AI-Radiomics Can Improve Inclusion Criteria and Clinical Trial Performance
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patient Populations
2.2. Patient Data and CT Images
2.3. Radiomic Feature Extraction
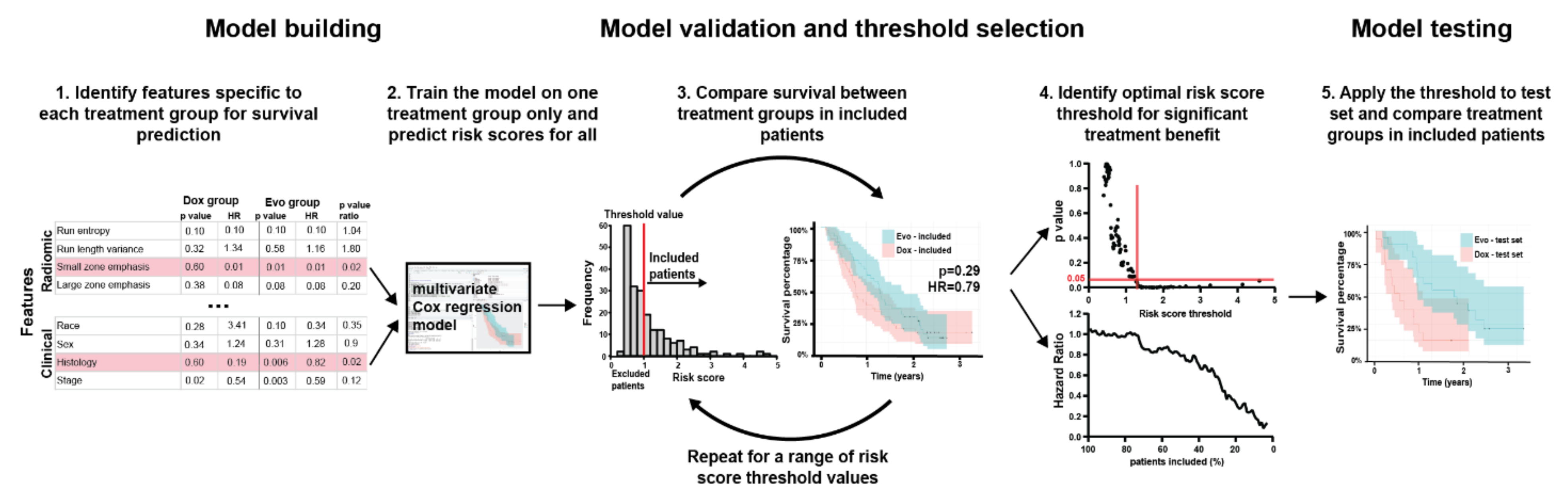
2.4. Feature Selection
2.5. Final Model Construction
3. Results
3.1. Patients
3.2. Feature Stability
3.3. Feature Selection
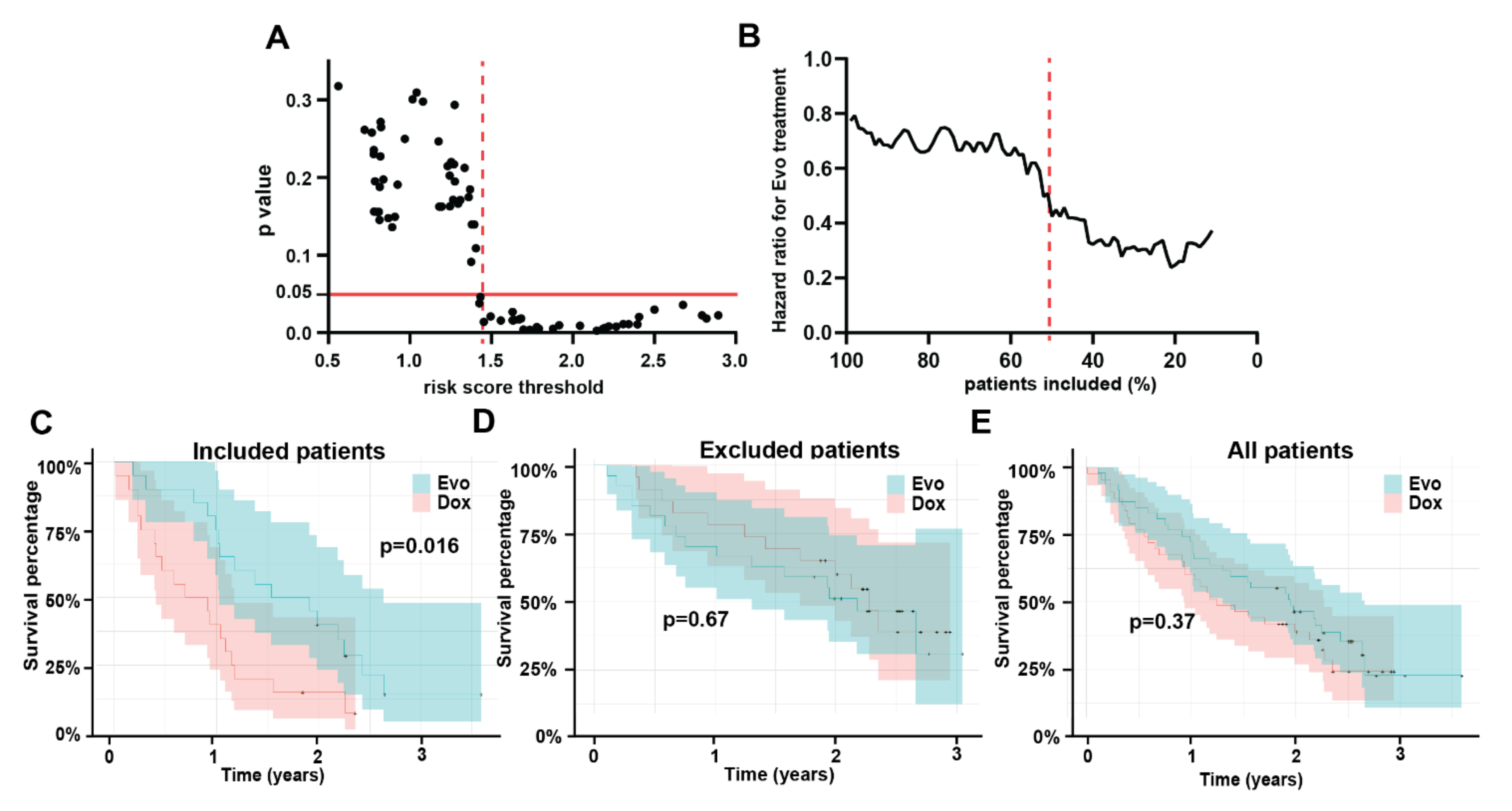
3.4. Multivariable Model
3.5. Model Testing
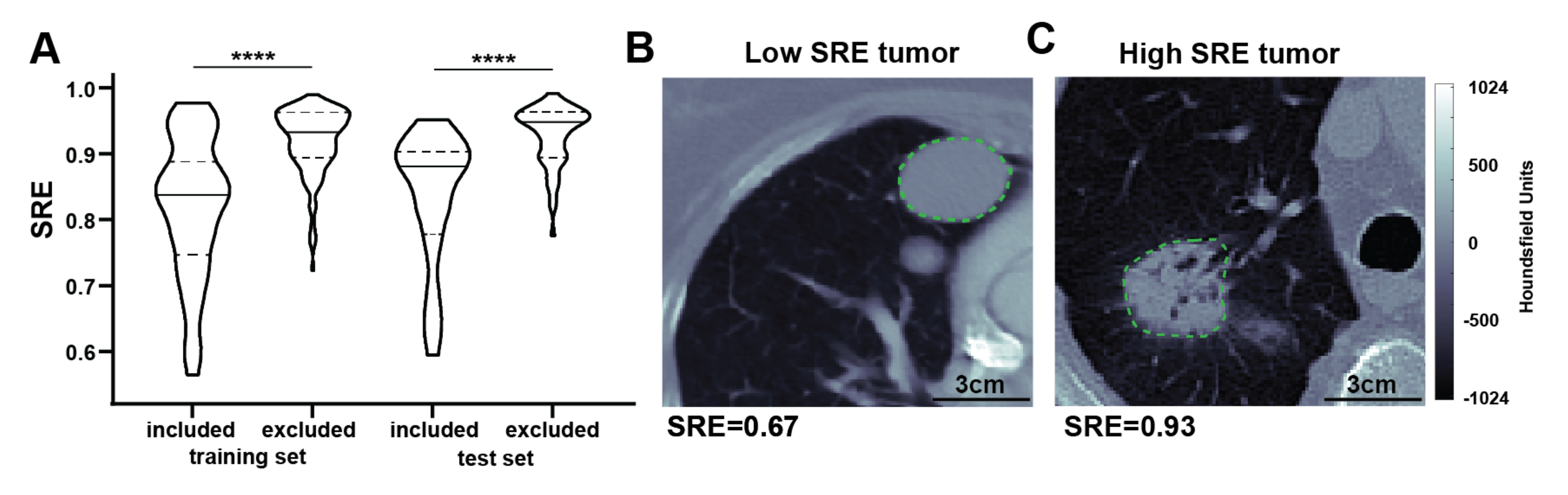
3.6. Model Interpretation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lambin, P.; Leijenaar, R.T.H.; Deist, T.M.; Peerlings, J.; de Jong, E.E.C.; van Timmeren, J.; Sanduleanu, S.; Larue, R.; Even, A.J.G.; Jochems, A.; et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef] [PubMed]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mu, W.; Jiang, L.; Zhang, J.; Shi, Y.; Gray, J.E.; Tunali, I.; Gao, C.; Sun, Y.; Tian, J.; Zhao, X.; et al. Non-invasive decision support for NSCLC treatment using PET/CT radiomics. Nat. Commun. 2020, 11, 5228. [Google Scholar] [CrossRef] [PubMed]
- Tomaszewski, M.; Gillies, R.J. The biological meaning of radiomic features. Radiology 2020, in press. [Google Scholar]
