, 40, 90–95. [Google Scholar] [CrossRef]
- Zhang, F.; Carvalho, C.M.B.; Lupski, J.R. Complex human chromosomal and genomic rearrangements. Trends Genet. 2009, 25, 298–307. [Google Scholar] [CrossRef]
- Collins, R.L.; Talkowski, M.E. Diversity and consequences of structural variation in the human genome. Nat. Rev. Genet. 2025, 26, 443–462. [Google Scholar] [CrossRef]
- Shaikh, T.H. Copy Number Variation Disorders. Curr. Genet. Med. Rep. 2017, 5, 183–190. [Google Scholar] [CrossRef]
- Komlósi, K.; Gyenesei, A.; Bene, J. Editorial: Copy Number Variation in Rare Disorders. Front. Genet. 2022, 13, 898059. [Google Scholar] [CrossRef] [PubMed]
- Truty, R.; Paul, J.; Kennemer, M.; Lincoln, S.E.; Olivares, E.; Nussbaum, R.L.; Aradhya, S. Prevalence and properties of intragenic copy-number variation in Mendelian disease genes. Genet. Med. 2019, 21, 114–123. [Google Scholar] [CrossRef] [PubMed]
- Hahn, E.; Dharmadhikari, A.V.; Markowitz, A.L.; Estrine, D.; Quindipan, C.; Maggo, S.D.S.; Sharma, A.; Lee, B.; Maglinte, D.T.; Shams, S.; et al. Copy number variant analysis improves diagnostic yield in a diverse pediatric exome sequencing cohort. NPJ Genomic Med. 2025, 10, 16. [Google Scholar] [CrossRef]
- Fang, H.; Wu, Y.; Narzisi, G.; ORawe, J.A.; Barrón, L.T.J.; Rosenbaum, J.; Ronemus, M.; Iossifov, I.; Schatz, M.C.; Lyon, G.J. Reducing INDEL calling errors in whole genome and exome sequencing data. Genome Med. 2014, 6, 89. [Google Scholar] [CrossRef]
- Goldfeder, R.L.; Priest, J.R.; Zook, J.M.; Grove, M.E.; Waggott, D.; Wheeler, M.T.; Salit, M.; Ashley, E.A. Medical implications of technical accuracy in genome sequencing. Genome Med. 2016, 8, 24. [Google Scholar] [CrossRef]
- NIHR BioResource—Rare Disease; Next Generation Children Project; French, C.E.; Delon, I.; Dolling, H.; Sanchis-Juan, A.; Shamardina, O.; Mégy, K.; Abbs, S.; Austin, T.; et al. Whole genome sequencing reveals that genetic conditions are frequent in intensively ill children. Intensive Care Med. 2019, 45, 627–636. [Google Scholar] [CrossRef]
- Taylor, J.C.; Martin, H.C.; Lise, S.; Broxholme, J.; Cazier, J.-B.; Rimmer, A.; Kanapin, A.; Lunter, G.; Fiddy, S.; Allan, C.; et al. Factors influencing success of clinical genome sequencing across a broad spectrum of disorders. Nat. Genet. 2015, 47, 717–726. [Google Scholar] [CrossRef]
- Royer-Bertrand, B.; Cisarova, K.; Niel-Butschi, F.; Mittaz-Crettol, L.; Fodstad, H.; Superti-Furga, A. CNV Detection from Exome Sequencing Data in Routine Diagnostics of Rare Genetic Disorders: Opportunities and Limitations. Genes 2021, 12, 1427. [Google Scholar] [CrossRef]
- Jegathisawaran, J.; Tsiplova, K.; Hayeems, R.; Ungar, W.J. Determining accurate costs for genomic sequencing technologies—A necessary prerequisite. J. Community Genet. 2020, 11, 235–238. [Google Scholar] [CrossRef]
- Schwarze, K.; Buchanan, J.; Taylor, J.C.; Wordsworth, S. Are whole-exome and whole-genome sequencing approaches cost-effective? A systematic review of the literature. Genet. Med. 2018, 20, 1122–1130. [Google Scholar] [CrossRef] [PubMed]
- Nurchis, M.C.; Radio, F.C.; Salmasi, L.; Heidar Alizadeh, A.; Raspolini, G.M.; Altamura, G.; Tartaglia, M.; Dallapiccola, B.; Pizzo, E.; Gianino, M.M.; et al. Cost-Effectiveness of Whole-Genome vs Whole-Exome Sequencing Among Children With Suspected Genetic Disorders. JAMA Netw. Open 2024, 7, e2353514. [Google Scholar] [CrossRef]
