Deep Learning Automation of Kidney, Liver, and Spleen Segmentation for Organ Volume Measurements in Autosomal Dominant Polycystic Kidney Disease
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. MR Imaging
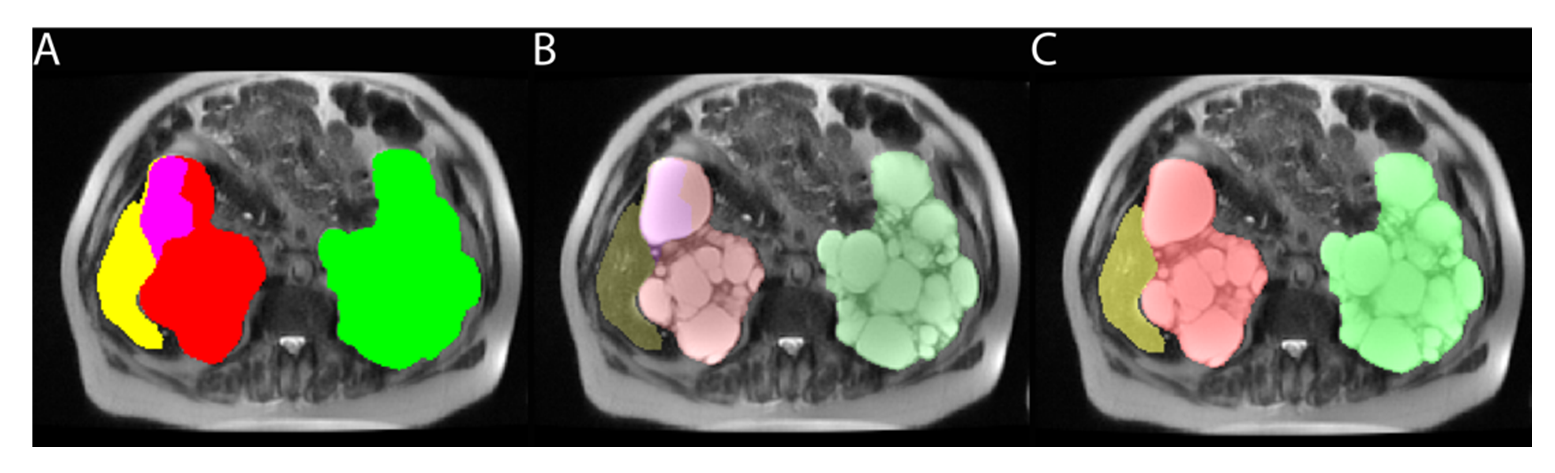
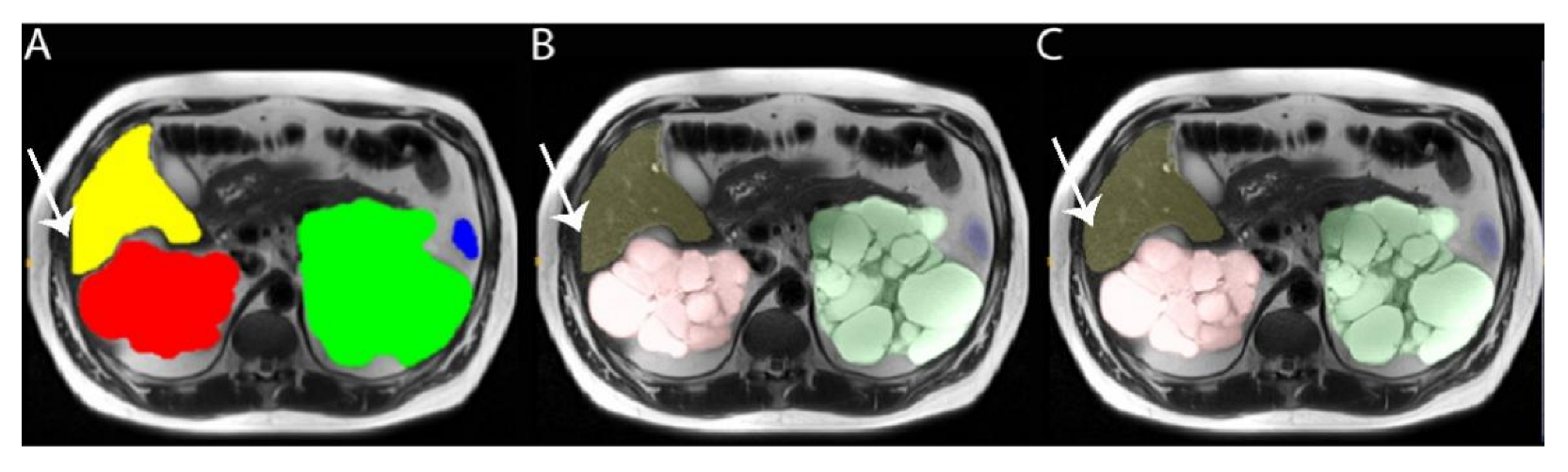
2.3. Labeling Training Data
2.4. Data Preparation
2.5. Stratification
2.6. Deep Learning Model Training
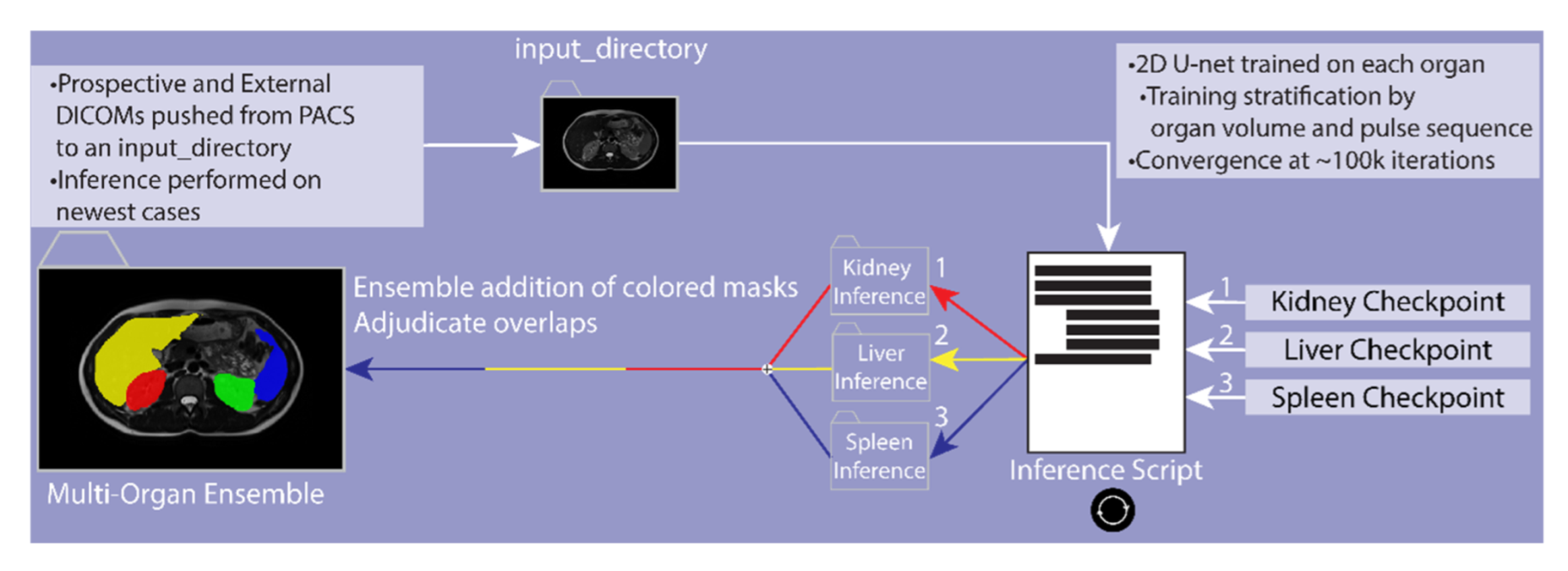
2.7. Resolving Conflicts between Different Organs
2.8. Image Cropping
2.9. External and Prospective Validation
2.10. Time Savings and Reproducibility with Model Assisted Contouring
2.11. Statistical Analysis
3. Results
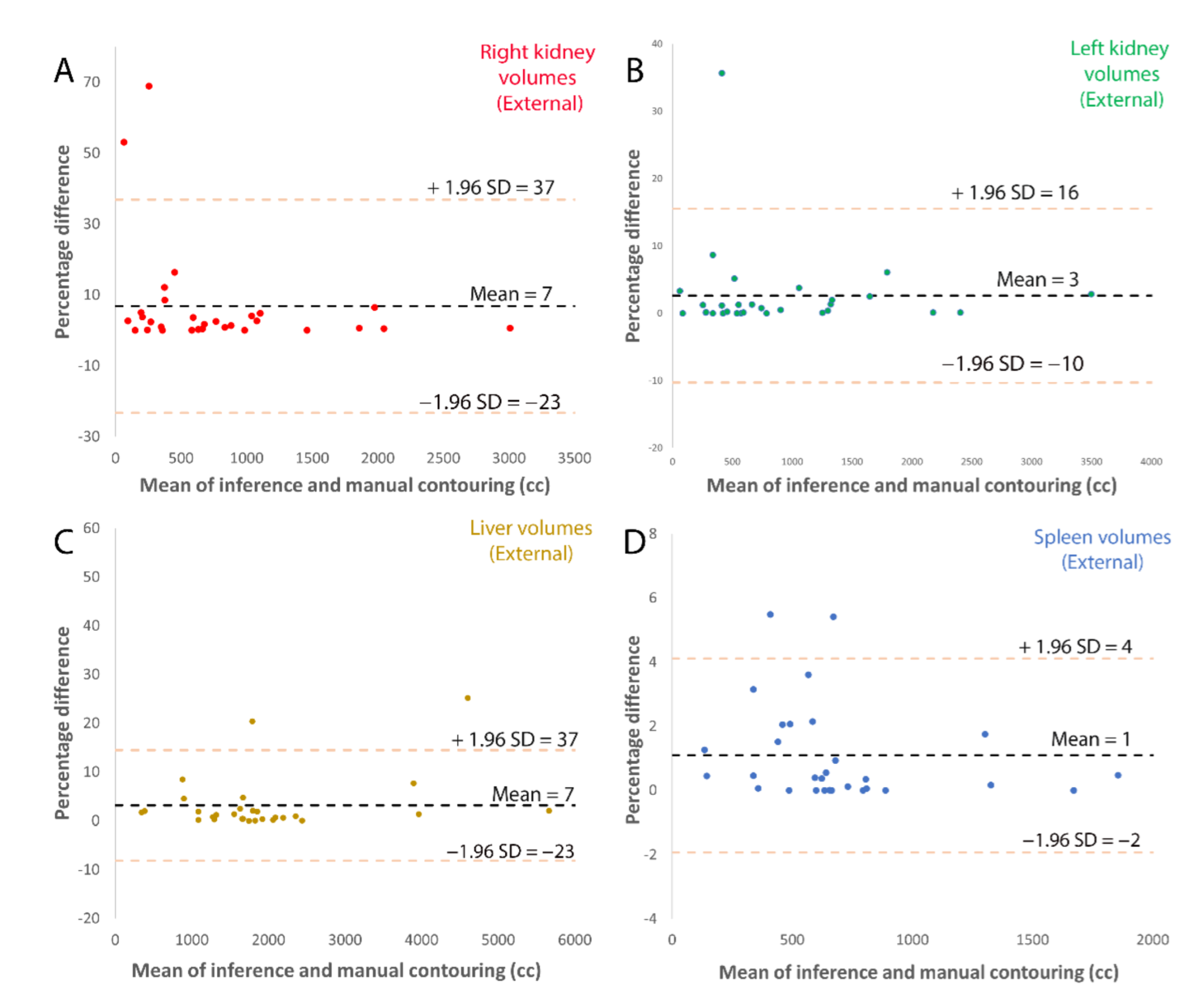
3.1. Model Accuracy
3.2. Time Savings with Model-Assisted Annotation
3.3. Interobserver Variability Improvement with Model Assisted Annotations
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Table of Abbreviations
| ADPKD | Autosomal Dominant Polycystic Kidney Disease |
| CKD | Chronic Kidney Disease |
| CKD-EPI | Chronic Kidney Disease Epidemiology Collaboration |
| CNN | Convolutional Neural Network |
| CT | Computerized Tomography |
| DICOM | Digital Imaging and Communications in Medicine |
| DSC | Dice Similarity Coefficient |
| eGFR | Estimated Glomerular Filtration Rate (based on CKD-EPI method) |
| ESKD | End Stage Kidney Disease |
| HASTE | Half Fourier Single-shot Turbo Spin-Echo |
| MRI | Magnetic Resonance Imaging |
| NIfTI | Neuroimaging Informatics Technology Initiative |
| PACS | Picture Archiving and Communication System |
| SSFP | Steady State Free Precession |
| SSFSE | Single-Shot Fast Spin Echo |
| TE | Time to Echo |
| TKV | Total Kidney Volume |
| ht-TKV | Height Adjusted Total Kidney Volume |
| TR | Time to Repeat |
References
- Chapman, A.B.; Devuyst, O.; Eckardt, K.U.; Gansevoort, R.T.; Harris, T.; Horie, S.; Kasiske, B.L.; Odland, D.; Pei, Y.; Perrone, R.D.; et al. Autosomal-dominant polycystic kidney disease (ADPKD): Executive summary from a Kidney Disease: Improving Global Outcomes (KDIGO) Controversies Conference. Kidney Int. 2015, 88, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.A.; Blumenfeld, J.D.; Chhabra, S.; Dutruel, S.P.; Thimmappa, N.D.; Bobb, W.O.; Donahue, S.; Rennert, H.E.; Tan, A.Y.; Giambrone, A.E.; et al. Pancreatic Cysts in Autosomal Dominant Polycystic Kidney Disease: Prevalence and Association with PKD2 Gene Mutations. Radiology 2016, 280, 762–770. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Stephens, C.J.; Blumenfeld, J.D.; Behzadi, A.H.; Donahue, S.; Bobb, W.O.; Newhouse, J.H.; Rennert, H.; Zhao, Y.; Prince, M.R. Relationship of Seminal Megavesicles, Prostate Median Cysts, and Genotype in Autosomal Dominant Polycystic Kidney Disease. J. Magn. Reson Imaging 2019, 49, 894–903. [Google Scholar] [CrossRef] [PubMed]