- Tap, W.D.; Papai, Z.; Van Tine, B.A.; Attia, S.; Ganjoo, K.N.; Jones, R.L.; Schuetze, S.; Reed, D.; Chawla, S.P.; Riedel, R.F.; et al. Doxorubicin plus evofosfamide versus doxorubicin alone in locally advanced, unresectable or metastatic soft-tissue sarcoma (TH CR-406/SARC021): An international, multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2017, 18, 1089–1103. [Google Scholar] [CrossRef]
- Chawla, S.P.; Cranmer, L.D.; Van Tine, B.A.; Reed, D.R.; Okuno, S.H.; Butrynski, J.E.; Adkins, D.R.; Hendifar, A.E.; Kroll, S.; Ganjoo, K.N. Phase II study of the safety and antitumor activity of the hypoxia-activated prodrug TH-302 in combination with doxorubicin in patients with advanced soft tissue sarcoma. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2014, 32, 3299–3306. [Google Scholar] [CrossRef]
- Billingsley, K.G.; Burt, M.E.; Jara, E.; Ginsberg, R.J.; Woodruff, J.M.; Leung, D.H.; Brennan, M.F. Pulmonary metastases from soft tissue sarcoma: Analysis of patterns of diseases and postmetastasis survival. Ann. Surg. 1999, 229, 602. [Google Scholar] [CrossRef]
- Tap, W.D.; Wagner, A.J.; Schoffski, P.; Martin-Broto, J.; Krarup-Hansen, A.; Ganjoo, K.N.; Yen, C.C.; Abdul Razak, A.R.; Spira, A.; Kawai, A.; et al. Effect of Doxorubicin Plus Olaratumab vs. Doxorubicin Plus Placebo on Survival in Patients with Advanced Soft Tissue Sarcomas: The ANNOUNCE Randomized Clinical Trial. JAMA 2020, 323, 1266–1276. [Google Scholar] [CrossRef]
