Medical Image Segmentation: A Comprehensive Review of Deep Learning-Based Methods
Abstract
1. Introduction
- (1)
- Diversity of Medical Image Modalities [5]:
- (2)
- Blurred Edges in Medical Images [7]:
- (3)
- Scarcity of Annotated Medical Image Data [8]:
- (4)
- Complex and Diverse Segmentation Targets in Medical Images [9]:
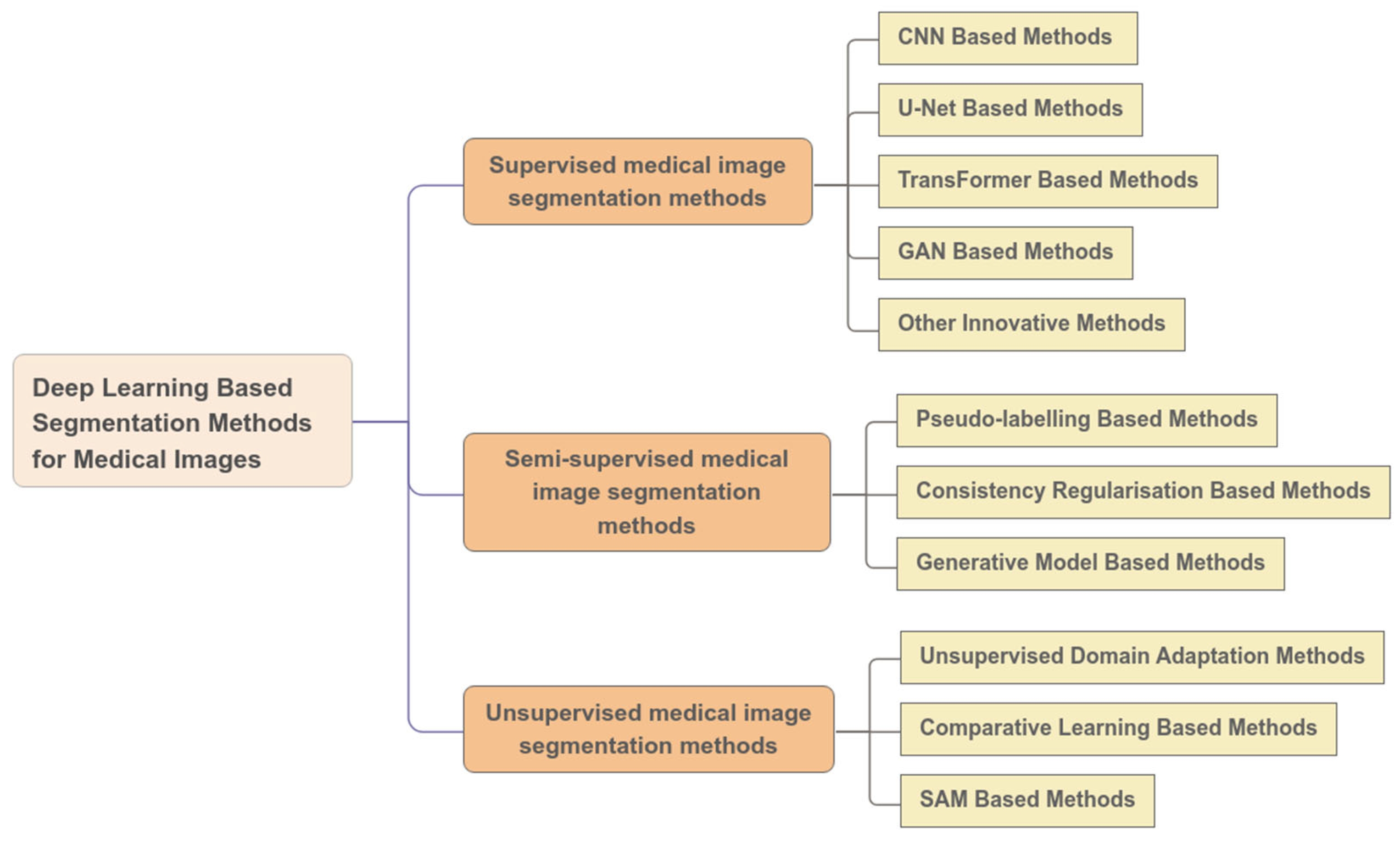
2. Supervised Learning Algorithms for Medical Image Segmentation
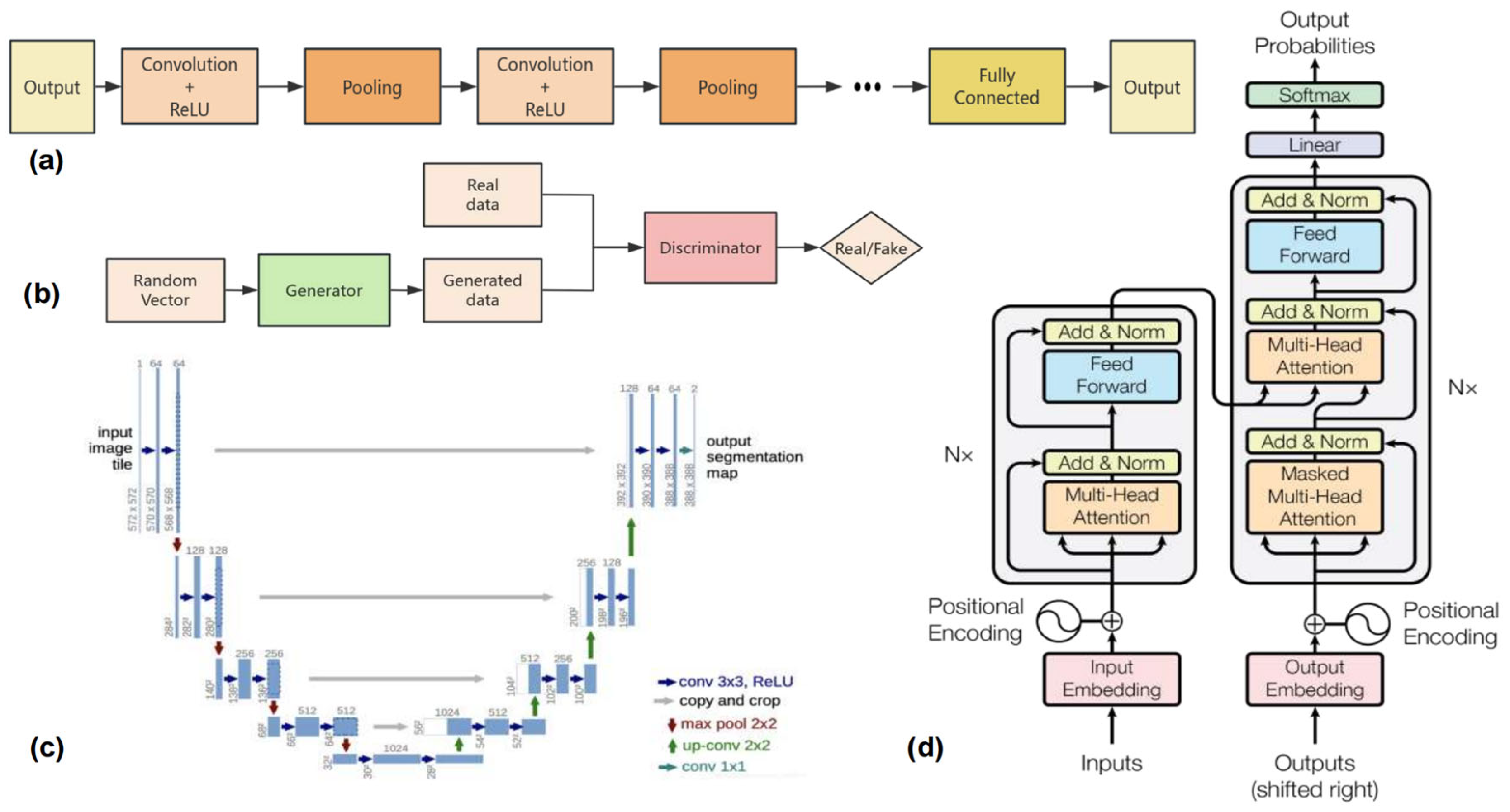
2.1. CNN-Based Methods
2.1.1. Colonoscopy Image Processing Methods
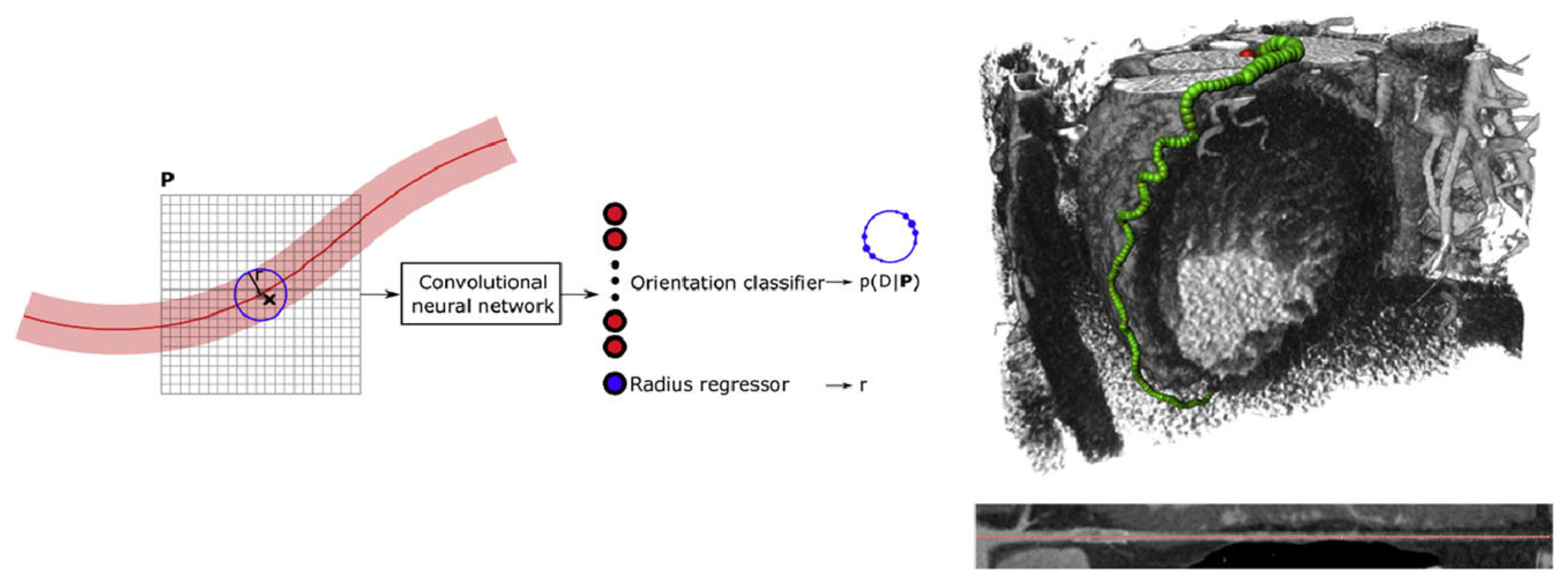
2.1.2. Coronary Artery Segmentation Methods
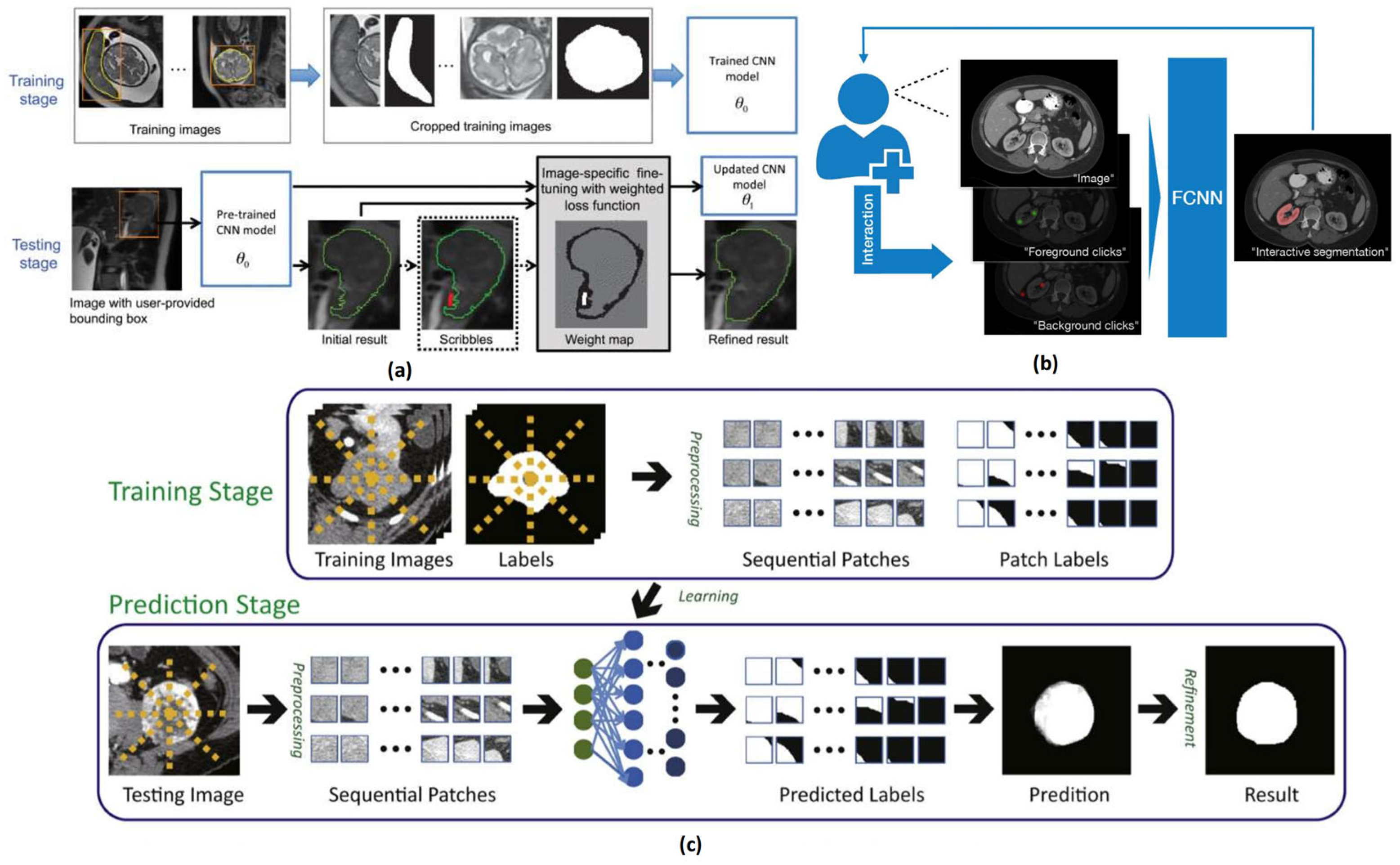
2.1.3. Interactive Medical Image Segmentation Methods
2.2. U-Net-Based Algorithms
2.3. Transformer-Based Methods
2.4. GAN-Based Methods
2.5. Other Innovative Methods
3. Semi-Supervised Medical Image Segmentation Methods
3.1. Pseudo-Labeling-Based Methods
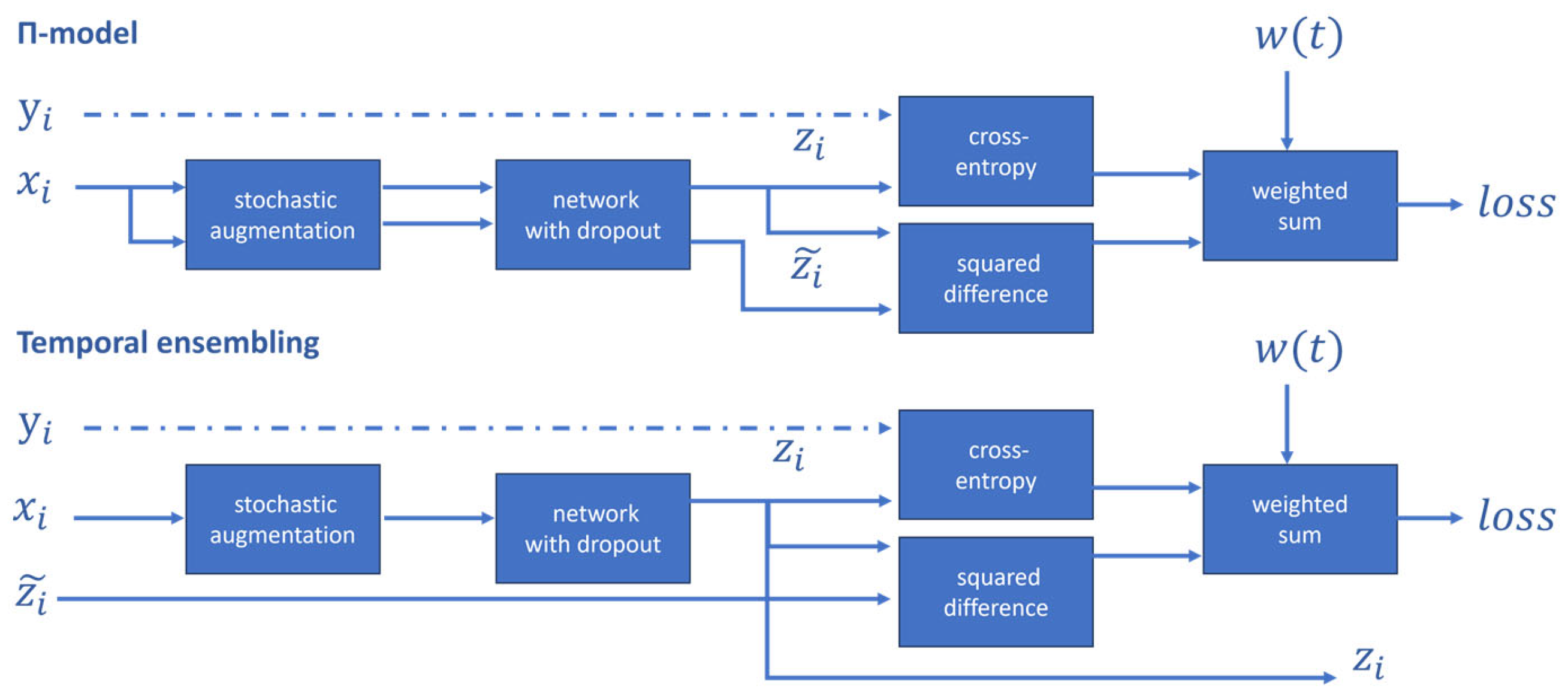
3.2. Consistency Regularization-Based Methods
3.3. Generative Model-Based Methods
4. Unsupervised Medical Image Segmentation Methods
4.1. Unsupervised Domain Adaptation Methods
4.1.1. Image Alignment-Based Unsupervised Domain Adaptation Methods
4.1.2. Fourier Transform-Based Image Style Transfer Methods
4.1.3. Unified Unsupervised Domain Adaptation Framework
4.2. Contrastive Learning-Based Unsupervised Segmentation Methods
4.3. SAM-Based Segmentation Methods
5. Commonly Used Datasets, Evaluation Metrics, and Loss Functions
5.1. Common Medical Image Datasets
5.2. Evaluation Metrics
5.3. Loss Functions
6. Discussion
6.1. Summary of Deep Learning-Based Medical Image Segmentation Methods
6.1.1. Supervised Deep Learning-Based Medical Image Segmentation Methods
6.1.2. Deep Learning-Based Semi-Supervised Medical Image Segmentation Methods
6.1.3. Deep Learning-Based Unsupervised Medical Image Segmentation Methods
6.2. Challenges in Current Medical Image Segmentation Methods
6.2.1. Limited Generalization Across Domains
6.2.2. Challenge of Overfitting in Medical Image Segmentation
6.2.3. The Computational Cost of the Proposed Methods
6.3. Development Trends in Deep Learning-Based Medical Image Segmentation Methods
6.3.1. Deepening of Semi-Supervised and Unsupervised Learning
6.3.2. Exploration of Lightweight and Efficient Models
6.3.3. Enhancing Interpretability and Clinical Trustworthiness
6.3.4. Collaborative Development of Federated Learning and Privacy Protection
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Otsu, N. A Threshold Selection Method from Gray-Level Histograms. Automatica 1975, 11, 23–27. [Google Scholar] [CrossRef]
- Muthukrishnan, R.; Radha, M. Edge Detection Techniques for Image Segmentation. Int. J. Comput. Sci. Inf. Technol. 2011, 3, 259. [Google Scholar] [CrossRef]
- Gong, M.; Liang, Y.; Shi, J.; Ma, W.; Ma, J. Fuzzy C-Means Clustering with Local Information and Kernel Metric for Image Segmentation. IEEE Trans. Image Process. 2012, 22, 573–584. [Google Scholar] [CrossRef] [PubMed]
- Haralick, R.M.; Shapiro, L.G. Image Segmentation Techniques. Comput. Vis. Graph. Image Process. 1985, 29, 100–132. [Google Scholar] [CrossRef]
- Lee, J.-G.; Jun, S.; Cho, Y.-W.; Lee, H.; Kim, G.B.; Seo, J.B.; Kim, N. Deep Learning in Medical Imaging: General Overview. Korean J. Radiol. 2017, 18, 570–584. [Google Scholar] [CrossRef] [PubMed]
- Biersmith, M.A.; Tong, M.S.; Guha, A.; Simonetti, O.P.; Addison, D. Multimodality Cardiac Imaging in the Era of Emerging Cancer Therapies. J. Am. Heart Assoc. 2020, 9, e013755. [Google Scholar] [CrossRef]
- Jiao, Y. Research on Medical Image Segmentation Algorithm Basedon Enhanced Edge Region Learning. Ph.D. Thesis, Xiamen University, Xiamen, China, 2021. [Google Scholar]
- Despotović, I.; Goossens, B.; Philips, W. MRI Segmentation of the Human Brain: Challenges, Methods, and Applications. Comput. Math. Methods Med. 2015, 2015, 1–23. [Google Scholar] [CrossRef]
- Li, Q.; Bai, K.; Zhao, L.; Guan, X. Progresss and challenges of MRI brain tumor image segmentation. J. Image Graph. 2020, 25, 419–431. [Google Scholar] [CrossRef]
- Hinton, G.E.; Salakhutdinov, R.R. Reducing the Dimensionality of Data with Neural Networks. Science 2006, 313, 504–507. [Google Scholar] [CrossRef]
- Mao, Z.; Kobayashi, R.; Nabae, H.; Suzumori, K. Multimodal Strain Sensing System for Shape Recognition of Tensegrity Structures by Combining Traditional Regression and Deep Learning Approaches. IEEE Robot. Autom. Lett. 2024, 9, 10050–10056. [Google Scholar] [CrossRef]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-Cnn: Towards Real-Time Object Detection with Region Proposal Networks. Adv. Neural Inf. Process. Syst. 2015, 28, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet Classification with Deep Convolutional Neural Networks. Adv. Neural Inf. Process. Syst. 2012, 25, 84–90. [Google Scholar] [CrossRef]
- Sutskever, I.; Vinyals, O.; Le, Q.V. Sequence to Sequence Learning with Neural Networks. Adv. Neural Inf. Process. Syst. 2014, 2, 3104–3112. [Google Scholar]
- Devlin, J.; Chang, M.-W.; Lee, K.; Toutanova, K. Bert: Pre-Training of Deep Bidirectional Transformers for Language Understanding. In Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long and Short Papers), Minneapolis, MN, USA, 2–7 June 2019; pp. 4171–4186. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention Is All You Need. Adv. Neural Inf. Process. Syst. 2017, 30, 6000–6010. [Google Scholar]
- Wang, R.-F.; Su, W.-H. The Application of Deep Learning in the Whole Potato Production Chain: A Comprehensive Review. Agriculture 2024, 14, 1225. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, R.; Wang, M.; Lai, T.; Zhang, M. Self-Supervised Transformer-Based Pre-Training Method with General Plant Infection Dataset. In Pattern Recognition and Computer Vision; Lin, Z., Cheng, M.-M., He, R., Ubul, K., Silamu, W., Zha, H., Zhou, J., Liu, C.-L., Eds.; Lecture Notes in Computer Science; Springer Nature Singapore: Singapore, Singapore, 2025; Volume 15032, pp. 189–202. ISBN 978-981-97-8489-9. [Google Scholar]
- Chang-Tao, Z.; Rui-Feng, W.; Yu-Hao, T.; Xiao-Xu, P.; Wen-Hao, S. Automatic Lettuce Weed Detection and Classification Based on Optimized Convolutional Neural Networks for Robotic Weed Control. Agronomy 2024, 14, 2838. [Google Scholar] [CrossRef]
- Tu, Y.-H.; Wang, R.-F.; Su, W.-H. Active Disturbance Rejection Control—New Trends in Agricultural Cybernetics in the Future: A Comprehensive Review. Machines 2025, 13, 111. [Google Scholar] [CrossRef]
- Peng, Y.; Yang, X.; Li, D.; Ma, Z.; Liu, Z.; Bai, X.; Mao, Z. Predicting Flow Status of a Flexible Rectifier Using Cognitive Computing. Expert Syst. Appl. 2025, 264, 125878. [Google Scholar] [CrossRef]
- Mao, Z.; Peng, Y.; Hu, C.; Ding, R.; Yamada, Y.; Maeda, S. Soft Computing-Based Predictive Modeling of Flexible Electrohydrodynamic Pumps. Biomim. Intell. Robot. 2023, 3, 100114. [Google Scholar] [CrossRef]
