Mortality Prediction Analysis among COVID-19 Inpatients Using Clinical Variables and Deep Learning Chest Radiography Imaging Features
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Source
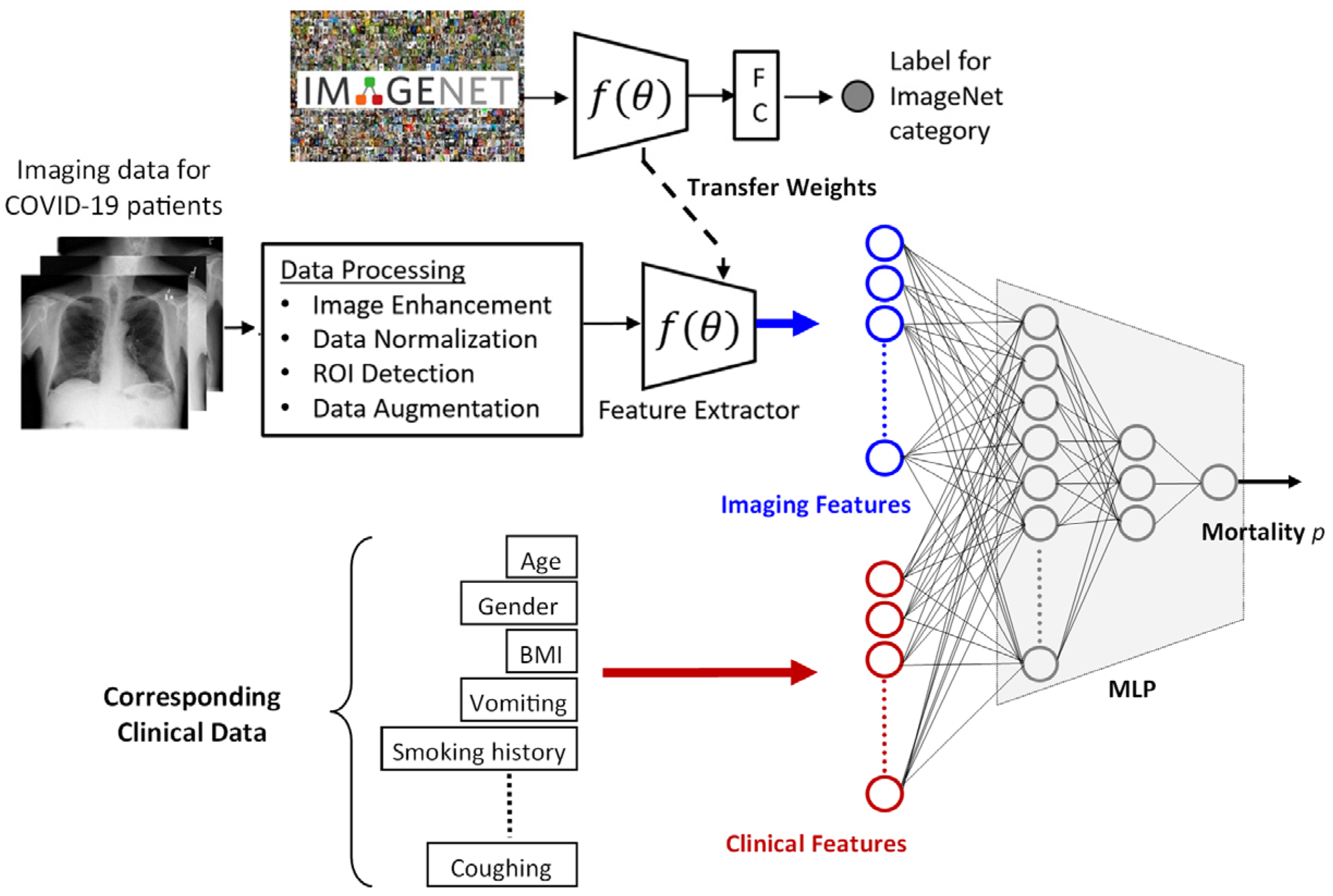
2.2. Prediction Framework