- Seddon, B.; Strauss, S.J.; Whelan, J.; Leahy, M.; Woll, P.J.; Cowie, F.; Rothermundt, C.; Wood, Z.; Benson, C.; Ali, N.; et al. Gemcitabine and docetaxel versus doxorubicin as first-line treatment in previously untreated advanced unresectable or metastatic soft-tissue sarcomas (GeDDiS): A randomised controlled phase 3 trial. Lancet Oncol. 2017, 18, 1397–1410. [Google Scholar] [CrossRef] [Green Version]
- Lorigan, P.; Verweij, J.; Papai, Z.; Rodenhuis, S.; Le Cesne, A.; Leahy, M.G.; Radford, J.A.; Van Glabbeke, M.M.; Kirkpatrick, A.; Hogendoorn, P.C.; et al. Phase III trial of two investigational schedules of ifosfamide compared with standard-dose doxorubicin in advanced or metastatic soft tissue sarcoma: A European Organisation for Research and Treatment of Cancer Soft Tissue and Bone Sarcoma Group Study. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2007, 25, 3144–3150. [Google Scholar] [CrossRef]
- Lindner, L.H. Hypoxia-activated prodrug: An appealing preclinical concept yet lost in clinical translation. Lancet Oncol. 2017, 18, 991–993. [Google Scholar] [CrossRef]
- Papanikolaou, N.; Matos, C.; Koh, D.M. How to develop a meaningful radiomic signature for clinical use in oncologic patients. Cancer Imaging Off. Publ. Int. Cancer Imaging Soc. 2020, 20, 33. [Google Scholar] [CrossRef]
- Zwanenburg, A.; Leger, S.; Vallières, M.; Löck, S. Image Biomarker Standardisation Initiative. 2016. Available online: https://pubmed.ncbi.nlm.nih.gov/32154773/ (accessed on 4 June 2021).