- Fukushima, K. Neocognitron: A Self-Organizing Neural Network Model for a Mechanism of Pattern Recognition Unaffected by Shift in Position. Biol. Cybern. 1980, 36, 193–202. [Google Scholar] [CrossRef]
- Xi, C.; Yang, J.; Liang, X.; Ramli, R.B.; Tian, S.; Feng, G.; Zhen, D. An Improved Gated Convolutional Neural Network for Rolling Bearing Fault Diagnosis with Imbalanced Data. Int. J. Hosp. Manag. 2023, 6, 108. [Google Scholar] [CrossRef]
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global Cancer Statistics 2022: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef]
- Stryker, S.J.; Wolff, B.G.; Culp, C.E.; Libbe, S.D.; Ilstrup, D.M.; MacCarty, R.L. Natural History of Untreated Colonic Polyps. Gastroenterology 1987, 93, 1009–1013. [Google Scholar] [CrossRef]
- Cai, L.; Chen, L.; Huang, J.; Wang, Y.; Zhang, Y. Know Your Orientation: A Viewpoint-Aware Framework for Polyp Segmentation. Med. Image Anal. 2024, 97, 103288. [Google Scholar] [CrossRef]
- Du, X.; Xu, X.; Ma, K. ICGNet: Integration Context-Based Reverse-Contour Guidance Network for Polyp Segmentation. In Proceedings of the Thirty-First International Joint Conference on Artificial Intelligence, Vienna, Austria, 23–29 July 2022; pp. 877–883. [Google Scholar]
- Du, X.; Xu, X.; Chen, J.; Zhang, X.; Li, L.; Liu, H.; Li, S. UM-Net: Rethinking ICGNet for Polyp Segmentation with Uncertainty Modeling. Med. Image Anal. 2025, 99, 103347. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Li, G.; Li, Z.; Cui, S.; Qian, D.; Yu, Y. Adaptive Context Selection for Polyp Segmentation. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2020; Martel, A.L., Abolmaesumi, P., Stoyanov, D., Mateus, D., Zuluaga, M.A., Zhou, S.K., Racoceanu, D., Joskowicz, L., Eds.; Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2020; Volume 12266, pp. 253–262. ISBN 978-3-030-59724-5. [Google Scholar]
- Fan, D.-P.; Ji, G.-P.; Zhou, T.; Chen, G.; Fu, H.; Shen, J.; Shao, L. PraNet: Parallel Reverse Attention Network for Polyp Segmentation. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2020; Martel, A.L., Abolmaesumi, P., Stoyanov, D., Mateus, D., Zuluaga, M.A., Zhou, S.K., Racoceanu, D., Joskowicz, L., Eds.; Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2020; Volume 12266, pp. 263–273. [Google Scholar]
- Wei, J.; Hu, Y.; Zhang, R.; Li, Z.; Zhou, S.K.; Cui, S. Shallow Attention Network for Polyp Segmentation. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2021; De Bruijne, M., Cattin, P.C., Cotin, S., Padoy, N., Speidel, S., Zheng, Y., Essert, C., Eds.; Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2021; Volume 12901, pp. 699–708. ISBN 978-3-030-87192-5. [Google Scholar]
- Mao, Z.; Suzuki, S.; Wiranata, A.; Zheng, Y.; Miyagawa, S. Bio-Inspired Circular Soft Actuators for Simulating Defecation Process of Human Rectum. J. Artif. Organs 2024, 1–10. [Google Scholar] [CrossRef]
- Wolterink, J.M.; van Hamersvelt, R.W.; Viergever, M.A.; Leiner, T.; Išgum, I. Coronary Artery Centerline Extraction in Cardiac CT Angiography Using a CNN-Based Orientation Classifier. Med. Image Anal. 2019, 51, 46–60. [Google Scholar]
- Shahzad, R.; van Walsum, T.; Kirisli, H.; Tang, H.; Metz, C.; Schaap, M.; van Vliet, L.; Niessen, W. Automatic Stenoses Detection, Quantification and Lumen Segmentation of the Coronary Arteries Using a Two Point Centerline Extraction Scheme. In Proceedings of the MICCAI 2012 Workshop Proceedings, Nice, France, 1–5 October 2012. [Google Scholar]
- Kong, B.; Wang, X.; Bai, J.; Lu, Y.; Gao, F.; Cao, K.; Xia, J.; Song, Q.; Yin, Y. Learning Tree-Structured Representation for 3D Coronary Artery Segmentation. Comput. Med. Imaging Graph. 2020, 80, 101688. [Google Scholar] [CrossRef]
- Zreik, M.; Van Hamersvelt, R.W.; Wolterink, J.M.; Leiner, T.; Viergever, M.A.; Išgum, I. A Recurrent CNN for Automatic Detection and Classification of Coronary Artery Plaque and Stenosis in Coronary CT Angiography. IEEE Trans. Med. Imaging 2018, 38, 1588–1598. [Google Scholar]
- Wang, W.; Xia, Q.; Yan, Z.; Hu, Z.; Chen, Y.; Zheng, W.; Wang, X.; Nie, S.; Metaxas, D.; Zhang, S. AVDNet: Joint Coronary Artery and Vein Segmentation with Topological Consistency. Med. Image Anal. 2024, 91, 102999. [Google Scholar] [CrossRef]
- Boykov, Y.Y.; Jolly, M.-P. Interactive Graph Cuts for Optimal Boundary & Region Segmentation of Objects in ND Images. In Proceedings of the Proceedings Eighth IEEE International Conference on Computer Vision, Vancouver, BC, Canada, 7–14 July 2001; IEEE: New York, NY, USA, 2001; Volume 1, pp. 105–112. [Google Scholar]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; Van Der Laak, J.A.; Van Ginneken, B.; Sánchez, C.I. A Survey on Deep Learning in Medical Image Analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Li, W.; Zuluaga, M.A.; Pratt, R.; Patel, P.A.; Aertsen, M.; Doel, T.; David, A.L.; Deprest, J.; Ourselin, S. Interactive Medical Image Segmentation Using Deep Learning with Image-Specific Fine Tuning. IEEE Trans. Med. Imaging 2018, 37, 1562–1573. [Google Scholar] [CrossRef]
- Wang, G.; Zuluaga, M.A.; Li, W.; Pratt, R.; Patel, P.A.; Aertsen, M.; Doel, T.; David, A.L.; Deprest, J.; Ourselin, S. DeepIGeoS: A Deep Interactive Geodesic Framework for Medical Image Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2018, 41, 1559–1572. [Google Scholar] [CrossRef]
- Luo, X.; Wang, G.; Song, T.; Zhang, J.; Aertsen, M.; Deprest, J.; Ourselin, S.; Vercauteren, T.; Zhang, S. MIDeepSeg: Minimally Interactive Segmentation of Unseen Objects from Medical Images Using Deep Learning. Med. Image Anal. 2021, 72, 102102. [Google Scholar] [CrossRef]
- Zhang, J.; Shi, Y.; Sun, J.; Wang, L.; Zhou, L.; Gao, Y.; Shen, D. Interactive Medical Image Segmentation via a Point-Based Interaction. Artif. Intell. Med. 2021, 111, 101998. [Google Scholar] [CrossRef] [PubMed]
- Sakinis, T.; Milletari, F.; Roth, H.; Korfiatis, P.; Kostandy, P.; Philbrick, K.; Akkus, Z.; Xu, Z.; Xu, D.; Erickson, B.J. Interactive Segmentation of Medical Images through Fully Convolutional Neural Networks. arXiv 2019. [Google Scholar]
- Kaushal, C.; Islam, M.K.; Althubiti, S.A.; Alenezi, F.; Mansour, R.F. A Framework for Interactive Medical Image Segmentation Using Optimized Swarm Intelligence with Convolutional Neural Networks. Comput. Intell. Neurosci. 2022, 2022, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Xu, M. One-Prompt to Segment All Medical Images. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 17–21 June 2024; pp. 11302–11312. [Google Scholar]
- Ding, Y.; Li, L.; Wang, W.; Yang, Y. Clustering Propagation for Universal Medical Image Segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 17–21 June 2024; pp. 3357–3369. [Google Scholar]