- Shigemori, K.; Higashihara, E.; Itoh, M.; Yoshida, H.; Yamamoto, K.; Nutahara, K.; Shiokawa, Y.; Kaname, S.; Tambo, M.; Yamaguchi, T.; et al. PKD1-Associated Arachnoid Cysts in Autosomal Dominant Polycystic Kidney Disease. J. Stroke Cereb. Dis. 2021, 30, 105943. [Google Scholar] [CrossRef]
- Reig, B.; Blumenfeld, J.; Donahue, S.; Prince, M.R. Seminal megavesicle in autosomal dominant polycystic kidney disease. Clin. Imaging 2015, 39, 289–292. [Google Scholar] [CrossRef]
- Yin, X.; Prince, W.K.; Blumenfeld, J.D.; Zhang, W.; Donahue, S.; Bobb, W.O.; Rennert, H.; Askin, G.; Barash, I.; Prince, M.R. Spleen phenotype in autosomal dominant polycystic kidney disease. Clin. Radiol. 2019, 74, 975.e17–975.e24. [Google Scholar] [CrossRef]
- Liu, J.; Fujikura, K.; Dev, H.; Riyahi, S.; Blumenfeld, J.; Kim, J.; Rennert, H.; Prince, M.R. Pericardial Effusion on MRI in Autosomal Dominant Polycystic Kidney Disease. J. Clin. Med. 2022, 11, 1127. [Google Scholar] [CrossRef]
- Zhang, W.; Blumenfeld, J.D.; Prince, M.R. MRI in autosomal dominant polycystic kidney disease. J. Magn. Reson Imaging 2019, 50, 41–51. [Google Scholar] [CrossRef]
- Schnelldorfer, T.; Torres, V.E.; Zakaria, S.; Rosen, C.B.; Nagorney, D.M. Polycystic liver disease: A critical appraisal of hepatic resection, cyst fenestration, and liver transplantation. Ann. Surg. 2009, 250, 112–118. [Google Scholar] [CrossRef]
- Irazabal, M.V.; Rangel, L.J.; Bergstralh, E.J.; Osborn, S.L.; Harmon, A.J.; Sundsbak, J.L.; Bae, K.T.; Chapman, A.B.; Grantham, J.J.; Mrug, M.; et al. Imaging classification of autosomal dominant polycystic kidney disease: A simple model for selecting patients for clinical trials. J. Am. Soc. Nephrol. 2015, 26, 160–172. [Google Scholar] [CrossRef]
- Cadnapaphornchai, M.A.; Masoumi, A.; Strain, J.D.; McFann, K.; Schrier, R.W. Magnetic resonance imaging of kidney and cyst volume in children with ADPKD. Clin. J. Am. Soc. Nephrol. 2011, 6, 369–376. [Google Scholar] [CrossRef] [PubMed]
- Fick-Brosnahan, G.M.; Belz, M.M.; McFann, K.K.; Johnson, A.M.; Schrier, R.W. Relationship between renal volume growth and renal function in autosomal dominant polycystic kidney disease: A longitudinal study. Am. J. Kidney Dis. 2002, 39, 1127–1134. [Google Scholar] [CrossRef] [PubMed]