- Tunali, I.; Stringfield, O.; Guvenis, A.; Wang, H.; Liu, Y.; Balagurunathan, Y.; Lambin, P.; Gillies, R.J.; Schabath, M.B. Radial gradient and radial deviation radiomic features from pre-surgical CT scans are associated with survival among lung adenocarcinoma patients. Oncotarget 2017, 8, 96013–96026. [Google Scholar] [CrossRef] [Green Version]
- Tunali, I.; Hall, L.O.; Napel, S.; Cherezov, D.; Guvenis, A.; Gillies, R.J.; Schabath, M.B. Stability and reproducibility of computed tomography radiomic features extracted from peritumoral regions of lung cancer lesions. Med. Phys. 2019, 46, 5075–5085. [Google Scholar] [CrossRef]
- Yip, S.S.; Aerts, H.J. Applications and limitations of radiomics. Phys. Med. Biol. 2016, 61, R150. [Google Scholar] [CrossRef] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Judson, I.; Verweij, J.; Gelderblom, H.; Hartmann, J.T.; Schöffski, P.; Blay, J.Y.; Kerst, J.M.; Sufliarsky, J.; Whelan, J.; Hohenberger, P.; et al. Doxorubicin alone versus intensified doxorubicin plus ifosfamide for first-line treatment of advanced or metastatic soft-tissue sarcoma: A randomised controlled phase 3 trial. Lancet Oncol. 2014, 15, 415–423. [Google Scholar] [CrossRef]
- Ryan, C.W.; Merimsky, O.; Agulnik, M.; Blay, J.Y.; Schuetze, S.M.; Van Tine, B.A.; Jones, R.L.; Elias, A.D.; Choy, E.; Alcindor, T.; et al. PICASSO III: A Phase III, Placebo-Controlled Study of Doxorubicin with or without Palifosfamide in Patients with Metastatic Soft Tissue Sarcoma. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2016, 34, 3898–3905. [Google Scholar] [CrossRef]
- Gronchi, A.; Palmerini, E.; Quagliuolo, V.; Martin Broto, J.; Lopez Pousa, A.; Grignani, G.; Brunello, A.; Blay, J.Y.; Tendero, O.; Diaz Beveridge, R.; et al. Neoadjuvant Chemotherapy in High-Risk Soft Tissue Sarcomas: Final Results of a Randomized Trial From Italian (ISG), Spanish (GEIS), French (FSG), and Polish (PSG) Sarcoma Groups. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2020, 38, 2178–2186. [Google Scholar] [CrossRef]
- Brenner, A.J.; Floyd, J.; Fichtel, L.; Michalek, J.; Kanakia, K.P.; Huang, S.; Reardon, D.; Wen, P.Y.; Lee, E.Q. Phase 2 trial of hypoxia activated evofosfamide (TH302) for treatment of recurrent bevacizumab-refractory glioblastoma. Sci. Rep. 2021, 11, 2306. [Google Scholar] [CrossRef]
- Chetan, M.R.; Gleeson, F.V. Radiomics in predicting treatment response in non-small-cell lung cancer: Current status, challenges and future perspectives. Eur. Radiol. 2021, 31, 1049–1058. [Google Scholar] [CrossRef]
- Agrawal, V.; Coroller, T.P.; Hou, Y.; Lee, S.W.; Romano, J.L.; Baldini, E.H.; Chen, A.B.; Jackman, D.M.; Kozono, D.; Swanson, S.J.; et al. Radiologic-pathologic correlation of response to chemoradiation in resectable locally advanced NSCLC. Lung Cancer 2016, 102, 1–8. [Google Scholar] [CrossRef]