- Mao, Z.; Bai, X.; Peng, Y.; Shen, Y. Design, Modeling, and Characteristics of Ring-Shaped Robot Actuated by Functional Fluid. J. Intell. Mater. Syst. Struct. 2024, 35, 1459–1470. [Google Scholar] [CrossRef]
- Peng, Y.; Sakai, Y.; Funabora, Y.; Yokoe, K.; Aoyama, T.; Doki, S. Funabot-Sleeve: A Wearable Device Employing McKibben Artificial Muscles for Haptic Sensation in the Forearm. IEEE Robot. Autom. Lett. 2025, 10, 1–8. [Google Scholar] [CrossRef]
- Peng, Y.; Sakai, Y.; Nakagawa, K.; Funabora, Y.; Aoyama, T.; Yokoe, K.; Doki, S. Funabot-Suit: A Bio-Inspired and McKibben Muscle-Actuated Suit for Natural Kinesthetic Perception. Biomim. Intell. Robot. 2023, 3, 100127. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2015; Volume 9351, pp. 234–241. ISBN 978-3-319-24573-7. [Google Scholar]
- Oktay, O.; Schlemper, J.; Folgoc, L.L.; Lee, M.; Heinrich, M.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, N.Y.; Kainz, B.; et al. Attention U-Net: Learning Where to Look for the Pancreas. arXiv 2018. [Google Scholar]
- Zhou, Z.; Rahman Siddiquee, M.M.; Tajbakhsh, N.; Liang, J. UNet++: A Nested U-Net Architecture for Medical Image Segmentation. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support; Stoyanov, D., Taylor, Z., Carneiro, G., Syeda-Mahmood, T., Martel, A., Maier-Hein, L., Tavares, J.M.R.S., Bradley, A., Papa, J.P., Belagiannis, V., et al., Eds.; Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2018; Volume 11045, pp. 3–11. ISBN 978-3-030-00888-8. [Google Scholar]
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation. In Medical Image Computing and Computer-Assisted Intervention–MICCAI 2016; Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W., Eds.; Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2016; Volume 9901, pp. 424–432. ISBN 978-3-319-46722-1. [Google Scholar]
- Dai, D.; Dong, C.; Yan, Q.; Sun, Y.; Zhang, C.; Li, Z.; Xu, S. I2U-Net: A Dual-Path U-Net with Rich Information Interaction for Medical Image Segmentation. Med. Image Anal. 2024, 97, 103241. [Google Scholar] [CrossRef] [PubMed]
- Alom, M.Z.; Hasan, M.; Yakopcic, C.; Taha, T.M.; Asari, V.K. Recurrent Residual Convolutional Neural Network Based on U-Net (R2U-Net) for Medical Image Segmentation. arXiv 2018, arXiv:1802.06955. [Google Scholar]
- Isensee, F.; Petersen, J.; Klein, A.; Zimmerer, D.; Jaeger, P.F.; Kohl, S.; Wasserthal, J.; Koehler, G.; Norajitra, T.; Wirkert, S.; et al. nnU-Net: Self-Adapting Framework for U-Net-Based Medical Image Segmentation. Nat. Methods 2020, 18, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Chen, J.; Li, J.; Peng, Y.; Mao, Z. Large Language Models for Human–Robot Interaction: A Review. Biomim. Intell. Robot. 2023, 3, 100131. [Google Scholar] [CrossRef]
- Chen, J.; Lu, Y.; Yu, Q.; Luo, X.; Adeli, E.; Wang, Y.; Lu, L.; Yuille, A.L.; Zhou, Y. TransUNet: Transformers Make Strong Encoders for Medical Image Segmentation. arXiv 2021, arXiv:2102.04306. [Google Scholar]
- Zheng, S.; Lu, J.; Zhao, H.; Zhu, X.; Luo, Z.; Wang, Y.; Fu, Y.; Feng, J.; Xiang, T.; Torr, P.H. Rethinking Semantic Segmentation from a Sequence-to-Sequence Perspective with Transformers. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021; pp. 6881–6890. [Google Scholar]
- Hatamizadeh, A.; Tang, Y.; Nath, V.; Yang, D.; Myronenko, A.; Landman, B.; Roth, H.R.; Xu, D. Unetr: Transformers for 3d Medical Image Segmentation. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Waikoloa, HI, USA, 3–8 January 2022; pp. 574–584. [Google Scholar]
- Xie, Y.; Zhang, J.; Shen, C.; Xia, Y. CoTr: Efficiently Bridging CNN and Transformer for 3D Medical Image Segmentation. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2021; De Bruijne, M., Cattin, P.C., Cotin, S., Padoy, N., Speidel, S., Zheng, Y., Essert, C., Eds.; Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2021; Volume 12903, pp. 171–180. ISBN 978-3-030-87198-7. [Google Scholar]
- Qin, Y.-M.; Tu, Y.-H.; Li, T.; Ni, Y.; Wang, R.-F.; Wang, H. Deep Learning for Sustainable Agriculture: A Systematic Review on Applications in Lettuce Cultivation. Sustainability 2025, 17, 3190. [Google Scholar] [CrossRef]
- Cao, H.; Wang, Y.; Chen, J.; Jiang, D.; Zhang, X.; Tian, Q.; Wang, M. Swin-Unet: Unet-Like Pure Transformer for Medical Image Segmentation. In Computer Vision–ECCV 2022 Workshops; Karlinsky, L., Michaeli, T., Nishino, K., Eds.; Lecture Notes in Computer Science; Springer Nature Switzerland: Cham, Switzerland, 2023; Volume 13803, pp. 205–218. ISBN 978-3-031-25065-1. [Google Scholar]
- Wang, W.; Chen, C.; Ding, M.; Yu, H.; Zha, S.; Li, J. Transbts: Multimodal Brain Tumor Segmentation Using Transformer. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2021, Strasbourg, France, 27 September–1 October 2021; Springer: Cham, Switzerland, 2021; pp. 109–119. [Google Scholar]
- Liu, T.; Bai, Q.; Torigian, D.A.; Tong, Y.; Udupa, J.K. VSmTrans: A Hybrid Paradigm Integrating Self-Attention and Convolution for 3D Medical Image Segmentation. Med. Image Anal. 2024, 98, 103295. [Google Scholar] [CrossRef]
- Chu, M.; De Maria, G.L.; Dai, R.; Benenati, S.; Yu, W.; Zhong, J.; Kotronias, R.; Walsh, J.; Andreaggi, S.; Zuccarelli, V.; et al. DCCAT: Dual-Coordinate Cross-Attention Transformer for Thrombus Segmentation on Coronary OCT. Med. Image Anal. 2024, 97, 103265. [Google Scholar] [CrossRef]
- Li, H.; Chen, D.; Nailon, W.H.; Davies, M.E.; Laurenson, D.I. Signed Laplacian Deep Learning with Adversarial Augmentation for Improved Mammography Diagnosis. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2019; Shen, D., Liu, T., Peters, T.M., Staib, L.H., Essert, C., Zhou, S., Yap, P.-T., Khan, A., Eds.; Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2019; Volume 11769, pp. 486–494. ISBN 978-3-030-32225-0. [Google Scholar]
- Nie, D.; Shen, D. Adversarial Confidence Learning for Medical Image Segmentation and Synthesis. Int. J. Comput. Vis. 2020, 128, 2494–2513. [Google Scholar] [CrossRef]
- SegAN: Adversarial Network with Multi-Scale L1 Loss for Medical Image Segmentation|Neuroinformatics. Available online: https://link.springer.com/article/10.1007/s12021-018-9377-x (accessed on 8 February 2025).
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Isola, P.; Zhu, J.-Y.; Zhou, T.; Efros, A.A. Image-to-Image Translation with Conditional Adversarial Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1125–1134. [Google Scholar]
- Ning, Y.; Han, Z.; Zhong, L.; Zhang, C. Automated Pancreas Segmentation Using Recurrent Adversarial Learning. In Proceedings of the 2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Madrid, Spain, 3–6 December 2018; IEEE: New York, NY, USA, 2018; pp. 927–934. [Google Scholar]