- Chapman, A.B.; Wei, W. Imaging approaches to patients with polycystic kidney disease. Semin. Nephrol. 2011, 31, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Yu, A.S.L.; Shen, C.; Landsittel, D.P.; Grantham, J.J.; Cook, L.T.; Torres, V.E.; Chapman, A.B.; Bae, K.T.; Mrug, M.; Harris, P.C.; et al. Long-term trajectory of kidney function in autosomal-dominant polycystic kidney disease. Kidney Int. 2019, 95, 1253–1261. [Google Scholar] [CrossRef]
- Liebau, M.C.; Serra, A.L. Looking at the (w)hole: Magnet resonance imaging in polycystic kidney disease. Pediatr. Nephrol. 2013, 28, 1771–1783. [Google Scholar] [CrossRef]
- Grantham, J.J.; Torres, V.E. The importance of total kidney volume in evaluating progression of polycystic kidney disease. Nat. Rev. Nephrol. 2016, 12, 667–677. [Google Scholar] [CrossRef]
- Bae, K.T.; Commean, P.K.; Lee, J. Volumetric measurement of renal cysts and parenchyma using MRI: Phantoms and patients with polycystic kidney disease. J. Comput. Assist. Tomogr. 2000, 24, 614–619. [Google Scholar] [CrossRef]
- Higashihara, E.; Nutahara, K.; Okegawa, T.; Tanbo, M.; Hara, H.; Miyazaki, I.; Kobayasi, K.; Nitatori, T. Kidney Volume Estimations with Ellipsoid Equations by Magnetic Resonance Imaging in Autosomal Dominant Polycystic Kidney Disease. Nephron 2015, 129, 253–262. [Google Scholar] [CrossRef]
- Kistler, A.D.; Poster, D.; Krauer, F.; Weishaupt, D.; Raina, S.; Senn, O.; Binet, I.; Spanaus, K.; Wüthrich, R.P.; Serra, A.L. Increases in kidney volume in autosomal dominant polycystic kidney disease can be detected within 6 months. Kidney Int. 2009, 75, 235–241. [Google Scholar] [CrossRef][Green Version]
- Chapman, A.B.; Guay-Woodford, L.M.; Grantham, J.J.; Torres, V.E.; Bae, K.T.; Baumgarten, D.A.; Kenney, P.J.; King, B.F., Jr.; Glockner, J.F.; Wetzel, L.H.; et al. Renal structure in early autosomal-dominant polycystic kidney disease (ADPKD): The Consortium for Radiologic Imaging Studies of Polycystic Kidney Disease (CRISP) cohort. Kidney Int. 2003, 64, 1035–1045. [Google Scholar] [CrossRef]
- O’Neill, W.C.; Robbin, M.L.; Bae, K.T.; Grantham, J.J.; Chapman, A.B.; Guay-Woodford, L.M.; Torres, V.E.; King, B.F.; Wetzel, L.H.; Thompson, P.A.; et al. Sonographic assessment of the severity and progression of autosomal dominant polycystic kidney disease: The Consortium of Renal Imaging Studies in Polycystic Kidney Disease (CRISP). Am. J. Kidney Dis. 2005, 46, 1058–1064. [Google Scholar] [CrossRef] [PubMed]