- Horvat, N.; Veeraraghavan, H.; Khan, M.; Blazic, I.; Zheng, J.; Capanu, M.; Sala, E.; Garcia-Aguilar, J.; Gollub, M.J.; Petkovska, I. MR Imaging of Rectal Cancer: Radiomics Analysis to Assess Treatment Response after Neoadjuvant Therapy. Radiology 2018, 287, 833–843. [Google Scholar] [CrossRef] [Green Version]
- Kickingereder, P.; Götz, M.; Muschelli, J.; Wick, A.; Neuberger, U.; Shinohara, R.T.; Sill, M.; Nowosielski, M.; Schlemmer, H.P.; Radbruch, A.; et al. Large-scale Radiomic Profiling of Recurrent Glioblastoma Identifies an Imaging Predictor for Stratifying Anti-Angiogenic Treatment Response. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2016, 22, 5765–5771. [Google Scholar] [CrossRef] [Green Version]
- Ganeshan, B.; Goh, V.; Mandeville, H.C.; Ng, Q.S.; Hoskin, P.J.; Miles, K.A. Non-small cell lung cancer: Histopathologic correlates for texture parameters at CT. Radiology 2013, 266, 326–336. [Google Scholar] [CrossRef]
- Rooney, M.; Govindan, R. The era of big trials is over. Lancet Oncol. 2013, 14, 12–14. [Google Scholar] [CrossRef]
- Salem, A.; Asselin, M.C.; Reymen, B.; Jackson, A.; Lambin, P.; West, C.M.L.; O’Connor, J.P.B.; Faivre-Finn, C. Targeting Hypoxia to Improve Non-Small Cell Lung Cancer Outcome. J. Natl. Cancer Inst. 2018, 110, 14–30. [Google Scholar] [CrossRef] [Green Version]
- Aerts, H.J.; Velazquez, E.R.; Leijenaar, R.T.; Parmar, C.; Grossmann, P.; Cavalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef]

| Training Cohort | Test Cohort | |||||
|---|---|---|---|---|---|---|
| Dox + Evo (n = 105) | Dox (n = 101) | p-Value | Dox + Evo (n = 47) | Dox (n = 43) | p-Value | |
| Age (years) | 60 (47–73) | 55 (33–78) | 0.06 | 60 (44–75) | 57 (38–76) | 0.82 |
| Sex | 1.00 | 1.00 | ||||
| Female | 59 (56%) | 57 (56%) | 26 (60%) | 24 (51%) | ||
| Male | 46 (44%) | 44 (44%) | 21 (49%) | 19 (40%) | ||
| Smoking history | 0.91 | 0.46 | ||||
| Never smoker | 59 (56%) | 55 (54%) | 26 (60%) | 28 (60%) | ||
| Ever smoker | 46 (44%) | 46 (46%) | 21 (49%) | 15 (32%) | ||
| Primary Tumor Site | 0.89 | 0.25 | ||||
| Extremity | 35 (33%) | 40 (40%) | 17 (40%) | 20 (43%) | ||
| Head/Neck | 7 (7%) | 5 (5%) | 0 (0%) | 3 (6%) | ||
| Retroperitoneum | 15 (14%) | 12 (12%) | 8 (19%) | 4 (9%) | ||
| Visceral | 19 (18%) | 17 (17%) | 9 (21%) | 7 (15%) | ||
| Other | 29 (28%) | 27 (27%) | 13 (30%) | 9 (19%) | ||
| Metastatic sites number | 1.00 | 0.46 | ||||
| ≥2 | 73 (70%) | 71 (70%) | 36 (84%) | 29 (62%) | ||
| <2 | 32 (30%) | 30 (30%) | 11 (26%) | 14 (30%) | ||
| Lung lesions number | 1.00 | 0.62 | ||||
| >1 | 82 (78%) | 78 (77%) | 35 (81%) | 29 (62%) | ||
| 1 | 23 (22%) | 23 (23%) | 12 (28%) | 14 (30%) | ||
| Stage | 0.21 | 0.46 | ||||
| 0 | 4 (4%) | 0 (0%) | 1 (2%) | 0 (0%) | ||
| Stage I | 3 (3%) | 6 (6%) | 2 (5%) | 2 (4%) | ||