- Lau, F.; Hendriks, T.; Lieman-Sifry, J.; Sall, S.; Golden, D. ScarGAN: Chained Generative Adversarial Networks to Simulate Pathological Tissue on Cardiovascular MR Scans. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support; Stoyanov, D., Taylor, Z., Carneiro, G., Syeda-Mahmood, T., Martel, A., Maier-Hein, L., Tavares, J.M.R.S., Bradley, A., Papa, J.P., Belagiannis, V., et al., Eds.; Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2018; Volume 11045, pp. 343–350. ISBN 978-3-030-00888-8. [Google Scholar]
- Xing, J.; Li, Z.; Wang, B.; Qi, Y.; Yu, B.; Zanjani, F.G.; Zheng, A.; Duits, R.; Tan, T. Lesion Segmentation in Ultrasound Using Semi-Pixel-Wise Cycle Generative Adversarial Nets. IEEE/ACM Trans. Comput. Biol. Bioinform. 2020, 18, 2555–2565. [Google Scholar] [CrossRef] [PubMed]
- Huo, Y.; Xu, Z.; Moon, H.; Bao, S.; Assad, A.; Moyo, T.K.; Savona, M.R.; Abramson, R.G.; Landman, B.A. Synseg-Net: Synthetic Segmentation without Target Modality Ground Truth. IEEE Trans. Med. Imaging 2018, 38, 1016–1025. [Google Scholar] [CrossRef]
- Zhang, L.; Ning, G.; Liang, H.; Han, B.; Liao, H. One-Shot Neuroanatomy Segmentation through Online Data Augmentation and Confidence Aware Pseudo Label. Med. Image Anal. 2024, 95, 103182. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, C. MixSegNet: Fusing Multiple Mixed-Supervisory Signals with Multiple Views of Networks for Mixed-Supervised Medical Image Segmentation. Eng. Appl. Artif. Intell. 2024, 133, 108059. [Google Scholar] [CrossRef]
- Zhu, Z.; Ma, X.; Wang, W.; Dong, S.; Wang, K.; Wu, L.; Luo, G.; Wang, G.; Li, S. Boosting Knowledge Diversity, Accuracy, and Stability via Tri-Enhanced Distillation for Domain Continual Medical Image Segmentation. Med. Image Anal. 2024, 94, 103112. [Google Scholar] [CrossRef] [PubMed]
- Jha, D.; Smedsrud, P.H.; Johansen, D.; De Lange, T.; Johansen, H.D.; Halvorsen, P.; Riegler, M.A. A Comprehensive Study on Colorectal Polyp Segmentation with ResUNet++, Conditional Random Field and Test-Time Augmentation. IEEE J. Biomed. Health Inform. 2021, 25, 2029–2040. [Google Scholar] [CrossRef]
- Karani, N.; Erdil, E.; Chaitanya, K.; Konukoglu, E. Test-Time Adaptable Neural Networks for Robust Medical Image Segmentation. Med. Image Anal. 2021, 68, 101907. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Tao, Y.; Zhang, Y.; Ji, Z.; Zhang, Y.; Chen, Q. Test-Time Generative Augmentation for Medical Image Segmentation. arXiv 2024. [Google Scholar]
- Luo, X.; Chen, J.; Song, T.; Wang, G. Semi-Supervised Medical Image Segmentation through Dual-Task Consistency. In Proceedings of the AAAI Conference on Artificial Intelligence, Online, 2–9 February 2021; Volume 35, pp. 8801–8809. [Google Scholar]
- Wang, Y.; Yang, Y. Improved Co-Training-Based Lung CT Image COVID-19 Lesion Segmentation Method. Comput. Eng. Des. 2023, 44, 2447–2453. [Google Scholar]
- Lee, D.-H. Pseudo-Label: The Simple and Efficient Semi-Supervised Learning Method for Deep Neural Networks. In Proceedings of the Workshop on challenges in representation learning, ICML, Atlanta, GA, USA, 16–21 June 2013; Volume 3, p. 896. [Google Scholar]
- Shen, N.; Wang, Z.; Li, J. Semi-supervised Abdominal Multi-organ CT Image Segmentation Method Based on Mean Teacher. Ind. Control. Comput. 2023, 36, 107–108. [Google Scholar]
- Kervadec, H.; Dolz, J.; Granger, É.; Ben Ayed, I. Curriculum Semi-Supervised Segmentation. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2019; Shen, D., Liu, T., Peters, T.M., Staib, L.H., Essert, C., Zhou, S., Yap, P.-T., Khan, A., Eds.; Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2019; Volume 11765, pp. 568–576. ISBN 978-3-030-32244-1. [Google Scholar]
- Wu, H.; Zhang, B.; Chen, C.; Qin, J. Federated Semi-Supervised Medical Image Segmentation via Prototype-Based Pseudo-Labeling and Contrastive Learning. IEEE Trans. Med. Imaging 2023, 43, 649–661. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Sun, Z.; Sun, Y.; Chen, H. Confidence-Aware Cross-Supervised Model for Semi-Supervised Skin Lesion Segmentation. J. Electron. Imaging 2023, 32, 013016. [Google Scholar] [CrossRef]
- Miao, J.; Zhou, S.-P.; Zhou, G.-Q.; Wang, K.-N.; Yang, M.; Zhou, S.; Chen, Y. SC-SSL: Self-Correcting Collaborative and Contrastive Co-Training Model for Semi-Supervised Medical Image Segmentation. IEEE Trans. Med. Imaging 2023, 43, 1347–1364. [Google Scholar] [CrossRef]
- Min, S.; Chen, X.; Zha, Z.-J.; Wu, F.; Zhang, Y. A Two-Stream Mutual Attention Network for Semi-Supervised Biomedical Segmentation with Noisy Labels. arXiv 2018. [Google Scholar] [CrossRef]
- Lei, T.; Zhang, D.; Du, X.; Wang, X.; Wan, Y.; Nandi, A.K. Semi-Supervised Medical Image Segmentation Using Adversarial Consistency Learning and Dynamic Convolution Network. IEEE Trans. Med. Imaging 2022, 42, 1265–1277. [Google Scholar] [CrossRef]
- Bai, Y.; Chen, D.; Li, Q.; Shen, W.; Wang, Y. Bidirectional Copy-Paste for Semi-Supervised Medical Image Segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Vancouver, BC, Canada, 17–24 June 2023; pp. 11514–11524. [Google Scholar]
- You, C.; Dai, W.; Min, Y.; Liu, F.; Clifton, D.; Zhou, S.K.; Staib, L.; Duncan, J. Rethinking Semi-Supervised Medical Image Segmentation: A Variance-Reduction Perspective. Adv. Neural Inf. Process. Syst. 2023, 36, 9984–10021. [Google Scholar]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A Simple Way to Prevent Neural Networks from Overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Huang, G.; Sun, Y.; Liu, Z.; Sedra, D.; Weinberger, K.Q. Deep Networks with Stochastic Depth. In Computer Vision–ECCV 2016; Leibe, B., Matas, J., Sebe, N., Welling, M., Eds.; Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2016; Volume 9908, pp. 646–661. ISBN 978-3-319-46492-3. [Google Scholar]
- Laine, S.; Aila, T. Temporal Ensembling for Semi-Supervised Learning. arXiv 2017. [Google Scholar]
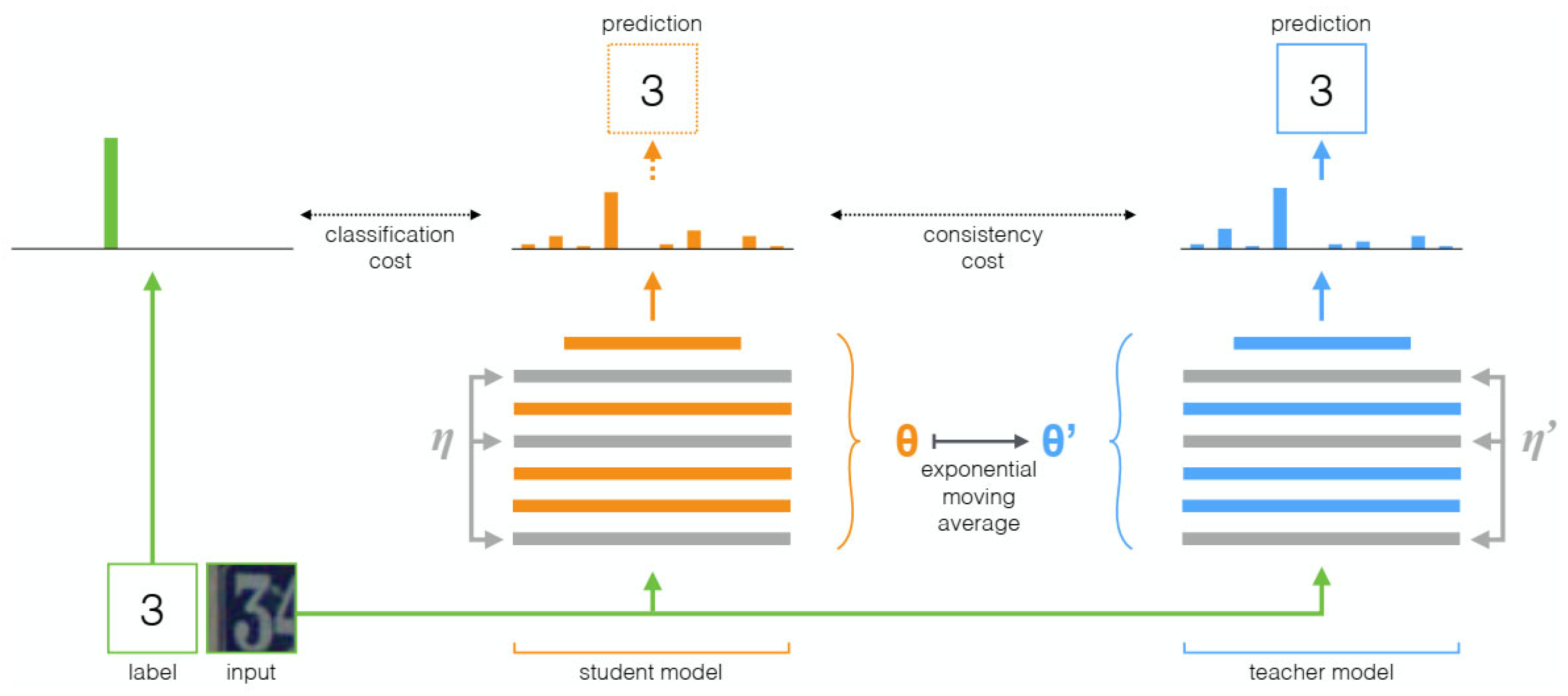
- Tarvainen, A.; Valpola, H. Mean Teachers Are Better Role Models: Weight-Averaged Consistency Targets Improve Semi-Supervised Deep Learning Results. Adv. Neural Inf. Process. Syst. 2017, 30. [Google Scholar]
- Yu, L.; Wang, S.; Li, X.; Fu, C.-W.; Heng, P.-A. Uncertainty-Aware Self-Ensembling Model for Semi-Supervised 3D Left Atrium Segmentation. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2019; Shen, D., Liu, T., Peters, T.M., Staib, L.H., Essert, C., Zhou, S., Yap, P.-T., Khan, A., Eds.; Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2019; Volume 11765, pp. 605–613. ISBN 978-3-030-32244-1. [Google Scholar]
- Ouali, Y.; Hudelot, C.; Tami, M. Semi-Supervised Semantic Segmentation with Cross-Consistency Training. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 12674–12684. [Google Scholar]
- Sohn, K.; Berthelot, D.; Carlini, N.; Zhang, Z.; Zhang, H.; Raffel, C.A.; Cubuk, E.D.; Kurakin, A.; Li, C.-L. Fixmatch: Simplifying Semi-Supervised Learning with Consistency and Confidence. Adv. Neural Inf. Process. Syst. 2020, 33, 596–608. [Google Scholar]