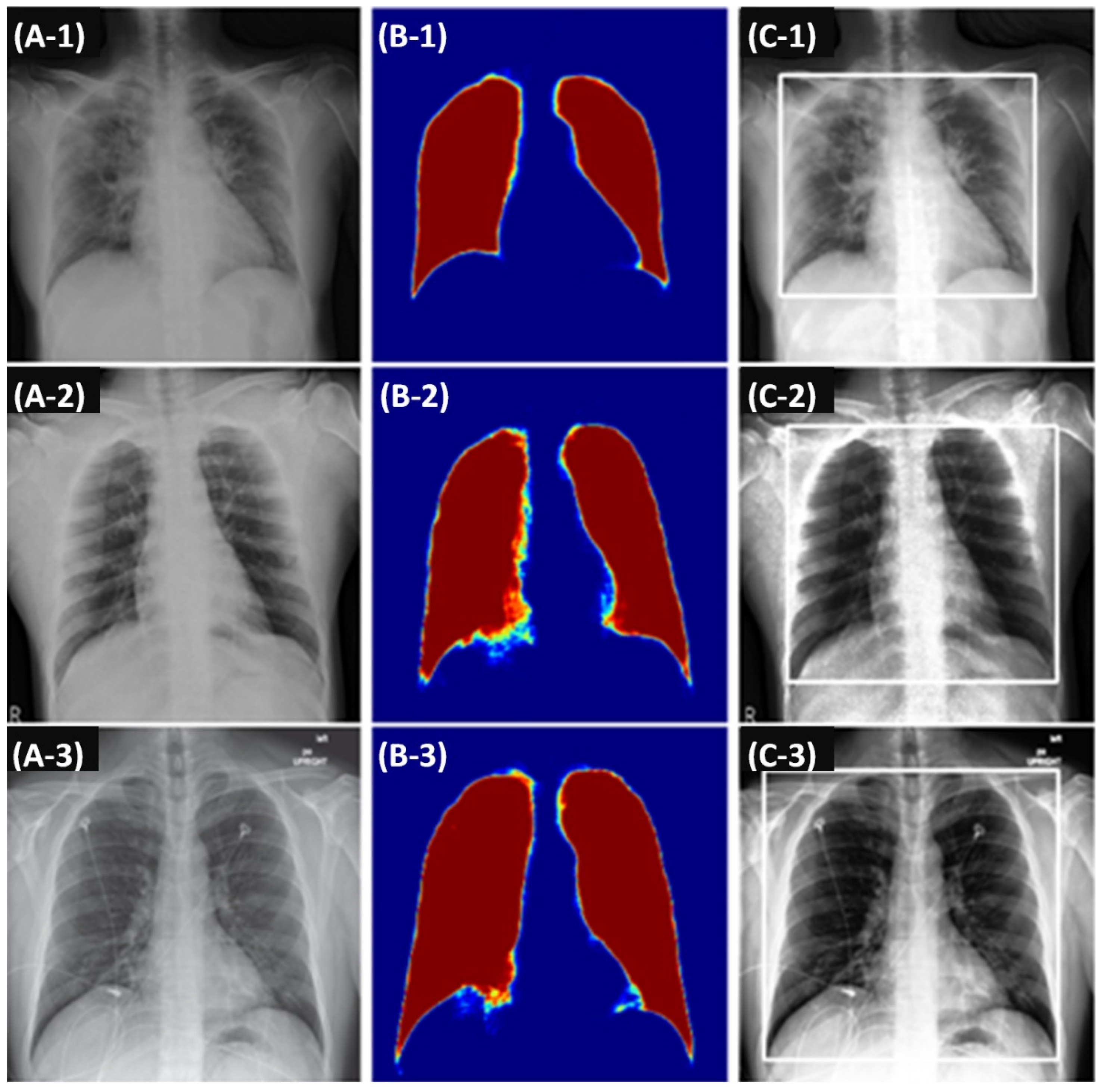
2.2.1. Imaging Features
2.2.2. Clinical Features
2.2.3. Processing Imaging and Clinical Features
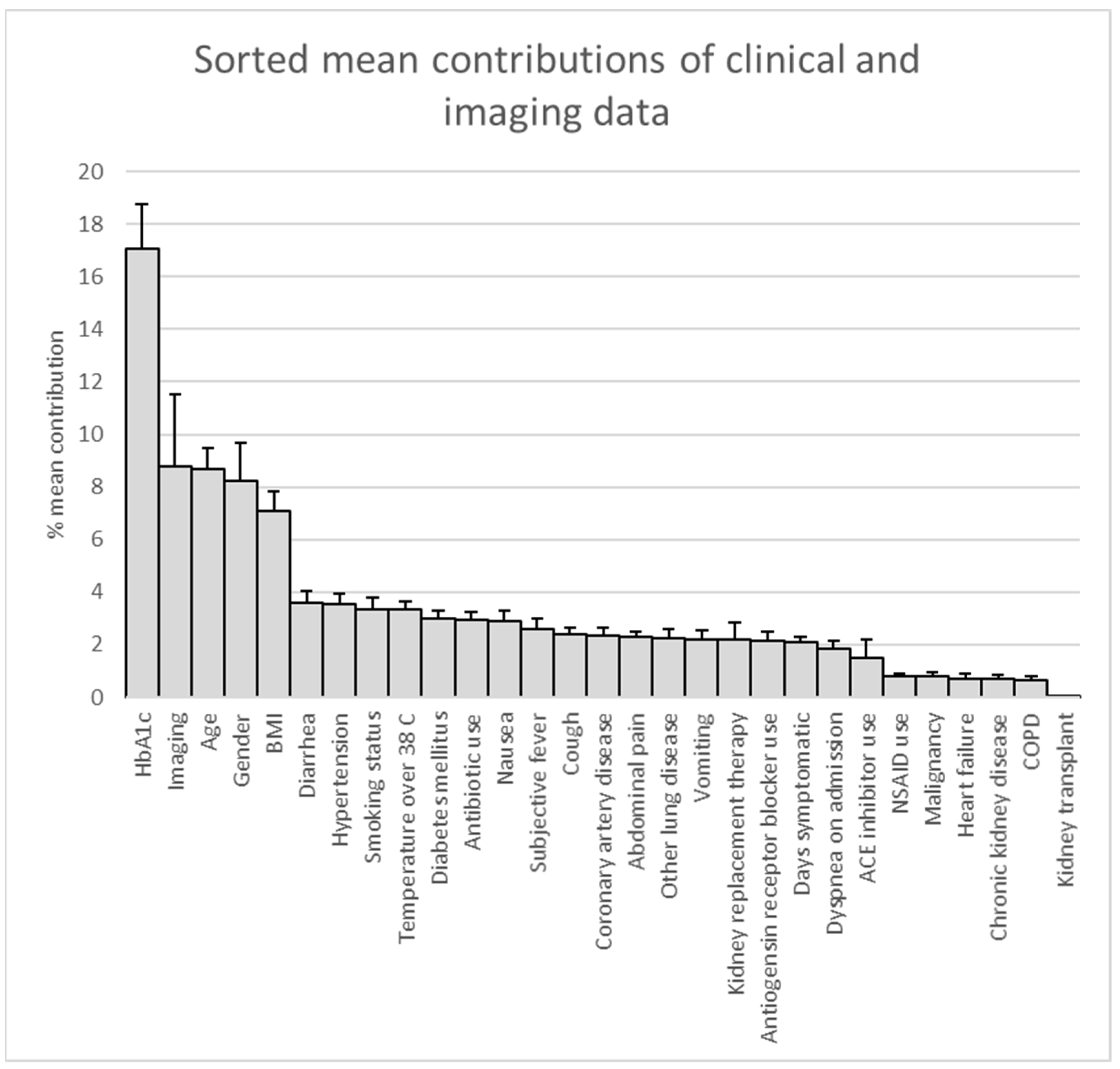
2.3. Deriving Prediction Explanations
2.4. Statistical Analysis
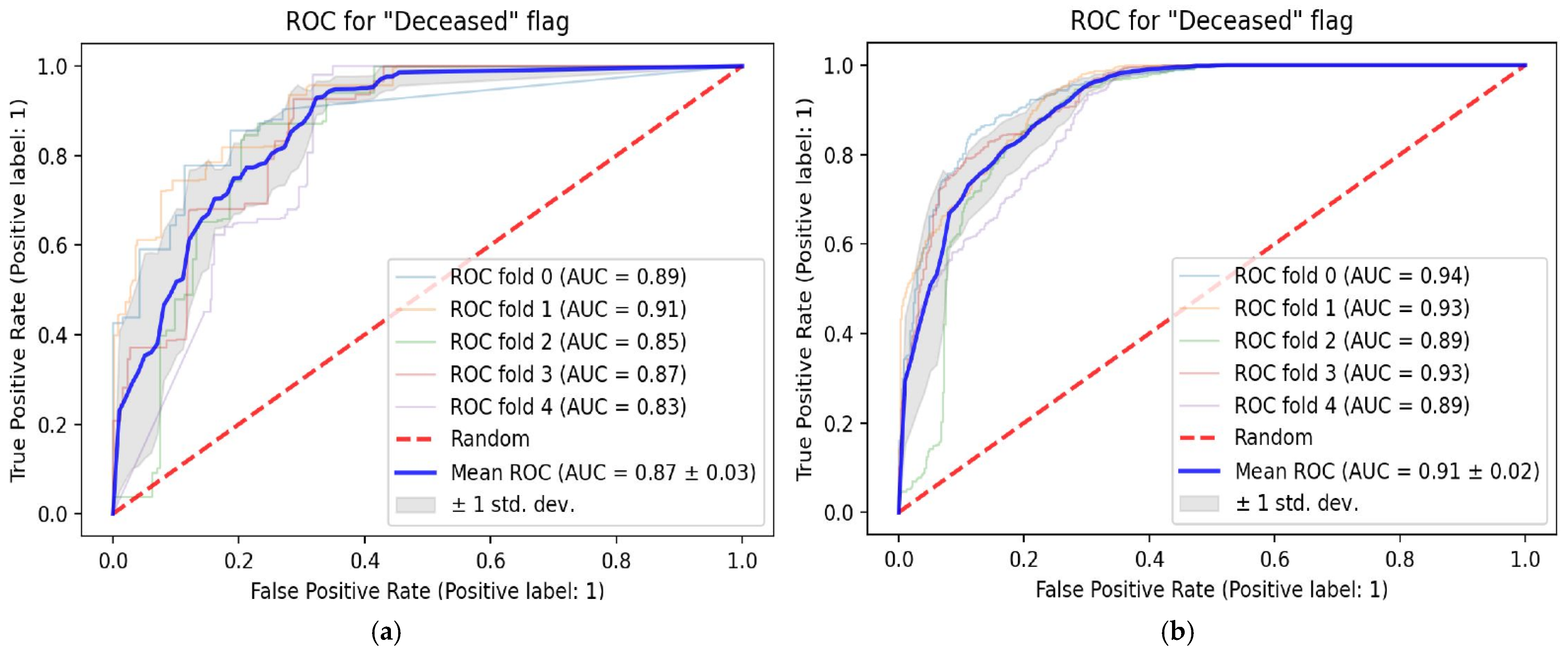