- Goel, A.; Shih, G.; Riyahi, S.; Jeph, S.; Dev, H.; Hu, R.; Romano, D.; Teichman, K.; Blumenfeld, J.D.; Barash, I.; et al. Deployed Deep Learning Kidney Segmentation for Polycystic Kidney Disease MRI. Radiol. Artif. Intell. 2022, 4, e210205. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, C.-H.; Lin, P.-C.; Chung, L.-A.; Lin, F.Y.-S.; Yang, F.-J.; Yang, S.-Y.; Wu, C.-H.; Huang, Y.; Sun, T.-L. A deep learning-based precision and automatic kidney segmentation system using efficient feature pyramid networks in computed tomography images. Comput. Methods Programs Biomed. 2022, 221, 106854. [Google Scholar] [CrossRef]
- Keshwani, D.; Kitamura, Y.; Li, Y. Computation of Total Kidney Volume from CT Images in Autosomal Dominant Polycystic Kidney Disease Using Multi-task 3D Convolutional Neural Networks. In International Workshop on Machine Learning in Medical Imaging; Spring: Cham, Switzerland, 2018; pp. 380–388. [Google Scholar]
- Onthoni, D.D.; Sheng, T.-W.; Sahoo, P.K.; Wang, L.-J.; Gupta, P. Deep Learning Assisted Localization of Polycystic Kidney on Contrast-Enhanced CT Images. Diagnostics 2020, 10, 1113. [Google Scholar] [CrossRef]
- Sharma, K.; Rupprecht, C.; Caroli, A.; Aparicio, M.C.; Remuzzi, A.; Baust, M.; Navab, N. Automatic Segmentation of Kidneys using Deep Learning for Total Kidney Volume Quantification in Autosomal Dominant Polycystic Kidney Disease. Sci. Rep. 2017, 7, 2049. [Google Scholar] [CrossRef]
- Shin, T.Y.; Kim, H.; Lee, J.H.; Choi, J.S.; Min, H.S.; Cho, H.; Kim, K.; Kang, G.; Kim, J.; Yoon, S.; et al. Expert-level segmentation using deep learning for volumetry of polycystic kidney and liver. Investig. Clin. Urol. 2020, 61, 555–564. [Google Scholar] [CrossRef] [PubMed]
- Jagtap, J.M.; Gregory, A.V.; Homes, H.L.; Wright, D.E.; Edwards, M.E.; Akkus, Z.; Erickson, B.J.; Kline, T.L. Automated measurement of total kidney volume from 3D ultrasound images of patients affected by polycystic kidney disease and comparison to MR measurements. Abdom. Radiol. 2022, 47, 2408–2419. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Ge, Y.; Tao, C.; Zhu, J.; Chapman, A.B.; Torres, V.E.; Yu, A.S.L.; Mrug, M.; Bennett, W.M.; Flessner, M.F.; et al. Automated Segmentation of Kidneys from MR Images in Patients with Autosomal Dominant Polycystic Kidney Disease. Clin. J. Am. Soc. Nephrol. 2016, 11, 576–584. [Google Scholar] [CrossRef]