- Wu, Y.; Wu, Z.; Wu, Q.; Ge, Z.; Cai, J. Exploring Smoothness and Class-Separation for Semi-Supervised Medical Image Segmentation. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2022; Wang, L., Dou, Q., Fletcher, P.T., Speidel, S., Li, S., Eds.; Lecture Notes in Computer Science; Springer Nature Switzerland: Cham, Switzerland, 2022; Volume 13435, pp. 34–43. ISBN 978-3-031-16442-2. [Google Scholar]
- You, C.; Dai, W.; Min, Y.; Staib, L.; Sekhon, J.; Duncan, J.S. ACTION++: Improving Semi-Supervised Medical Image Segmentation with Adaptive Anatomical Contrast. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2023; Greenspan, H., Madabhushi, A., Mousavi, P., Salcudean, S., Duncan, J., Syeda-Mahmood, T., Taylor, R., Eds.; Lecture Notes in Computer Science; Springer Nature Switzerland: Cham, Switzerland, 2023; Volume 14223, pp. 194–205. ISBN 978-3-031-43900-1. [Google Scholar]
- Mondal, A.K.; Dolz, J.; Desrosiers, C. Few-Shot 3D Multi-Modal Medical Image Segmentation Using Generative Adversarial Learning. arXiv 2018. [Google Scholar]
- Madani, A.; Moradi, M.; Karargyris, A.; Syeda-Mahmood, T. Chest X-Ray Generation and Data Augmentation for Cardiovascular Abnormality Classification. In Proceedings of the Medical Imaging 2018: Image Processing, Houston, TX, USA, 11–13 February 2018; SPIE: Bellingham, WA, USA, 2018; Volume 10574, pp. 415–420. [Google Scholar]
- Kugelman, J.; Alonso-Caneiro, D.; Read, S.A.; Vincent, S.J.; Collins, M.J. Enhancing OCT Patch-Based Segmentation with Improved GAN Data Augmentation and Semi-Supervised Learning. Neural Comput. Appl. 2024, 36, 18087–18105. [Google Scholar] [CrossRef]
- Zhu, J.-Y.; Park, T.; Isola, P.; Efros, A.A. Unpaired Image-to-Image Translation Using Cycle-Consistent Adversarial Networks. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2223–2232. [Google Scholar]
- Xu, Z.; Qi, C.; Xu, G. Semi-Supervised Attention-Guided Cyclegan for Data Augmentation on Medical Images. In Proceedings of the 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), San Diego, CA, USA, 18–21 November; IEEE: New York, NY, USA, 2019; pp. 563–568. [Google Scholar]
- Qi, C.; Chen, J.; Xu, G.; Xu, Z.; Lukasiewicz, T.; Liu, Y. SAG-GAN: Semi-Supervised Attention-Guided GANs for Data Augmentation on Medical Images. arXiv 2020. [Google Scholar]
- Zhang, D. Research on Medical Image Semantic Segmentation Basedon Semi-Supervised Learning. Master’s Thesis, Shaanxi University of Science and Technology, Xi’an, China, 2023. [Google Scholar]
- Du, W.; Huo, Y.; Zhou, R.; Li, G.; Li, Y.; Zhang, J.A. Weakly Supervised Medical Image Segmentation Method Based on Deep Generative Model. Chinese Patent CN116485816A, 25 July 2023. [Google Scholar]
- Peng, Y.; Nabae, H.; Funabora, Y.; Suzumori, K. Peristaltic Transporting Device Inspired by Large Intestine Structure. Sens. Actuators A Phys. 2024, 365, 114840. [Google Scholar] [CrossRef]
- Mao, Z.; Asai, Y.; Yamanoi, A.; Seki, Y.; Wiranata, A.; Minaminosono, A. Fluidic Rolling Robot Using Voltage-Driven Oscillating Liquid. Smart Mater. Struct. 2022, 31, 105006. [Google Scholar] [CrossRef]
- Chen, C.; Dou, Q.; Chen, H.; Qin, J.; Heng, P.A. Unsupervised Bidirectional Cross-Modality Adaptation via Deeply Synergistic Image and Feature Alignment for Medical Image Segmentation. IEEE Trans. Med. Imaging 2020, 39, 2494–2505. [Google Scholar] [CrossRef]
- Han, X.; Qi, L.; Yu, Q.; Zhou, Z.; Zheng, Y.; Shi, Y.; Gao, Y. Deep Symmetric Adaptation Network for Cross-Modality Medical Image Segmentation. IEEE Trans. Med. Imaging 2021, 41, 121–132. [Google Scholar] [CrossRef]
- Zou, D. Unsupervised Medical Image Segmentationalgorithm Based on Generative Adversarialnetwork. Ph.D. Thesis, Wuhan University, Wuhan, China, 2021. [Google Scholar]
- Luo, Q. Unsupervised Domain Adaptation for MedicalImage Segmentation. Ph.D. Thesis, Huazhong University of Science and Technology, Wuhan, China, 2021. [Google Scholar]
- Zhuang, Y.; Liu, H.; Song, E.; Xu, X.; Liao, Y.; Ye, G.; Hung, C.-C. A 3D Anatomy-Guided Self-Training Segmentation Framework for Unpaired Cross-Modality Medical Image Segmentation. IEEE Trans. Radiat. Plasma Med. Sci. 2023, 8, 33–52. [Google Scholar] [CrossRef]
- Yang, Y.; Soatto, S. Fda: Fourier Domain Adaptation for Semantic Segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 4085–4095. [Google Scholar]
- Oh, K.; Jeon, E.; Heo, D.-W.; Shin, Y.; Suk, H.-I. FIESTA: Fourier-Based Semantic Augmentation with Uncertainty Guidance for Enhanced Domain Generalizability in Medical Image Segmentation. arXiv 2024. [Google Scholar]
- Xian, J.; Li, X.; Tu, D.; Zhu, S.; Zhang, C.; Liu, X.; Li, X.; Yang, X. Unsupervised Cross-Modality Adaptation via Dual Structural-Oriented Guidance for 3D Medical Image Segmentation. IEEE Trans. Med. Imaging 2023, 42, 1774–1785. [Google Scholar] [CrossRef]
- Zhang, X.; Wu, Y.; Angelini, E.; Li, A.; Guo, J.; Rasmussen, J.M.; O’Connor, T.G.; Wadhwa, P.D.; Jackowski, A.P.; Li, H.; et al. MAPSeg: Unified Unsupervised Domain Adaptation for Heterogeneous Medical Image Segmentation Based on 3D Masked Autoencoding and Pseudo-Labeling. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 17–21 June 2024; pp. 5851–5862. [Google Scholar]
- Li, Z.; Sun, C.; Wang, H.; Wang, R.-F. Hybrid Optimization of Phase Masks: Integrating Non-Iterative Methods with Simulated Annealing and Validation via Tomographic Measurements. Symmetry 2025, 17, 530. [Google Scholar] [CrossRef]
- Jiang, Z.; Li, Y.; Yang, C.; Gao, P.; Wang, Y.; Tai, Y.; Wang, C. Prototypical Contrast Adaptation for Domain Adaptive Semantic Segmentation. In Computer Vision–ECCV 2022; Avidan, S., Brostow, G., Cissé, M., Farinella, G.M., Hassner, T., Eds.; Lecture Notes in Computer Science; Springer Nature Switzerland: Cham, Switzerland, 2022; Volume 13694, pp. 36–54. ISBN 978-3-031-19829-8. [Google Scholar]
- Gao, Z.; Jia, C.; Li, Y.; Zhang, X.; Hong, B.; Wu, J.; Gong, T.; Wang, C.; Meng, D.; Zheng, Y. Unsupervised Representation Learning for Tissue Segmentation in Histopathological Images: From Global to Local Contrast. IEEE Trans. Med. Imaging 2022, 41, 3611–3623. [Google Scholar] [CrossRef]
- Liu, L.; Aviles-Rivero, A.I.; Schönlieb, C.-B. Contrastive Registration for Unsupervised Medical Image Segmentation. IEEE Trans. Neural Netw. Learn. Syst. 2025, 36, 147–159. [Google Scholar] [CrossRef]
- Li, Z.; Yang, L.T.; Ren, B.; Nie, X.; Gao, Z.; Tan, C.; Li, S.Z. MLIP: Enhancing Medical Visual Representation with Divergence Encoder and Knowledge-Guided Contrastive Learning. In Proceedings of the 2024 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 17–21 June 2024; IEEE: Seattle, WA, USA, 2024; pp. 11704–11714. [Google Scholar]
- Kirillov, A.; Mintun, E.; Ravi, N.; Mao, H.; Rolland, C.; Gustafson, L.; Xiao, T.; Whitehead, S.; Berg, A.C.; Lo, W.-Y. Segment Anything. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Paris, France, 1–6 October 2023; pp. 4015–4026. [Google Scholar]