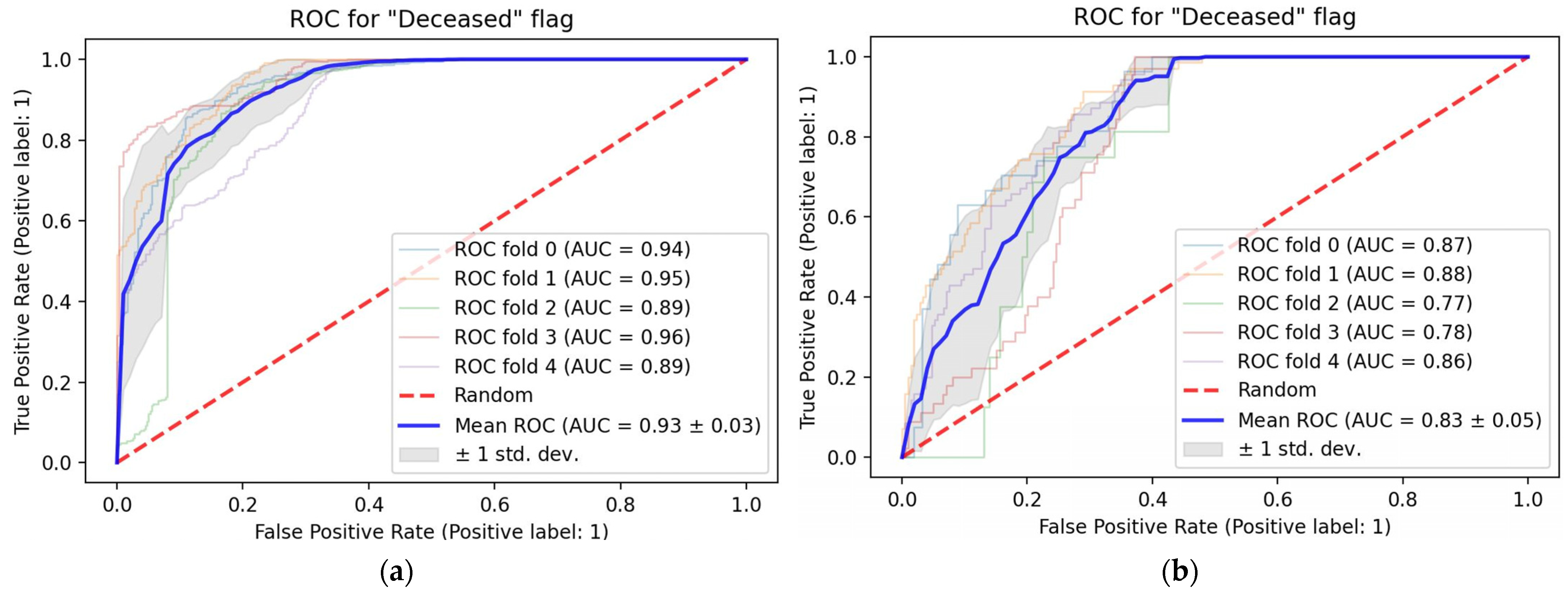
3. Results
Patient Population
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y.; Tao, Z.-W.; Tian, J.-H.; Pei, Y.-Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zu, Z.Y.; Di Jiang, M.; Xu, P.P.; Chen, W.; Ni, Q.Q.; Lu, G.M.; Zhang, L.J. Coronavirus Disease 2019 (COVID-19): A Perspective from China. Radiology 2020, 296, E15–E25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU). COVID-19 Dashboard. 2022. Available online: https://systems.jhu.edu/research/public-health/ncov/ (accessed on 19 April 2022).