- Kline, T.L.; Korfiatis, P.; Edwards, M.E.; Blais, J.D.; Czerwiec, F.S.; Harris, P.C.; King, B.F.; Torres, V.E.; Erickson, B.J. Performance of an Artificial Multi-observer Deep Neural Network for Fully Automated Segmentation of Polycystic Kidneys. J. Digit. Imaging 2017, 30, 442–448. [Google Scholar] [CrossRef]
- Mu, G.; Ma, Y.; Han, M.; Zhan, Y.; Zhou, X.; Gao, Y. Automatic MR Kidney Segmentation for Autosomal Dominant Polycystic Kidney Disease; SPIE: San Diego, CA, USA, 2019; Volume 10950. [Google Scholar]
- van Gastel, M.D.A.; Edwards, M.E.; Torres, V.E.; Erickson, B.J.; Gansevoort, R.T.; Kline, T.L. Automatic Measurement of Kidney and Liver Volumes from MR Images of Patients Affected by Autosomal Dominant Polycystic Kidney Disease. J. Am. Soc. Nephrol. 2019, 30, 1514–1522. [Google Scholar] [CrossRef]
- Kline, T.L.; Edwards, M.E.; Fetzer, J.; Gregory, A.V.; Anaam, D.; Metzger, A.J.; Erickson, B.J. Automatic semantic segmentation of kidney cysts in MR images of patients affected by autosomal-dominant polycystic kidney disease. Abdom. Imaging 2020, 46, 1053–1061. [Google Scholar] [CrossRef] [PubMed]
- Raj, A.; Tollens, F.; Hansen, L.; Golla, A.-K.; Schad, L.R.; Nörenberg, D.; Zöllner, F.G. Deep Learning-Based Total Kidney Volume Segmentation in Autosomal Dominant Polycystic Kidney Disease Using Attention, Cosine Loss, and Sharpness Aware Minimization. Diagnostics 2022, 12, 1159. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.; Thomas, R.; Metherall, P.; Ong, A.; Simms, R. MO012: Development of an Accurate Automated Segmentation Algorithm to Measure Total Kidney Volume in ADPKD Suitable for Clinical Application (The Cystvas Study). Nephrol. Dial. Transplant. 2022, 37, gfac061-007. [Google Scholar] [CrossRef]
- Levey, A.S.; Stevens, L.A.; Schmid, C.H.; Zhang, Y.L.; Castro, A.F., 3rd; Feldman, H.I.; Kusek, J.W.; Eggers, P.; Van Lente, F.; Greene, T.; et al. A new equation to estimate glomerular filtration rate. Ann. Intern. Med. 2009, 150, 604–612. [Google Scholar] [CrossRef] [PubMed]
- Brett, M.; Markiewicz, C.; Hanke, M.; Côté, M.-A.; Cipollini, B.; McCarthy, P.; Cheng, C. Neuroimaging in Python—NiBabel 3.2.0 Documentation. Available online: https://nipy.org/nibabel/ (accessed on 6 September 2021).
- Tan, M.; Le, Q. Albumentations-Team/Albumentations EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. In Proceedings of the International Conference on Machine Learning, Long Beach, CA, USA, 9–15 June 2019; pp. 6105–6114. [Google Scholar]