- Ma, J.; He, Y.; Li, F.; Han, L.; You, C.; Wang, B. Segment Anything in Medical Images. Nat. Commun. 2024, 15, 654. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Ye, J.; Deng, Z.; Chen, J.; Li, T.; Wang, H.; Su, Y.; Huang, Z.; Chen, J.; Jiang, L.; et al. SAM-Med2D. arXiv 2023. [Google Scholar]
- Wang, H.; Guo, S.; Ye, J.; Deng, Z.; Cheng, J.; Li, T.; Chen, J.; Su, Y.; Huang, Z.; Shen, Y.; et al. SAM-Med3D: Towards General-Purpose Segmentation Models for Volumetric Medical Images. arXiv 2024. [Google Scholar]
- Gibson, E.; Giganti, F.; Hu, Y.; Bonmati, E.; Bandula, S.; Gurusamy, K.; Davidson, B.; Pereira, S.P.; Clarkson, M.J.; Barratt, D.C. Multi-Organ Abdominal CT Reference Standard Segmentations. This data set was developed as part of independent research supported by Cancer Research UK (Multidisciplinary C28070/A19985) and the National Institute for Health Research UCL/UCL Hospitals Biomedical Research Centre. arXiv 2018. [Google Scholar]
- Ji, Y.; Bai, H.; Ge, C.; Yang, J.; Zhu, Y.; Zhang, R.; Li, Z.; Zhanng, L.; Ma, W.; Wan, X. Amos: A Large-Scale Abdominal Multi-Organ Benchmark for Versatile Medical Image Segmentation. Adv. Neural Inf. Process. Syst. 2022, 35, 36722–36732. [Google Scholar]
- Roth, H.R.; Lu, L.; Farag, A.; Shin, H.-C.; Liu, J.; Turkbey, E.B.; Summers, R.M. DeepOrgan: Multi-Level Deep Convolutional Networks for Automated Pancreas Segmentation. In Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A., Eds.; Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2015; Volume 9349, pp. 556–564. ISBN 978-3-319-24552-2. [Google Scholar]
- Simpson, A.L.; Antonelli, M.; Bakas, S.; Bilello, M.; Farahani, K.; van Ginneken, B.; Kopp-Schneider, A.; Landman, B.A.; Litjens, G.; Menze, B.; et al. A Large Annotated Medical Image Dataset for the Development and Evaluation of Segmentation Algorithms. arXiv 2019. [Google Scholar]
- Bernal, J.; Sánchez, F.J.; Fernández-Esparrach, G.; Gil, D.; Rodríguez, C.; Vilariño, F. WM-DOVA Maps for Accurate Polyp Highlighting in Colonoscopy: Validation vs. Saliency Maps from Physicians. Comput. Med. Imaging Graph. 2015, 43, 99–111. [Google Scholar] [CrossRef]
- Jha, D.; Smedsrud, P.H.; Riegler, M.A.; Halvorsen, P.; De Lange, T.; Johansen, D.; Johansen, H.D. Kvasir-SEG: A Segmented Polyp Dataset. In MultiMedia Modeling; Ro, Y.M., Cheng, W.-H., Kim, J., Chu, W.-T., Cui, P., Choi, J.-W., Hu, M.-C., De Neve, W., Eds.; Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2020; Volume 11962, pp. 451–462. ISBN 978-3-030-37733-5. [Google Scholar]
- Bernard, O.; Lalande, A.; Zotti, C.; Cervenansky, F.; Yang, X.; Heng, P.-A.; Cetin, I.; Lekadir, K.; Camara, O.; Ballester, M.A.G. Deep Learning Techniques for Automatic MRI Cardiac Multi-Structures Segmentation and Diagnosis: Is the Problem Solved? IEEE Trans. Med. Imaging 2018, 37, 2514–2525. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, X.; Li, L.; Payer, C.; Štern, D.; Urschler, M.; Heinrich, M.P.; Oster, J.; Wang, C.; Smedby, Ö.; Bian, C. Evaluation of Algorithms for Multi-Modality Whole Heart Segmentation: An Open-Access Grand Challenge. Med. Image Anal. 2019, 58, 101537. [Google Scholar] [CrossRef]
- Shiraishi, J.; Katsuragawa, S.; Ikezoe, J.; Matsumoto, T.; Kobayashi, T.; Komatsu, K.; Matsui, M.; Fujita, H.; Kodera, Y.; Doi, K. Development of a Digital Image Database for Chest Radiographs With and Without a Lung Nodule: Receiver Operating Characteristic Analysis of Radiologists’ Detection of Pulmonary Nodules. Am. J. Roentgenol. 2000, 174, 71–74. [Google Scholar] [CrossRef]
- Allaouzi, I.; Ahmed, M.B. A Novel Approach for Multi-Label Chest X-Ray Classification of Common Thorax Diseases. IEEE Access 2019, 7, 64279–64288. [Google Scholar] [CrossRef]
- Setio, A.A.A.; Traverso, A.; De Bel, T.; Berens, M.S.; Van Den Bogaard, C.; Cerello, P.; Chen, H.; Dou, Q.; Fantacci, M.E.; Geurts, B. Validation, Comparison, and Combination of Algorithms for Automatic Detection of Pulmonary Nodules in Computed Tomography Images: The LUNA16 Challenge. Med. Image Anal. 2017, 42, 1–13. [Google Scholar] [CrossRef]
- Lambert, Z.; Petitjean, C.; Dubray, B.; Kuan, S. Segthor: Segmentation of Thoracic Organs at Risk in Ct Images. In Proceedings of the 2020 Tenth International Conference on Image Processing Theory, Tools and Applications (IPTA), Paris, France, 9–12 November 2020; IEEE: New York, NY, USA, 2020; pp. 1–6. [Google Scholar]
- Menze, B.H.; Jakab, A.; Bauer, S.; Kalpathy-Cramer, J.; Farahani, K.; Kirby, J.; Burren, Y.; Porz, N.; Slotboom, J.; Wiest, R. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans. Med. Imaging 2014, 34, 1993–2024. [Google Scholar] [CrossRef] [PubMed]
- Klein, A.; Tourville, J. 101 Labeled Brain Images and a Consistent Human Cortical Labeling Protocol. Front. Neurosci. 2012, 6, 171. [Google Scholar] [CrossRef]
- Staal, J.; Abràmoff, M.D.; Niemeijer, M.; Viergever, M.A.; Van Ginneken, B. Ridge-Based Vessel Segmentation in Color Images of the Retina. IEEE Trans. Med. Imaging 2004, 23, 501–509. [Google Scholar] [CrossRef]
- Orlando, J.I.; Fu, H.; Breda, J.B.; Van Keer, K.; Bathula, D.R.; Diaz-Pinto, A.; Fang, R.; Heng, P.-A.; Kim, J.; Lee, J. Refuge Challenge: A Unified Framework for Evaluating Automated Methods for Glaucoma Assessment from Fundus Photographs. Med. Image Anal. 2020, 59, 101570. [Google Scholar] [CrossRef]
- Porwal, P.; Pachade, S.; Kokare, M.; Deshmukh, G.; Son, J.; Bae, W.; Liu, L.; Wang, J.; Liu, X.; Gao, L. Idrid: Diabetic Retinopathy–Segmentation and Grading Challenge. Med. Image Anal. 2020, 59, 101561. [Google Scholar] [CrossRef]
- Fraz, M.M.; Remagnino, P.; Hoppe, A.; Uyyanonvara, B.; Rudnicka, A.R.; Owen, C.G.; Barman, S.A. An Ensemble Classification-Based Approach Applied to Retinal Blood Vessel Segmentation. IEEE Trans. Biomed. Eng. 2012, 59, 2538–2548. [Google Scholar] [CrossRef]
- Heller, N.; Sathianathen, N.; Kalapara, A.; Walczak, E.; Moore, K.; Kaluzniak, H.; Rosenberg, J.; Blake, P.; Rengel, Z.; Oestreich, M.; et al. The KiTS19 Challenge Data: 300 Kidney Tumor Cases with Clinical Context, CT Semantic Segmentations, and Surgical Outcomes. arXiv 2020. [Google Scholar]
- Clark, K.; Vendt, B.; Smith, K.; Freymann, J.; Kirby, J.; Koppel, P.; Moore, S.; Phillips, S.; Maffitt, D.; Pringle, M.; et al. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository. J. Digit. Imaging 2013, 26, 1045–1057. [Google Scholar] [CrossRef]
- Soler, L.; Hostettler, A.; Agnus, V.; Charnoz, A.; Fasquel, J.-B.; Moreau, J.; Osswald, A.-B.; Bouhadjar, M.; Marescaux, J. 3D Image Reconstruction for Comparison of Algorithm Database. Available online: https://www.ircad.fr/research/data-sets/liver-segmentation-3d-ircadb-01 (accessed on 27 April 2025).