- Damiani, S.; Fiorentino, M.; De Palma, A.; Foschini, M.P.; Lazzarotto, T.; Gabrielli, L.; Viale, P.L.; Attard, L.; Riefolo, M.; D’Errico, A. Pathological post-mortem findings in lungs infected with SARS-CoV-2. J. Pathol. 2021, 253, 31–40. [Google Scholar] [CrossRef]
- Ai, T.; Yang, Z.; Hou, H.; Zhan, C.; Chen, C.; Lv, W.; Tao, Q.; Sun, Z.; Xia., L. Correlation of Chest CT and RT-PCR Testing for Coronavirus Disease 2019 (COVID-19) in China: A Report of 1014 Cases. Radiology 2020, 296, E32–E40. [Google Scholar] [CrossRef] [Green Version]
- Shi, F.; Wang, J.; Shi, J.; Wu, Z.; Wang, Q.; Tang, Z.; He, K.; Shi, Y.; Shen, D. Review of Artificial Intelligence Techniques in Imaging Data Acquisition, Segmentation, and Diagnosis for COVID-19. IEEE Rev. Biomed. Eng. 2021, 14, 4–15. [Google Scholar] [CrossRef] [Green Version]
- Jain, G.; Mittal, D.; Thakur, D.; Mittal, M.K. A deep learning approach to detect COVID-19 coronavirus with X-Ray images. Biocybern. Biomed. Eng. 2020, 40, 1391–1405. [Google Scholar] [CrossRef]
- Wang, L.; Lin, Z.Q.; Wong, A. COVID-Net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. Sci. Rep. 2020, 10, 19549. [Google Scholar] [CrossRef]
- Mei, X.; Lee, H.-C.; Diao, K.-Y.; Huang, M.; Lin, B.; Liu, C.; Xie, Z.; Ma, Y.; Robson, P.M.; Chung, M.; et al. Artificial intelligence–enabled rapid diagnosis of patients with COVID-19. Nat. Med. 2020, 26, 1224–1228. [Google Scholar] [CrossRef]
- Oh, Y.; Park, S.; Ye, J.C. Deep Learning COVID-19 Features on CXR Using Limited Training Data Sets. IEEE Trans. Med Imaging 2020, 39, 2688–2700. [Google Scholar] [CrossRef]
- Summers, R.M. Artificial Intelligence of COVID-19 Imaging: A Hammer in Search of a Nail. Radiology 2021, 298, E162–E164. [Google Scholar] [CrossRef]
- Weiss, K.; Khoshgoftaar, T.M.; Wang, D. A survey of transfer learning. J. Big Data 2016, 3, 1–40. [Google Scholar] [CrossRef] [Green Version]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Saltz, J.; Saltz, M.; Prasanna, P.; Moffitt, R.; Hajagos, J.; Bremer, E.; Balsamo, J.; Kurc, T. Stony Brook University COVID-19 Positive Cases [Data Set]. 2021. Available online: https://wiki.cancerimagingarchive.net/pages/viewpage.action?pageId=89096912 (accessed on 10 May 2022).