- Green, R.C.; Berg, J.S.; Grody, W.W.; Kalia, S.S.; Korf, B.R.; Martin, C.L.; McGuire, A.L.; Nussbaum, R.L.; O’Daniel, J.M.; Ormond, K.E.; et al. Correction: Corrigendum: ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet. Med. 2017, 19, 606. [Google Scholar] [CrossRef]
- Islam, M.R.; Ahmed, M.U.; Barua, S.; Begum, S. A Systematic Review of Explainable Artificial Intelligence in Terms of Different Application Domains and Tasks. Appl. Sci. 2022, 12, 1353. [Google Scholar] [CrossRef]
- De Paoli, F.; Nicora, G.; Berardelli, S.; Gazzo, A.; Bellazzi, R.; Magni, P.; Rizzo, E.; Limongelli, I.; Zucca, S. Digenic variant interpretation with hypothesis-driven explainable AI. NAR Genomics Bioinforma. 2025, 7, lqaf029. [Google Scholar] [CrossRef]
- Wratten, L.; Wilm, A.; Göke, J. Reproducible, scalable, and shareable analysis pipelines with bioinformatics workflow managers. Nat. Methods 2021, 18, 1161–1168. [Google Scholar] [CrossRef]
- Di Tommaso, P.; Chatzou, M.; Floden, E.W.; Barja, P.P.; Palumbo, E.; Notredame, C. Nextflow enables reproducible computational workflows. Nat. Biotechnol. 2017, 35, 316–319. [Google Scholar] [CrossRef]
- Mölder, F.; Jablonski, K.P.; Letcher, B.; Hall, M.B.; Tomkins-Tinch, C.H.; Sochat, V.; Forster, J.; Lee, S.; Twardziok, S.O.; Kanitz, A.; et al. Sustainable data analysis with Snakemake. F1000Research 2021, 10, 33. [Google Scholar] [CrossRef] [PubMed]
- Kurtzer, G.M.; Cclerget; Bauer, M.; Kaneshiro, I.; Trudgian, D.; Godlove, D. Hpcng/Singularity: Singularity, 3.7.3; Zenodo: Geneva, Switzerland, 2021. [Google Scholar] [CrossRef]
- Kurtzer, G.M.; Sochat, V.; Bauer, M.W. Singularity: Scientific containers for mobility of compute. PLoS ONE 2017, 12, e0177459. [Google Scholar] [CrossRef] [PubMed]
- Merkel, D. Docker: Lightweight Linux containers for consistent development and deployment. Linux J. 2014, 2014, 2. [Google Scholar]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Published by MDPI on behalf of the International Society for Neonatal Screening. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hovhannesyan, K.; Helou, L.; Charloteaux, B.; Jacquemin, V.; Piazzon, F.; Mni, M.; Flohimont, C.; Fasquelle, C.; Mashhadizadeh, D.; Dangouloff, T.; et al. Analytical Validation of a Genomic Newborn Screening Workflow. Int. J. Neonatal Screen. 2025, 11, 91. https://doi.org/10.3390/ijns11040091
Hovhannesyan K, Helou L, Charloteaux B, Jacquemin V, Piazzon F, Mni M, Flohimont C, Fasquelle C, Mashhadizadeh D, Dangouloff T, et al. Analytical Validation of a Genomic Newborn Screening Workflow. International Journal of Neonatal Screening. 2025; 11(4):91. https://doi.org/10.3390/ijns11040091
Chicago/Turabian StyleHovhannesyan, Kristine, Laura Helou, Benoit Charloteaux, Valerie Jacquemin, Flavia Piazzon, Myriam Mni, Charlotte Flohimont, Corinne Fasquelle, Davood Mashhadizadeh, Tamara Dangouloff, and et al. 2025. "Analytical Validation of a Genomic Newborn Screening Workflow" International Journal of Neonatal Screening 11, no. 4: 91. https://doi.org/10.3390/ijns11040091
APA StyleHovhannesyan, K., Helou, L., Charloteaux, B., Jacquemin, V., Piazzon, F., Mni, M., Flohimont, C., Fasquelle, C., Mashhadizadeh, D., Dangouloff, T., Bours, V., Servais, L., Palmeira, L., & Boemer, F. (2025). Analytical Validation of a Genomic Newborn Screening Workflow. International Journal of Neonatal Screening, 11(4), 91. https://doi.org/10.3390/ijns11040091