- Li, Z.; Wang, W. Research Progress of Medical Image Segmentation Method Based onDeep Learning. Electron. Sci. Technol. 2024, 37, 72–80. [Google Scholar] [CrossRef]
- Bai, X.; Peng, Y.; Li, D.; Liu, Z.; Mao, Z. Novel Soft Robotic Finger Model Driven by Electrohydrodynamic (EHD) Pump. J. Zhejiang Univ. Sci. A 2024, 25, 596–604. [Google Scholar] [CrossRef]
- Mao, Z.; Hosoya, N.; Maeda, S. Flexible Electrohydrodynamic Fluid-Driven Valveless Water Pump via Immiscible Interface. Cyborg Bionic Syst. 2024, 5, 91. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Wang, G.; Jia, J.; Li, W.; Zhang, C.; Wang, X. Compliance Control Method for Robot Joint with Variable Stiffness. Int. J. Hydromechatronics 2023, 6, 45–58. [Google Scholar] [CrossRef]
- Peng, Y.; Nabae, H.; Funabora, Y.; Suzumori, K. Controlling a Peristaltic Robot Inspired by Inchworms. Biomim. Intell. Robot. 2024, 4, 100146. [Google Scholar] [CrossRef]
- Ait Nasser, A.; Akhloufi, M.A. A Review of Recent Advances in Deep Learning Models for Chest Disease Detection Using Radiography. Diagnostics 2023, 13, 159. [Google Scholar] [CrossRef]
- Papandrianos, N.I.; Feleki, A.; Papageorgiou, E.I.; Martini, C. Deep Learning-Based Automated Diagnosis for Coronary Artery Disease Using SPECT-MPI Images. J. Clin. Med. 2022, 11, 3918. [Google Scholar] [CrossRef]
- Liu, C.Y.; Tang, C.X.; Zhang, X.L.; Chen, S.; Xie, Y.; Zhang, X.Y.; Qiao, H.Y.; Zhou, C.S.; Xu, P.P.; Lu, M.J.; et al. Deep Learning Powered Coronary CT Angiography for Detecting Obstructive Coronary Artery Disease: The Effect of Reader Experience, Calcification and Image Quality. Eur. J. Radiol. 2021, 142, 109835. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Kweon, J.; Roh, J.-H.; Lee, J.-H.; Kang, H.; Park, L.-J.; Kim, D.J.; Yang, H.; Hur, J.; Kang, D.-Y.; et al. Deep Learning Segmentation of Major Vessels in X-Ray Coronary Angiography. Sci. Rep. 2019, 9, 16897. [Google Scholar] [CrossRef] [PubMed]
| Datasets | Methods | Rec | Spec | Prec | Dice | MAE | Sα | Eϕ |
|---|---|---|---|---|---|---|---|---|
| Kvasir-SEG | UNet | 87.89 | 97.96 | 83.89 | 82.85 | n/a | n/a | n/a |
| U-Net++ | 88.67 | 97.49 | 83.17 | 82.80 | n/a | n/a | n/a | |
| ACSNet | 93.14 | 91.59 | 97.64 | 91.30 | 3.70 | 89.30 | 92.80 | |
| PraNet | 91.41 | 89.56 | 97.25 | 90.75 | 2.90 | 88.20 | 90.80 | |
| SANet | 93.24 | 91.55 | 96.58 | 91.57 | 3.80 | 89.30 | 92.10 | |
| ICGNet | 93.70 | 98.31 | 92.63 | 92.35 | 2.70 | 93.15 | 96.24 | |
| VANet | - | - | - | - | 2.50 | 92.30 | 96.10 | |
| UMNet | 94.65 | 92.81 | 97.87 | 93.04 | 2.31 | 93.82 | 96.66 | |
| EndoScene | UNet | 85.54 | 98.75 | 83.56 | 80.31 | n/a | n/a | n/a |
| U-Net++ | 5978.90 | 99.15 | 86.17 | 77.38 | n/a | n/a | n/a | |
| ACSNet | 87.96 | 99.16 | 90.99 | 86.59 | 2.84 | 90.45 | 94.07 | |
| PraNet | 82.94 | 99.03 | 90.52 | 83.34 | 2.31 | 90.39 | 92.91 | |
| SANet | 89.63 | - | 90.34 | 87.32 | 1.97 | 92.11 | 94.24 | |
| ICGNet | 88.45 | 88.45 | 91.24 | 87.93 | 1.89 | 92.42 | 95.04 | |
| VANet | - | - | - | - | - | - | - | |
| UMNet | 91.29 | - | 90.19 | 89.26 | 1.38 | 93.14 | 95.81 |
| Datasets | ClinicDB | Kvasir | EndoScene | |||
|---|---|---|---|---|---|---|
| Metrics | IoU | Dice | IoU | Dice | IoU | Dice |
| U-Net | 84.32 | 89.28 | 77.58 | 82.31 | 75.23 | 84.36 |
| AttU-Net | 84.24 | 89.51 | 76.62 | 81.95 | 75.07 | 84.98 |
| U-Net++ | 83.33 | 88.94 | 80.05 | 84.16 | 77.31 | 86.31 |
| Deeplabv3+ | 84.75 | 90.33 | 81.67 | 85.70 | 72.88 | 84.14 |
| nnU-Net | 84.43 | 89.77 | 84.33 | 87.27 | 76.05 | 85.09 |
| Trans-Unet | 84.98 | 90.30 | 83.34 | 86.64 | 73.44 | 84.63 |
| Swin-Unet | 83.78 | 89.47 | 83.65 | 87.50 | 75.56 | 85.30 |
| U-Net | 87.60 | 92.32 | 84.45 | 87.75 | 77.87 | 87.41 |
| Task/Modality | Spleen Segmentation (CT) | Brain Tumor Segmentation (MRI) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| WT | ET | TC | All | |||||||
| Metrics | Dice | HD95 | Dice | HD95 | Dice | HD95 | Dice | HD95 | Dice | HD95 |
| UNet | 0.953 | 4.087 | 0.766 | 9.205 | 0.561 | 11.122 | 0.665 | 10.243 | 0.664 | 10.190 |
| AttUNet | 0.951 | 4.091 | 0.767 | 9.004 | 0.543 | 10.447 | 0.683 | 10.463 | 0.665 | 9.971 |
| SETR NUP | 0.947 | 4.124 | 0.697 | 14.419 | 0.544 | 11.723 | 0.669 | 15.192 | 0.637 | 13.778 |
| SETR PUP | 0.949 | 4.107 | 0.696 | 15.245 | 0.549 | 11.759 | 0.670 | 15.023 | 0.638 | 14.009 |
| SETR MLA | 0.950 | 4.091 | 0.698 | 15.503 | 0.554 | 10.237 | 0.665 | 14.716 | 0.639 | 13.485 |
| TransUNet | 0.950 | 4.031 | 0.706 | 14.027 | 0.542 | 10.421 | 0.684 | 14.501 | 0.644 | 12.983 |
| TransBTS | - | - | 0.779 | 10.030 | 0.574 | 9.969 | 0.735 | 8.950 | 0.696 | 9.650 |
| CoTr w/ oCNN encoder | 0.946 | 4.748 | 0.712 | 11.492 | 0.523 | 9.592 | 0.698 | 12.581 | 0.6444 | 11.221 |
| CoTr | 0.954 | 3.860 | 0.746 | 9.198 | 0.557 | 9.447 | 0.748 | 10.445 | 0.683 | 9.697 |
| UNETR | 0.964 | 1.333 | 0.789 | 8.266 | 0.585 | 9.354 | 0.761 | 8.845 | 0.711 | 8.822 |
| Method | Core Features | Advantages | Limitations | Applicable Scenarios |
|---|---|---|---|---|
| RITM | Achieves high-quality image segmentation without prior mask information | Capable of segmenting multiple complex structures across different imaging modalities | Requires significant computational resources and time | Multi-modal brain image structure segmentation |
| S2VNet | Achieves continuous prediction by compressing target information to centroids and passing it between adjacent slices | Achieves volumetric image segmentation using only a 2D network and can handle multiple categories simultaneously | Only handles multi-class interactions of the same category | Volumetric image segmentation of multiple targets within the same class, such as lung nodule segmentation |
| VANet | Introduces self-attention mechanisms and CVT architecture | Enhances feature representation of polyps | Struggles to distinguish polyps from other tissues, prone to misclassification | Colonoscopic polyp segmentation, where boundary accuracy is not extremely critical |
| ICGNet | RCG addresses low-contrast boundaries and missed detection issues; ALGM provides a larger acceptable range | Improves segmentation performance | Ignores inconsistencies in image color distribution, leading to overfitting and difficulty focusing on valuable image content | Boundary detection and feature fusion required, with relatively consistent color distribution, such as in normal tissue boundary segmentation |
| UM-Net | Introduces color transfer operations to weaken the relationship between color and polyps, making the model focus on shape | Addresses issues like inconsistent color distribution, low contrast, and misdiagnosis | Requires further model design and training improvements for more complex scenarios, such as handling background brightness variations | Polyp segmentation with inconsistent color distribution but relatively stable structure, such as under varying lighting conditions |
| AVDNet | Proposes two distinct types of neural networks: image feature recognition network and topology optimization network | Enables segmentation of both coronary arteries and veins with high accuracy and reliability | Currently limited to coronary artery and vein segmentation, with performance in other vascular types yet to be validated | Coronary artery and vein segmentation scenarios |
| Attention U-Net | Introduces attention modules on the classic U-Net architecture to guide the model’s focus on target region features | Improves segmentation accuracy and model robustness, enhancing decision interpretability | Relies on high-quality annotated data, lacks global context information mining | Scenarios requiring high accuracy in target region segmentation, with sufficient hardware support and high-quality labeled data, such as tumor segmentation |
| U-Net++ | Uses nested skip connections on top of U-Net to fully integrate features from different depths, enhancing feature expression | Strengthens the model’s ability to capture subtle structures and boundary information in medical images | Long training time and high hardware resource requirements | Scenarios with high demand for fine structure and boundary segmentation in medical images, such as fine segmentation of neural images |
| R2U-Net | Incorporates recurrent structures and residual blocks into U-Net, using the recurrent structure to capture temporal information and residual blocks to mitigate vanishing gradients | Better handles medical images with complex textures and contextual information | Recurrent structure increases computational complexity and training time; improper design may make the model more sensitive to noise | Segmentation of medical images with complex textures and contextual information, such as liver regions with intricate textures |
| I2U-Net | Enhances information interaction mechanisms to capture comprehensive features during feature extraction | Accurately identifies subtle differences between various tissues and lesions in complex textured and diverse structure medical images | Increases model design and training complexity, requiring more resources for parameter optimization to achieve optimal segmentation | Complex textured and diverse structured medical image segmentation, such as chest images containing various tissues and lesions |
| nnU-Net | Automatically adapts to different datasets | Can be quickly deployed and achieve good results in various medical image segmentation tasks | May not perform as well on specific datasets or complex tasks compared to manually fine-tuned models | Rapid deployment in various medical image segmentation tasks, where accuracy requirements for specific datasets and complex tasks are not extremely high |
| TransUNet | Introduces Transformer into medical image segmentation | Significantly improves segmentation accuracy and robustness, reducing training time and data requirements | High computational resource demand, slower inference speed | Scenarios requiring high segmentation accuracy and robustness, with some computational resources available and less emphasis on inference speed, such as fine brain image segmentation |
| Swin-UNet | Introduces Swin Transformer as the backbone, with a hierarchical window attention mechanism | Enhances computational efficiency while maintaining Transformer’s global modeling capability | Poor interpretability of decision-making process | Medical image segmentation scenarios requiring computational efficiency and Transformer’s global modeling capability, such as mid-sized organ segmentation |
| Unetr | Uses a pure Transformer architecture, directly feeding medical images as sequences into the Transformer encoder | Precisely handles high-resolution medical images and complex structures | High computational resource demand, long training and inference time | Medical image segmentation scenarios requiring computational efficiency and Transformer’s global modeling capability, such as mid-sized organ segmentation |
| MedFormer | Proposes a multi-scale window attention module combined with local and global context information | Accurately segments vessels of varying sizes, performing well on medical image segmentation with complex scale variations | May overlook details when handling small targets due to global attention | Medical image segmentation scenarios with complex scale variations, such as segmenting vessels of different sizes |
| SegAN | Introduces the adversarial training mechanism of GAN into medical image segmentation tasks | Learns complex features and distributions from medical image data | Complex training process, high computational cost, less effective on small targets or boundary details | Medical image data feature learning scenarios, where high accuracy in small target or boundary detail segmentation is not critical, such as coarse organ segmentation |
| cGAN | Introduces conditional information into both the generator and discriminator, allowing the generator to produce segmentation results relevant to the input image | Increases the alignment of generated results with actual needs | Highly dependent on the quality and selection of conditional information | Scenarios requiring high alignment of generated results with specific conditions, such as lesion segmentation based on specific conditions |
| pix2pix | Based on conditional GAN, implements precise mapping from input image to target image by introducing conditional inputs | Generates high-quality images with excellent visual effects, maintaining image structure and semantic information | Requires large amounts of paired labeled data for training, high labeling cost, and relatively complex model architecture | Suitable for image-to-image translation tasks |
| Methods | Scans Used | Metrics | ||||
|---|---|---|---|---|---|---|
| Labeled | Unlabeled | Dice | Jaccard | 95HD | ASD | |
| UA-MT | 82.26 | 70.98 | 13.71 | 3.82 | ||
| SASSNet | 81.6 | 69.63 | 16.16 | 3.58 | ||
| DTC | 81.25 | 69.33 | 14.9 | 3.99 | ||
| URPC | 4 (5%) | 76 (95%) | 82.48 | 71.35 | 14.65 | 3.65 |
| MC-Net | 83.59 | 72.36 | 14.07 | 2.7 | ||
| SS-Net | 86.33 | 76.15 | 9.97 | 2.31 | ||
| BCP | 88.02 | 78.72 | 7.9 | 2.15 | ||
| UA-MT | 87.79 | 78.39 | 8.68 | 2.12 | ||
| SASSNet | 87.54 | 78.05 | 9.84 | 2.59 | ||
| DTC | 87.51 | 78.17 | 8.23 | 2.36 | ||
| URPC | 8 (10%) | 72 (90%) | 86.92 | 77.03 | 11.13 | 2.28 |
| MC-Net | 87.62 | 78.25 | 10.03 | 1.82 | ||
| SS-Net | 88.55 | 79.62 | 7.49 | 1.9 | ||
| BCP | 89.62 | 81.31 | 6.81 | 1.76 | ||
| Method | Core Features | Advantages | Limitations | Applicable Scenarios |
|---|---|---|---|---|