- Clark, K.; Vendt, B.; Smith, K.; Freymann, J.; Kirby, J.; Koppel, P.; Moore, S.; Phillips, S.; Maffitt, D.; Pringle, M.; et al. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository. J. Digit. Imaging 2013, 26, 1045–1057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional networks for biomedical image segmentation. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015; Navab, N., Hornegger, J., Wells, W., Frangi, A., Eds.; MICCAI 2015, Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2015; p. 9351. [Google Scholar] [CrossRef] [Green Version]
- Shiraishi, J.; Katsuragawa, S.; Ikezoe, J.; Matsumoto, T.; Kobayashi, T.; Komatsu, K.I.; Matsui, M.; Fujita, H.; Fujita, Y.; Doi, K.; et al. Development of a digital image database for chest radiographs with and without a lung nodule: Receiver operating characteristic analysis of radiologists’ detection of pulmonary nodules. Am. J. Roentgenol. 2000, 174, 71–74. [Google Scholar] [CrossRef] [PubMed]
- Van Ginneken, B.; Stegmann, M.B.; Loog, M. Segmentation of anatomical structures in chest radiographs using supervised methods: A comparative study on a public database. Med. Image Anal. 2006, 10, 19–40. [Google Scholar] [CrossRef] [Green Version]
- Du, W.; Chen, H.; Wu, Z.; Sun, H.; Liao, P.; Zhang, Y. Stacked competitive networks for noise reduction in low-dose CT. PLoS ONE 2017, 12, e0190069. [Google Scholar] [CrossRef]
- Siddique, N.; Paheding, S.; Elkin, C.P.; Devabhaktuni, V. U-Net and Its Variants for Medical Image Segmentation: A Review of Theory and Applications. IEEE Access 2021, 9, 82031–82057. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. In Advances in Neural Information Processing Systems; Pereira, F., Burges, C.J., Bottou, L., Weinberger, K.Q., Eds.; The MIT Press: Cambridge, MA, USA, 2012; Volume 25. [Google Scholar]
- Glorot, X.; Bengio, Y. Understanding the difficulty of training deep feedforward neural networks. In Proceedings of the Thirteenth International Conference on Artificial Intelligence and Statistics, Sardinia, Italy, 13–15 May 2010; Volume 9, pp. 249–256. [Google Scholar]
- Roscher, R.; Bohn, B.; Duarte, M.F.; Garcke, J. Explainable Machine Learning for Scientific Insights and Discoveries. IEEE Access 2020, 8, 42200–42216. [Google Scholar] [CrossRef]
- Ribeiro, M.T.; Singh, S.; Guestrin, C. Why should i trust you? Explaining the predictions of any classifier. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 1135–1144. [Google Scholar]
- Jiao, Z.; Choi, J.W.; Halsey, K.; Tran, T.M.L.; Hsieh, B.; Wang, D.; Eweje, F.; Wang, R.; Chang, K.; Wu, J.; et al. Prognostication of patients with COVID-19 using artificial intelligence based on chest X-rays and clinical data: A retrospective study. Lancet Digit. Health 2021, 3, e286–e294. [Google Scholar] [CrossRef]
- Cheng, J.; Sollee, J.; Hsieh, C.; Yue, H.; Vandal, N.; Shanahan, J.; Choi, J.W.; Tran, T.M.L.; Halsey, K.; Iheanacho, F.; et al. COVID-19 mortality prediction in the intensive care unit with deep learning based on longitudinal chest X-rays and clinical data. Eur. Radiol. 2022, 32, 4446–4456. [Google Scholar] [CrossRef]
- Fan, G.; Tu, C.; Zhou, F.; Liu, Z.; Wang, Y.; Song, B.; Gu, X.; Wang, Y.; Wei, Y.; Li, H.; et al. Comparison of severity scores for COVID-19 patients with pneumonia: A retrospective study. Eur. Respir. J. 2020, 56, 2002113. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ge, P.; Zhu, J.; Li, H.; Graham, J.; Singer, A.; Richman, P.S.; Duong, T.Q. Deep learning prediction of likelihood of ICU admission and mortality in COVID-19 patients using clinical variables. PeerJ 2020, 8, e10337. [Google Scholar] [CrossRef] [PubMed]
- Moulaei, K.; Shanbehzadeh, M.; Mohammadi-Taghiabad, Z.; Kazemi-Arpanahi, H. Comparing machine learning algorithms for predicting COVID-19 mortality. BMC Med. Inform. Decis. Mak. 2022, 22, 2. [Google Scholar] [CrossRef]
- Nazir, A.; Ampadu, H.K. Interpretable deep learning for the prediction of ICU admission likelihood and mortality of COVID-19 patients. PeerJ Comput. Sci. 2022, 8, e889. [Google Scholar] [CrossRef] [PubMed]