| Stage II | 24 (23%) | 20 (20%) | 10 (23%) | 16 (34%) | ||
| Stage III | 44 (42%) | 40 (40%) | 16 (37%) | 12 (26%) | ||
| Stage IV | 30 (29%) | 35 (35%) | 18 (42%) | 13 (28%) | ||
| Histology | 0.78 | 0.44 | ||||
| Leiomyosarcoma | 44 (42%) | 39 (39%) | 25 (58%) | 17 (36%) | ||
| Epitheloid | 1 (1%) | 3 (3%) | 0 (0%) | 0 (0%) | ||
| Liposarcoma | 7 (7%) | 6 (6%) | 0 (0%) | 1 (2%) | ||
| Malignant peripheral nerve sheath tumor | 4 (4%) | 4 (4%) | 1 (2%) | 4 (9%) | ||
| Myxofibrosarcoma | 3 (3%) | 4 (4%) | 2 (5%) | 3 (6%) | ||
| Pleomorphic rhabdomyosarcoma | 0 (0%) | 2 (2%) | 0 (0%) | 1 (2%) | ||
| Pleomorphic sarcoma/Malignant fibrous histicytoma | 17 (16%) | 13 (13%) | 9 (21%) | 7 (15%) | ||
| Other | 29 (28%) | 30 (30%) | 0 (0%) | 1 (2%) | ||
| Histology Grade | 0.83 | 0.08 | ||||
| Intermediate | 29 (28%) | 28 (28%) | 21 (49%) | 13 (28%) | ||
| Intermediate/High | 1 (1%) | 2 (2%) | 0 (0%) | 4 (9%) | ||
| High | 75 (71%) | 71 (70%) | 26 (60%) | 25 (53%) | ||
| Unknown | 0 (0%) | 0 (0%) | 0 (0%) | 1 (2%) | ||
| ECOG score | 0.51 | 0.90 | ||||
| 0 | 58 (55%) | 59 (58%) | 29 (67%) | 25 (53%) | ||
| 1 | 47 (45%) | 41 (41%) | 18 (42%) | 18 (38%) | ||
| 2 | 0 (0%) | 1 (1%) | 0 (0%) | 0 (0%) | ||
| Prior radiotherapy | 0.55 | 0.06 | ||||
| No | 56 (53%) | 59 (58%) | 32 (74%) | 20 (43%) | ||
| Yes | 49 (47%) | 42 (42%) | 15 (35%) | 23 (49%) | ||
| Prior systemic therapy | 0.41 | 0.76 | ||||
| No | 98 (93%) | 90 (89%) | 43 (100%) | 41 (87%) | ||
| Yes | 7 (7%) | 11 (11%) | 4 (9%) | 2 (4%) | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tomaszewski, M.R.; Fan, S.; Garcia, A.; Qi, J.; Kim, Y.; Gatenby, R.A.; Schabath, M.B.; Tap, W.D.; Reinke, D.K.; Makanji, R.J.; et al. AI-Radiomics Can Improve Inclusion Criteria and Clinical Trial Performance. Tomography 2022, 8, 341-355. https://doi.org/10.3390/tomography8010028
Tomaszewski MR, Fan S, Garcia A, Qi J, Kim Y, Gatenby RA, Schabath MB, Tap WD, Reinke DK, Makanji RJ, et al. AI-Radiomics Can Improve Inclusion Criteria and Clinical Trial Performance. Tomography. 2022; 8(1):341-355. https://doi.org/10.3390/tomography8010028
Chicago/Turabian StyleTomaszewski, Michal R., Shuxuan Fan, Alberto Garcia, Jin Qi, Youngchul Kim, Robert A. Gatenby, Matthew B. Schabath, William D. Tap, Denise K. Reinke, Rikesh J. Makanji, and et al. 2022. "AI-Radiomics Can Improve Inclusion Criteria and Clinical Trial Performance" Tomography 8, no. 1: 341-355. https://doi.org/10.3390/tomography8010028
APA StyleTomaszewski, M. R., Fan, S., Garcia, A., Qi, J., Kim, Y., Gatenby, R. A., Schabath, M. B., Tap, W. D., Reinke, D. K., Makanji, R. J., Reed, D. R., & Gillies, R. J. (2022). AI-Radiomics Can Improve Inclusion Criteria and Clinical Trial Performance. Tomography, 8(1), 341-355. https://doi.org/10.3390/tomography8010028