| Pseudo-Labeling with Confidence Thresholding | Uses confidence thresholding to filter out noise | Reduces the interference of incorrect labels in model training, allowing more effective use of unlabeled data | High confidence thresholds may lead to an imbalanced class distribution in pseudo-labels | Semi-supervised classification of common medical images with broad disease categories |
| Curriculum Semi-Supervised Learning | Introduces additional constraints to enhance pseudo-label confidence | Effectively prevents the accumulation of training errors due to incorrect pseudo-labels | Longer training times | Semi-supervised medical image segmentation where pseudo-label accuracy is critical and sufficient training time is available |
| CCSM | Uses a confidence calculation module to generate pseudo-labels | Generates more reliable pseudo-labels | Complex model structure, sensitive to parameters and hyperparameters | High accuracy cardiac structure segmentation tasks |
| SC-SSL | Improves learning confidence of unlabeled data via self-correction modules | Effectively reduces noise in pseudo-labels | Performance is highly dependent on data quality | Semi-supervised medical image segmentation scenarios with high data quality |
| DAN | Adaptive noise label correction | Improves pseudo-label quality | Sensitive to the choice of data transformation methods | Semi-supervised medical image segmentation requiring high pseudo-label quality |
| BCP | Proposes a bidirectional copy–paste method to address label distribution imbalance in semi-supervised medical image segmentation | Utilizes unlabeled data to improve model performance | Difficulty in determining suitable copy–paste regions | Semi-supervised medical image segmentation with significant label data distribution imbalance |
| ARCO | Proposes a group sampling-based semi-supervised learning framework | Improves model performance, reduces training time | Requires manual selection of group sampling strategies | Semi-supervised scenarios with limited labeled data |
| Π-model | Applies the same or different dropout perturbations to the same input | Enhances model generalization capabilities | Difficulty in determining appropriate hyperparameters and consistency loss weight during training | Semi-supervised scenarios with limited labeled data |
| Temporal Ensembling | Uses an exponential moving average of historical predictions as a consistency target to constrain current predictions | Reduces reliance on labeled data to improve model performance | Requires storing predictions from multiple time steps, increasing memory overhead | Semi-supervised scenarios with limited labeled data |
| CCT | Applies consistency constraints to model predictions under different perturbations | Enhances model performance by leveraging unlabeled data and can be extended to other weakly supervised tasks | May lead to overfitting in cases of imbalanced data distributions | Tasks requiring a large amount of unlabeled data to enhance model performance |
| FixMatch | Applies varying intensities of data augmentation to the same unlabeled sample | Reduces the risk of incorrect label propagation | Sensitive to hyperparameter settings | Tasks requiring large amounts of unlabeled data to improve model performance |
| Mean Teacher | Uses the average model weights as targets to improve semi-supervised learning effectiveness | Improves test accuracy, trains with fewer labeled data, and does not require changes to network architecture | Targets generated by the teacher model may contain noise and unreliability | Tasks requiring large amounts of unlabeled data to improve model performance |
| UA-MT | Proposes an uncertainty-aware self-supervised learning framework | Effectively utilizes unlabeled data to improve segmentation accuracy | May overfit with limited data availability | Tasks requiring large amounts of unlabeled data to improve model performance |
| CCT | Enforces consistency of perturbations on the encoder’s output | Improves the encoder’s representational ability | Requires significant computational resources for training | Tasks lacking large labeled data |
| SS-NET | Considers pixel-level smoothness and class-level separability simultaneously | Effectively utilizes unlabeled data for semi-supervised learning, improving model performance | Requires manual setting of some hyperparameters | Scenarios with difficult data annotation |
| ACTION++ | Proposes adaptive supervised contrastive loss | Effectively addresses the long-tail distribution and class imbalance in medical image data | High model complexity, poor interpretability | Scenarios requiring extremely high result accuracy |
| Method | Core Features | Advantages | Limitations | Applicable Scenarios |
|---|---|---|---|---|
| SIFA | Adaptively learns from both image and feature perspectives for cross-modal medical image segmentation tasks | Offers good generalizability and scalability | Requires large computational resources to train the model, and may have limitations for certain specific application scenarios | Cross-modal image segmentation tasks in the medical field |
| DSAN | Implements bidirectional alignment of source/target domain feature distributions via shared encoders and private decoders | Fully leverages information from images with different styles | Requires significant computational resources to train the model | Cross-modal image segmentation tasks in the medical field |
| DSFN | Achieves collaborative alignment of source and target domains from both image-level and feature-level perspectives | Effectively narrows domain gaps and utilizes task complementarity | Requires significant computational resources to train the model | Medical image segmentation scenarios with domain shift challenges, such as brain tumor and heart structure segmentation |
| SIDA | Introduces a baseline model combining image and feature alignment, innovatively adding image translation degree prediction and contrastive learning self-supervised tasks | Effectively enhances domain adaptation performance | Not well adapted to cases with large data distribution differences | Unsupervised domain adaptation tasks in medical image segmentation |
| FDA | Reduces differences between source and target images by exchanging low-frequency information without any training process | Simple, intuitive, and highly efficient | Cannot handle high-frequency information, potentially losing some detailed information | Scenarios with significant differences between source and target datasets |
| FIESTA | Uses a Fourier-domain adaptation approach combined with uncertainty-guided data augmentation to enhance model generalization | Effectively handles detail and uncertainty issues | Limited to single-source domain generalization, may not perform well for multi-source domains | Single-source dataset tasks |
| DAG-Net | Proposes FCSA and RSA modules based on Fourier transform to achieve efficient cross-modal domain adaptation | Outperforms existing domain adaptation methods in cross-modal transfer tasks | Requires high computational resources and longer training times | Cross-modal transfer tasks in 3D medical image segmentation |
| MAPSeg | Proposes a joint learning framework based on 3D mask autoencoders, global–local context, and large-scale pre-training | Capable of handling various domain adaptation tasks, enhancing model generalization | Requires a large amount of labeled data for pre-training | Medical image segmentation tasks requiring handling of multi-source heterogeneous data |
| ProCA | Combines prototype contrastive learning and domain adaptation for unsupervised domain adaptation | No target domain labels required, enhances feature discriminability, significantly improves performance on the target domain | Relies on the quality of source domain labels, pseudo-label noise may affect prototype computation accuracy | Unsupervised domain adaptation tasks such as cross-domain image classification and semantic segmentation |
| CLMorph | Combines contrastive learning with image registration | Highly versatile, applicable to multiple medical image modalities | Dependent on registration accuracy when handling complex anatomical structures | Segmentation of CT, MRI, and other modalities in scenarios with scarce labeled data |
| MLIP | Combines medical domain expertise with contrastive learning to enhance medical visual representation | Improves model generalization capabilities | Relies on medical domain expertise | Medical image classification, object detection, and semantic segmentation tasks |
| MedSAM | Introduces SAM into the field of medical image segmentation for the first time | High generalizability and flexibility | Requires reliance on medical domain expertise | Accurate and rapid localization and segmentation of various tissues, organs, or lesion areas |
| Part | Imaging Modality | Name | Size | Format | Area | Address |
|---|---|---|---|---|---|---|
| Abdominal Organ | CT | BTCV [133] | 50 | NIFIT | Spleen, right kidney, left kidney, gallbladder, esophagus, liver, stomach, aorta, inferior vena cava, portal and splenic veins, pancreas, right adrenal gland, left adrenal gland | https://aistudio.baidu.com/datasetdetail/107078 (accessed on 27 April 2025) |
| CT | AMOS [134] | 600 | NIFIT | Spleen, right kidney, left kidney, gallbladder, esophagus, liver, stomach, aorta, inferior vena cava, pancreas, right adrenal gland, left adrenal gland, duodenum, bladder, prostate/uterus | https://zenodo.org/records/7155725#.Y0OOCOxBztM (accessed on 27 April 2025) | |
| CT | NIH Pancreas-CT [135] | 82 | NIFIT | Pancreatic | https://www.cancerimagingarchive.net/collection/pancreas-ct/ (accessed on 27 April 2025) | |
| CT | Task07_Pancreas [136] | 420 | NIFIT | Pancreas, Pancreatic tumors | https://pan.baidu.com/s/1fNRLPJuwGQWbwquSfrM1pw?pwd=2024 (accessed on 27 April 2025) | |
| Endoscopy | CVC-ClinicDB [137] | 612 | PNG | Colorectal | https://aistudio.baidu.com/datasetdetail/65816/1 (accessed on 27 April 2025) | |
| Endoscopy | Kvasir-SEG [138] | 1000 | JPG | Colon | https://datasets.simula.no/downloads/kvasir-seg.zip (accessed on 27 April 2025) | |
| Endoscopy | EndoScene [30] | 912 | JPEG, PNG | Colon | - | |
| Chests | MRI | ACDC [139] | 150 | NIFIT | Heart | https://aistudio.baidu.com/datasetdetail/267540 (accessed on 27 April 2025) |
| MRI | LA [104] | 154 | nrrd | Left atrium | https://www.cardiacatlas.org/atriaseg2018-challenge/atria-seg-data/ (accessed on 27 April 2025) | |
| CT MRI | MM-WHS [140] | 120 | NIFIT | Seven cardiac substructures | https://mega.nz/folder/UNMF2YYI#1cqJVzo4p_wESv9P_pc8uA (accessed on 27 April 2025) | |
| Chest X-ray | JSRT [141] | 247 | PNG | Lung | http://db.jsrt.or.jp/eng.php (accessed on 27 April 2025) | |
| Chest X-ray | ChestX-ray14 [142] | 112,120 | PNG | Lung, Heart | https://aistudio.baidu.com/aistudio/data (accessed on 27 April 2025) | |
| Chest X-ray | LUNA16 [143] | 888 | mhd | Lung/lung nodules | https://luna16.grand-challenge.org/Download/ (accessed on 27 April 2025) | |
| CT | SegTHOR [144] | 60 | NIFIT | Heart, Trachea, Aorta, Esophagus | https://competitions.codalab.org/competitions/21145#participate-get_starting_kit (accessed on 27 April 2025) | |
| Brain | MRI | BraTs2018 [145] | 285 | NIFIT | Glioma | https://aistudio.baidu.com/aistudio/datasetdetail/64660 (accessed on 27 April 2025) |
| MRI | Mindboggle [146] | 101 | NIFIT | Brain structure | https://mindboggle.info/data.html (accessed on 27 April 2025) | |
| Eye | Color Fundus Photography | DRIVE [147] | 40 | TIFF | Retinal vessels | https://gitee.com/zongfang/retina-unet/tree/master/DRIVE (accessed on 27 April 2025) |
| Color Fundus Photography | REFUGE [148] | 1200 | JPEG | Optic disc and Optic cup | https://refuge.grand-challenge.org/ (accessed on 27 April 2025) | |
| Color Fundus Photography | IDRiD [149] | 516 | JPG | Areas of lesions associated with diabetic retinopathy | https://idrid.grand-challenge.org/Data_Download/ (accessed on 27 April 2025) | |
| Color Fundus Photography | CHASE_DB1 [150] | 1200 | JPEG | Pathological myopia Vascular lesions | https://blogs.kingston.ac.uk/retinal/chasedb1/ (accessed on 27 April 2025) | |
| Kidney | CT | KiTS19 [151] | 300 | NIFIT | Renal tumor | https://github.com/neheller/kits19 (accessed on 27 April 2025) |
| CT MRI | TCIA [152] | - | DICOM | Renal parenchyma, renal cysts, renal tumors, etc. | http://www.cancerimagingarchive.net/ (accessed on 27 April 2025) | |
| Pancreas | CT | 3D-IRCADb [153] | 22 | DICOM | Liver, liver vessels | https://aistudio.baidu.com/datasetdetail/107717 (accessed on 27 April 2025) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, Y.; Jiang, Y.; Peng, Y.; Yuan, F.; Zhang, X.; Wang, J. Medical Image Segmentation: A Comprehensive Review of Deep Learning-Based Methods. Tomography 2025, 11, 52. https://doi.org/10.3390/tomography11050052
Gao Y, Jiang Y, Peng Y, Yuan F, Zhang X, Wang J. Medical Image Segmentation: A Comprehensive Review of Deep Learning-Based Methods. Tomography. 2025; 11(5):52. https://doi.org/10.3390/tomography11050052
Chicago/Turabian StyleGao, Yuxiao, Yang Jiang, Yanhong Peng, Fujiang Yuan, Xinyue Zhang, and Jianfeng Wang. 2025. "Medical Image Segmentation: A Comprehensive Review of Deep Learning-Based Methods" Tomography 11, no. 5: 52. https://doi.org/10.3390/tomography11050052
APA StyleGao, Y., Jiang, Y., Peng, Y., Yuan, F., Zhang, X., & Wang, J. (2025). Medical Image Segmentation: A Comprehensive Review of Deep Learning-Based Methods. Tomography, 11(5), 52. https://doi.org/10.3390/tomography11050052