- Jamshidi, E.; Asgary, A.; Tavakoli, N.; Zali, A.; Setareh, S.; Esmaily, H.; Jamaldini, S.H.; Daaee, A.; Babajani, A.; Kashi, M.A.S.; et al. Using Machine Learning to Predict Mortality for COVID-19 Patients on Day 0 in the ICU. Front. Digit. Heal. 2022, 3, 681608. [Google Scholar] [CrossRef] [PubMed]
- Aktar, S.; Talukder, A.; Ahamad, M.; Kamal, A.; Khan, J.; Protikuzzaman, M.; Hossain, N.; Azad, A.; Quinn, J.; Summers, M.; et al. Machine Learning Approaches to Identify Patient Comorbidities and Symptoms That Increased Risk of Mortality in COVID-19. Diagnostics 2021, 11, 1383. [Google Scholar] [CrossRef]
- Prattichizzo, F.; de Candia, P.; Nicolucci, A.; Ceriello, A. Elevated HbA1c levels in pre-COVID-19 infection increases the risk of mortality: A sistematic review and meta-analysis. Diabetes Metab. Res. Rev. 2022, 38, e3476. [Google Scholar] [CrossRef]
| Field Type | Variable Description | |
|---|---|---|
| Age | Categorical: [18, 59], (59, 74], (74, 90] | Age intervals (in years) at admission; truncated for patients > 90 years of age. |
| Gender | Categorical: Male, Female, Unknown/Missing | Documented gender in the electronic health record; dropped in select cases for de-identification. |
| Kidney replacement therapy | Categorical: Yes, No, Unknown/Missing | Documented renal replacement therapy. |
| Kidney transplant | Categorical: Yes, No, Unknown/Missing | History of kidney transplant. |
| Hypertension | Categorical: Yes, No, Unknown/Missing | Documented ICD-10 code for hypertension, taking anti-hypertensive medications, and/or documented blood pressure > 140/90. |
| Diabetes mellitus | Categorical: Yes, No, Unknown/Missing | Documented ICD-10 code for diabetes type 1 or 2 or taking insulin or oral medications for diabetes. |
| Coronary artery disease | Categorical: Yes, No, Unknown/Missing | Documented ICD-10 code for coronary artery disease, history of stent placement, or existing catheter report documenting disease. |
| Heart failure | Categorical: HFpEF, HFrEF, No, Unknown/Missing | For HFrEF, documented ICD-10 code for HFrEF or echocardiogram documenting reduced ejection fraction (reduced EF is <40%, 40% or higher is preserved EF). For HFpEF, documented ICD-10 code for HFpEF or echocardiogram documenting diastolic dysfunction. |
| Chronic kidney disease | Categorical: Yes, No, Unknown/Missing | Documented ICD-10 code for chronic kidney disease or reduced GFR on lab work. |
| Malignancy | Categorical: Yes, No, Unknown/Missing | Documented ICD-10 code for malignancies or receiving treatment for active malignancy. |
| COPD | Categorical: Yes, No, Unknown/Missing | Documented ICD-10 code for chronic obstructive pulmonary disease or pulmonary function tests documenting obstructive defect along with positive smoking history. |
| Other lung disease | Categorical: Yes, No, Unknown/Missing | Documented ICD-10 code for other lung diseases including asthma, interstitial lung disease, pulmonary hypertension, chronic pulmonary embolism, or lung resection. |
| Smoking status | Categorical: Current, Former, Never, Unknown/Missing | Patient’s smoking status as either Current, Former, Never Smoker, or Unknown. This referred only to cigarettes and cigars. E-cigarettes and marijuana were not counted. |
| ACE inhibitor use | Categorical: Yes, No, Unknown/Missing | Admission medication reconciliation documenting use of an ACE inhibitor as a home medication. |
| Angiotensin receptor blocker use | Categorical: Yes, No, Unknown/Missing | Admission medication reconciliation documenting use of an angiotensin receptor blocker as a home medication. |
| Antibiotic use | Categorical: Yes, No, Unknown/Missing | On an antibiotic prior to presentation. |
| NSAID use | Categorical: Yes, No, Unknown/Missing | Admission medication reconciliation documenting use of a non-steroidal anti-inflammatory drug as a home medication. |
| Cough | Categorical: Yes, No, Unknown/Missing | Reported cough on admission. |
| Dyspnea on admission | Categorical: Yes, No, Unknown/Missing | Reported shortness of breath on admission. |
| Nausea | Categorical: Yes, No, Unknown/Missing | Reported nausea on admission. |
| Vomiting | Categorical: Yes, No, Unknown/Missing | Reported vomiting on admission. |