- Neatrour, A.; Brunsvik, M.; Buckner, S.; McBride, B.; Myntti, J. The SIMP Tool: Facilitating Digital Library, Metadata, and Preservation Workflow at the University of Utah’s J. Willard Marriott Library. D-Lib Mag. 2014, 20, 7–8. [Google Scholar] [CrossRef]
- Zou, K.H.; Warfield, S.K.; Bharatha, A.; Tempany, C.M.; Kaus, M.R.; Haker, S.J.; Wells, W.M., 3rd; Jolesz, F.A.; Kikinis, R. Statistical validation of image segmentation quality based on a spatial overlap index. Acad. Radiol. 2004, 11, 178–189. [Google Scholar] [CrossRef]
- Lawrence, I.K.L. A Concordance Correlation Coefficient to Evaluate Reproducibility. Biometrics 1989, 45, 255–268. [Google Scholar] [CrossRef]
- Giavarina, D. Understanding Bland Altman analysis. Biochem. Med. 2015, 25, 141–151. [Google Scholar] [CrossRef]
- Grantham, J.J.; Torres, V.E.; Chapman, A.B.; Guay-Woodford, L.M.; Bae, K.T.; King, B.F., Jr.; Wetzel, L.H.; Baumgarten, D.A.; Kenney, P.J.; Harris, P.C.; et al. Volume progression in polycystic kidney disease. N. Engl. J. Med. 2006, 354, 2122–2130. [Google Scholar] [CrossRef]
- Zhang, Z.-Y.; Wang, Z.-M.; Huang, Y. Polycystic liver disease: Classification, diagnosis, treatment process, and clinical management. World J. Hepatol. 2020, 12, 72–83. [Google Scholar] [CrossRef]
- Riyahi, S.; Dev, H.; Blumenfeld, J.D.; Rennert, H.; Yin, X.; Attari, H.; Barash, I.; Chicos, I.; Bobb, W.; Donahue, S.; et al. Hemorrhagic Cysts and Other MR Biomarkers for Predicting Renal Dysfunction Progression in Autosomal Dominant Polycystic Kidney Disease. J. Magn. Reson. Imaging 2021, 53, 564–576. [Google Scholar] [CrossRef] [PubMed]
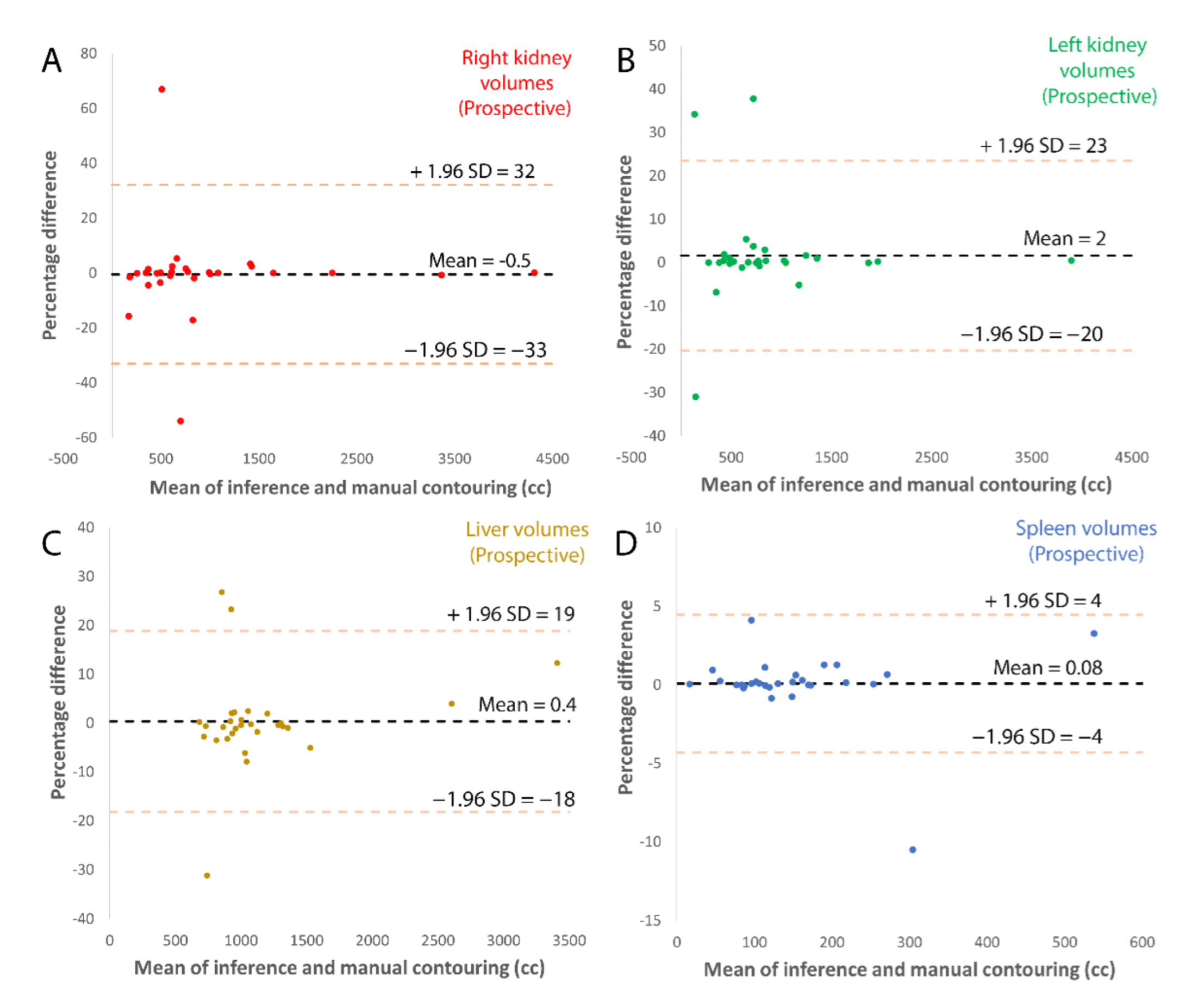
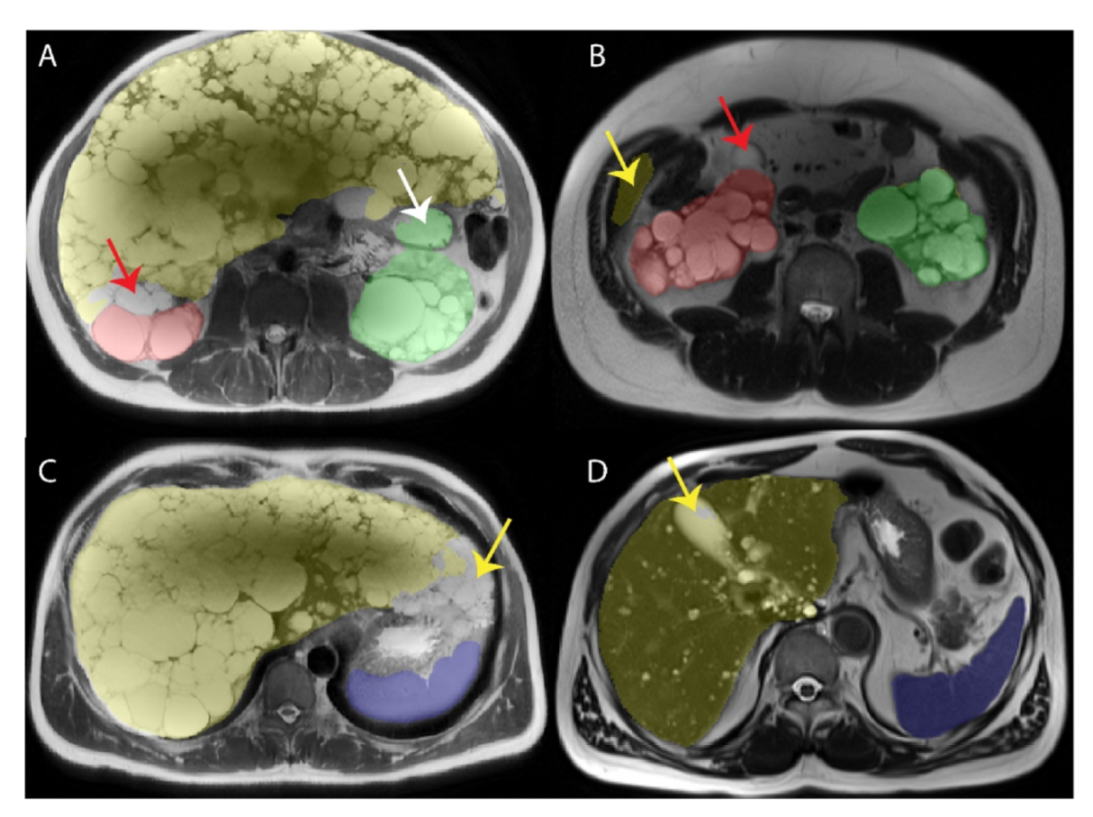
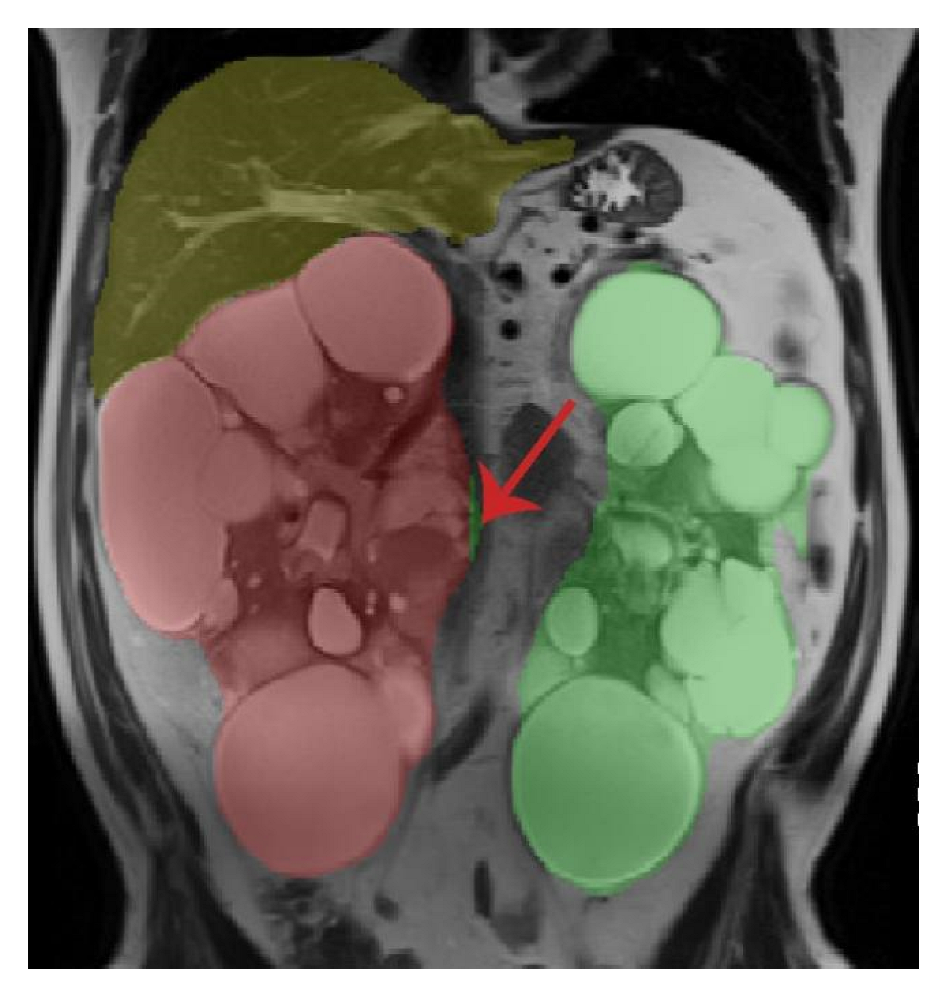
| First Author | Modality | Year | ADPKD subjects | Segmentation Methodology | Dice Score | Other Metrics | Organ |
|---|---|---|---|---|---|---|---|
| Sharma [26] | CT | 2017 | 125 | 2D VGG-16 FCN | 0.86 | 7.8% | Kidney |
| Keshwani [24] | CT | 2018 | 203 CT scans ** | Multi-task 3D FCN | 0.95 | 3.86% | Kidney |
| Shin [27] | CT | 2020 | 214 | 3D V-net | 0.961 * | 95% within 3% | Kidney + Liver |
| Onthoni [25] | CT | 2020 | 97 | 2D SSD Inception Network V2 | - | Images: mAP: 94% Subjects: mAP: 82% | Kidney |
| Hsiao [23] | CT | 2022 | 210 | FPN + EfficientNet | 0.969 | - | Kidney |
| Jagtap [28] | US | 2022 | 22 | 2D U-Net | 0.80 | 4.12% | Kidney |
| Kim [29] | MRI Cor T2 fatsat | 2016 | 60 | SPPM + PSC | 0.88 | MCC: 0.97 | Kidney |
| Kline [30] | MRI Cor T2 | 2017 | 2000 scans ** | 2D U-Net + ResNet-like encoder | 0.97 | 0.68% | Kidney |