| Diarrhea | Categorical: Yes, No, Unknown/Missing | Reported diarrhea on admission. |
| Abdominal pain | Categorical: Yes, No, Unknown/Missing | Reported abdominal pain on admission. |
| Subjective fever | Categorical: Yes, No, Unknown/Missing | Subjective or objective fever at home. Fever in ED was not counted. |
| Days symptomatic | Integer value or Unknown/Missing | The number of days prior to presentation that symptoms began. |
| BMI | Categorical: <30, [30, 35], >35, Unknown/Missing | Body mass index (kg/m2) |
| HbA1c | Categorical: <6.5, [6.5, 7.9], >7.9, Unknown/Missing | Hemoglobin A1c (%) |
| Temperature over 38 C | Categorical: Yes, No, Unknown/Missing | Temperature at time of admission over 38 degrees centigrade. |
| In-Hospital Mortality | ||||||||
|---|---|---|---|---|---|---|---|---|
| All | Column % | Yes | Column % | No | Column % | p Value 1 | ||
| Total patients | 841 | 180 | 661 | |||||
| Gender | <0.0001 | * | ||||||
| Male | 489 | 58% | 97 | 54% | 392 | 59% | ||
| Female | 321 | 38% | 54 | 30% | 267 | 40% | ||
| Not recorded | 31 | 4% | 29 | 16% | 2 | 0% | ||
| Age | <0.0001 | * | ||||||
| 18–59 | 380 | 45% | 29 | 16% | 351 | 53% | ||
| 60–74 | 252 | 30% | 65 | 36% | 187 | 28% | ||
| >75 | 209 | 25% | 86 | 48% | 123 | 19% | ||
| Comorbidities | ||||||||
| Hypertension | <0.0001 | * | ||||||
| Yes | 377 | 45% | 108 | 60% | 269 | 41% | ||
| No | 339 | 40% | 43 | 24% | 296 | 45% | ||
| Not recorded | 125 | 15% | 29 | 16% | 96 | 15% | ||
| Diabetes mellitus | 0.16 | NS | ||||||
| Yes | 215 | 26% | 55 | 31% | 160 | 24% | ||
| No | 503 | 60% | 97 | 54% | 406 | 61% | ||
| Not recorded | 123 | 15% | 28 | 16% | 95 | 14% | ||
| Coronary artery disease | <0.0001 | * | ||||||
| Yes | 131 | 16% | 49 | 27% | 82 | 12% | ||
| No | 582 | 69% | 99 | 55% | 483 | 73% | ||
| Not recorded | 128 | 15% | 32 | 18% | 96 | 15% | ||
| Heart failure | <0.0001 | * | ||||||
| Yes | 62 | 7% | 33 | 18% | 29 | 4% | ||
| No | 647 | 77% | 114 | 63% | 533 | 81% | ||
| Not recorded | 132 | 16% | 33 | 18% | 99 | 15% | ||
| Chronic kidney disease | 0.020 | NS | ||||||
| Yes | 69 | 8% | 23 | 13% | 46 | 7% | ||
| No | 645 | 77% | 126 | 70% | 519 | 79% | ||
| Not recorded | 127 | 15% | 31 | 17% | 96 | 15% | ||
| Malignancy | 0.034 | NS | ||||||
| Yes | 69 | 8% | 23 | 13% | 46 | 7% | ||
| No | 638 | 76% | 127 | 71% | 511 | 77% | ||
| Not recorded | 134 | 16% | 30 | 17% | 104 | 16% | ||
| Chronic obstructive pulmonary disease | 0.0053 | NS | ||||||
| Yes | 56 | 7% | 21 | 12% | 35 | 5% | ||
| No | 660 | 78% | 129 | 72% | 531 | 80% | ||
| Not recorded | 125 | 15% | 30 | 17% | 95 | 14% | ||
| Other lung disease | 0.77 | NS | ||||||
| Yes | 109 | 13% | 24 | 13% | 85 | 13% | ||
| No | 605 | 72% | 126 | 70% | 479 | 72% | ||
| Not recorded | 127 | 15% | 30 | 17% | 97 | 15% | ||
| Reduced Mortality | Increased Mortality | |
|---|---|---|
| Age | 18–59 | 74–90 |
| Gender | Female | Male |
| BMI | Below 30 | Over 30 |
| HbA1c | <6.6 | >6.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, X.V.; Dikici, E.; Candemir, S.; Ball, R.L.; Prevedello, L.M. Mortality Prediction Analysis among COVID-19 Inpatients Using Clinical Variables and Deep Learning Chest Radiography Imaging Features. Tomography 2022, 8, 1791-1803. https://doi.org/10.3390/tomography8040151
Nguyen XV, Dikici E, Candemir S, Ball RL, Prevedello LM. Mortality Prediction Analysis among COVID-19 Inpatients Using Clinical Variables and Deep Learning Chest Radiography Imaging Features. Tomography. 2022; 8(4):1791-1803. https://doi.org/10.3390/tomography8040151
Chicago/Turabian StyleNguyen, Xuan V., Engin Dikici, Sema Candemir, Robyn L. Ball, and Luciano M. Prevedello. 2022. "Mortality Prediction Analysis among COVID-19 Inpatients Using Clinical Variables and Deep Learning Chest Radiography Imaging Features" Tomography 8, no. 4: 1791-1803. https://doi.org/10.3390/tomography8040151
APA StyleNguyen, X. V., Dikici, E., Candemir, S., Ball, R. L., & Prevedello, L. M. (2022). Mortality Prediction Analysis among COVID-19 Inpatients Using Clinical Variables and Deep Learning Chest Radiography Imaging Features. Tomography, 8(4), 1791-1803. https://doi.org/10.3390/tomography8040151