| Guangrui [31] | MRI Axial + Cor T1 | 2019 | 305 | 3D VB-Net *** | RK-0.958 LK-0.965 | - | Kidney |
| Van Gastel [32] | MRI Cor T2 fatsat | 2019 | 145 | 2D U-Net | - | LK: 0.96 RK: 0.95 TKV: 0.96 Liver: 0.95 | Kidney + Liver |
| Kline [33] | MRI Cor T2 +/−fatsat | 2020 | 60 | 2D U-Net + ResNet-like encoder | 1st Reader: 0.86 2nd Reader: 0.84 | 1st Reader: 3.9% 2nd Reader: 8% | Kidney cysts |
| Goel [22] | MRI Axial T2 | 2022 | 173 | 2D U-Net + EfficientNet encoder | External: 0.98 Prospective: 0.97 | External: 2.6% Prospective: 3.6% | Kidney |
| Raj [34] | MRI Cor T1 | 2022 | 100 | 2D Attention U-Net | 0.922 | MSSD: 0.922 and 1.09 mm | Kidney |
| Taylor [35] | MRI | 2022 | 227 Scans | 3D U-Net | 0.96 each kidney | LK:1.8% RK:1.79% | Kidney |
| Parameter | Training/Validation Data | External Test Set | Prospective Test Set |
|---|---|---|---|
| Number of Patients | 215 | 30 | 30 |
| Number of MR exams | 260 | 30 | 30 |
| DICOM images | 9540 | 1368 | 2137 |
| Male:Female (%male) | 98:117 (46%) | 17:13 (57%) | 11:19 (37%) |
| Age at scan (years) | 49 ± 14 | 49 ± 16 | 46 ± 15 |
| eGFR (mL/min/1.73 m2) | 68 ± 28 | 85 ± 30 | 72 ± 34 |
| Total Kidney Volume (mL) * | 1287 (669–2213) | 1334 (693–2376) | 1444 (885–2020) |
| Ht-TKV (mL/m) * | 757 (415–1275) | 777 (393–1297) | 837 (550–1234) |
| Mayo class ** -Report N and % | |||
| A | 29 (13%) | 4 (13%) | 1 (3%) |
| B | 58 (27%) | 8 (27%) | 6 (20%) |
| C | 70 (33%) | 7 (24%) | 13 (44%) |
| D | 34 (16%) | 10 (33%) | 7 (23%) |
| E | 24 (11%) | 1 (3%) | 3 (10%) |
| Race-Report N and % | |||
| Asian | 10 (5%) | 1 (3%) | 4 (13%) |
| White | 148 (69%) | 23 (77%) | 16 (53%) |
| Black | 14 (6%) | 1 (3%) | 2 (67%) |
| Unknown | 43 (20%) | 5 (17%) | 8 (27%) |
| (A) | ||||
|---|---|---|---|---|
| External Test Set | Right Kidney | Left Kidney | Liver | Spleen |
| Model volume (mL) Corrected model volume (mL) | 617 (327–1009) 608 (316–1041) | 582 (416–1289) 582 (365–1285) | 1706 (1292–2087) 1684 (1287–2076) | 220 (145–274) 222 (157–280) |
| DSC | 0.96 | 0.98 | 0.97 | 0.96 |
| Concordance Coefficient | >0.99 | >0.99 | 0.98 | 0.99 |
| RMS error (mL) | 42 | 39 | 258 | 17 |
| Average % error | 7% | 3% | 3% | 1% |
| Number with zero error | 5 (17%) | 6 (20%) | 1 (3%) | 7 (23%) |
| (B) | ||||
| Prospective Test Set | Right Kidney | Left Kidney | Liver | Spleen |
| Model volume (mL) Corrected model volume (mL) | 625 (370–1000) 650 (394–998) | 729 (481–1039) 768 (485–1043) | 1711 (1489–2065) 1727 (1495–2051) | 244 (177–315) 241 (175–318) |
| DSC | 0.96 | 0.96 | 0.96 | 0.95 |
| Concordance Coefficient | >0.99 | >0.99 | >0.99 | 0.98 |
| RMS error (mL) | 112 | 65 | 112 | 37 |
| Average % error | 6% | 5% | 5% | 1% |
| # with zero error | 2 (7%) | 3 (20%) | 2 (7%) | 2 (7%) |
| Manual Segmentation | Model Assisted Segmentation | Time Savings | p-Value | |
|---|---|---|---|---|
| Right Kidney | 7:39 ± 2:26 | 4:31 ± 1:34 | 3:08 (41%) | 0.004 |
| Left Kidney | 7:34 ± 3:44 | 4:16 ± 1:35 | 3:19 (44%) | 0.01 |
| Liver | 12:49 ± 6:10 | 8:49 ± 3:52 | 3:59 (31%) | 0.007 |
| Spleen | 4:13 ± 0:48 | 2:04 ± 0:59 | 2:09 (51%) | 0.0003 |
| Total | 33:04 ± 8:05 | 19:17 ± 7:19 | 13:47 (42%) | 0.001 |
| Volume Measurement Standard Deviations | |||
|---|---|---|---|
| Manual Segmentation (mL) | Model Assisted Segmentation (mL) | p-Value | |
| Right Kidney Left Kidney | 14 10 | 7 5 | 0.02 0.07 |
| Liver | 55 | 11 | 0.001 |
| Spleen | 14 | 5 | 0.001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharbatdaran, A.; Romano, D.; Teichman, K.; Dev, H.; Raza, S.I.; Goel, A.; Moghadam, M.C.; Blumenfeld, J.D.; Chevalier, J.M.; Shimonov, D.; et al. Deep Learning Automation of Kidney, Liver, and Spleen Segmentation for Organ Volume Measurements in Autosomal Dominant Polycystic Kidney Disease. Tomography 2022, 8, 1804-1819. https://doi.org/10.3390/tomography8040152
Sharbatdaran A, Romano D, Teichman K, Dev H, Raza SI, Goel A, Moghadam MC, Blumenfeld JD, Chevalier JM, Shimonov D, et al. Deep Learning Automation of Kidney, Liver, and Spleen Segmentation for Organ Volume Measurements in Autosomal Dominant Polycystic Kidney Disease. Tomography. 2022; 8(4):1804-1819. https://doi.org/10.3390/tomography8040152
Chicago/Turabian StyleSharbatdaran, Arman, Dominick Romano, Kurt Teichman, Hreedi Dev, Syed I. Raza, Akshay Goel, Mina C. Moghadam, Jon D. Blumenfeld, James M. Chevalier, Daniil Shimonov, and et al. 2022. "Deep Learning Automation of Kidney, Liver, and Spleen Segmentation for Organ Volume Measurements in Autosomal Dominant Polycystic Kidney Disease" Tomography 8, no. 4: 1804-1819. https://doi.org/10.3390/tomography8040152
APA StyleSharbatdaran, A., Romano, D., Teichman, K., Dev, H., Raza, S. I., Goel, A., Moghadam, M. C., Blumenfeld, J. D., Chevalier, J. M., Shimonov, D., Shih, G., Wang, Y., & Prince, M. R. (2022). Deep Learning Automation of Kidney, Liver, and Spleen Segmentation for Organ Volume Measurements in Autosomal Dominant Polycystic Kidney Disease. Tomography, 8(4), 1804-1819. https://doi.org/10.3390/tomography8040152







