Stapled Peptides: An Innovative and Ultimate Future Drug Offering a Highly Powerful and Potent Therapeutic Alternative
Abstract
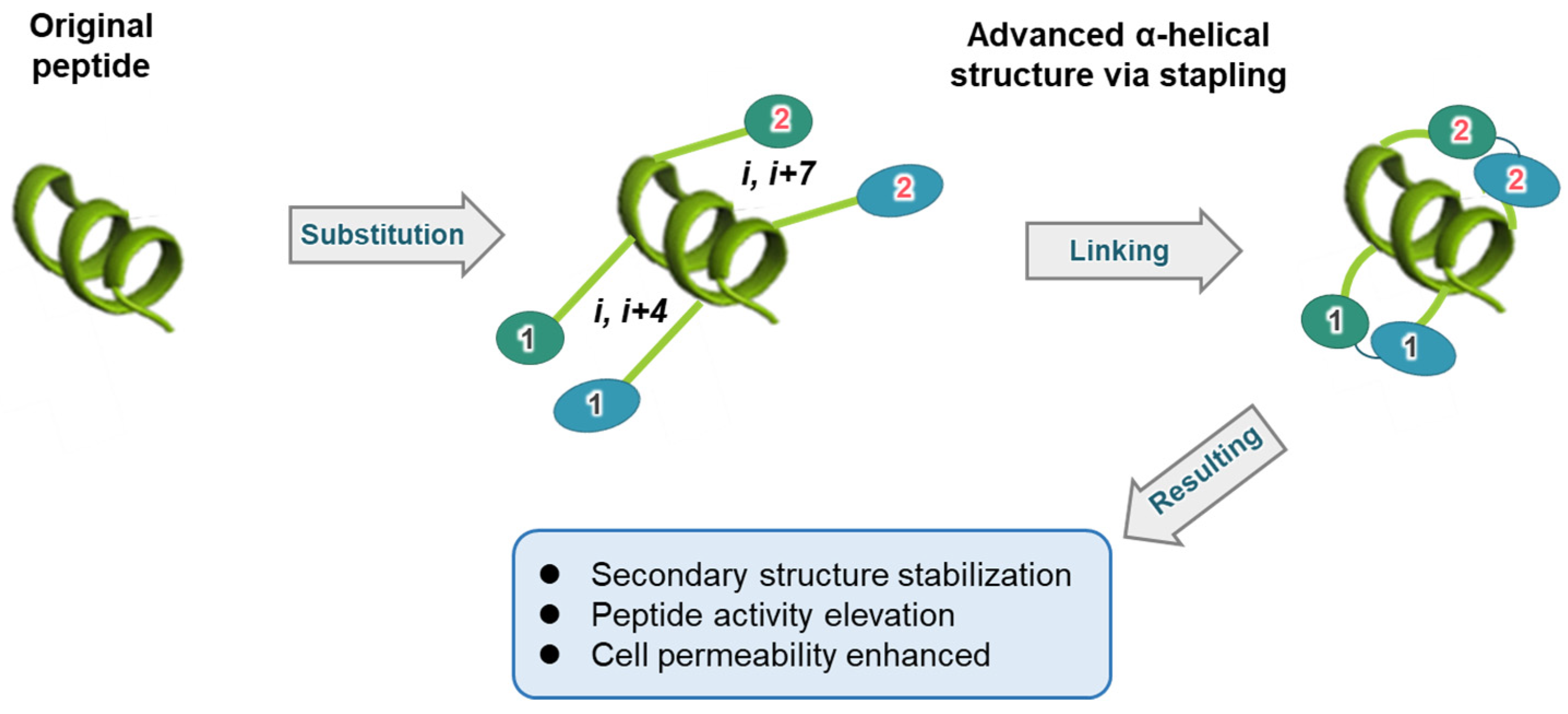
1. Introduction
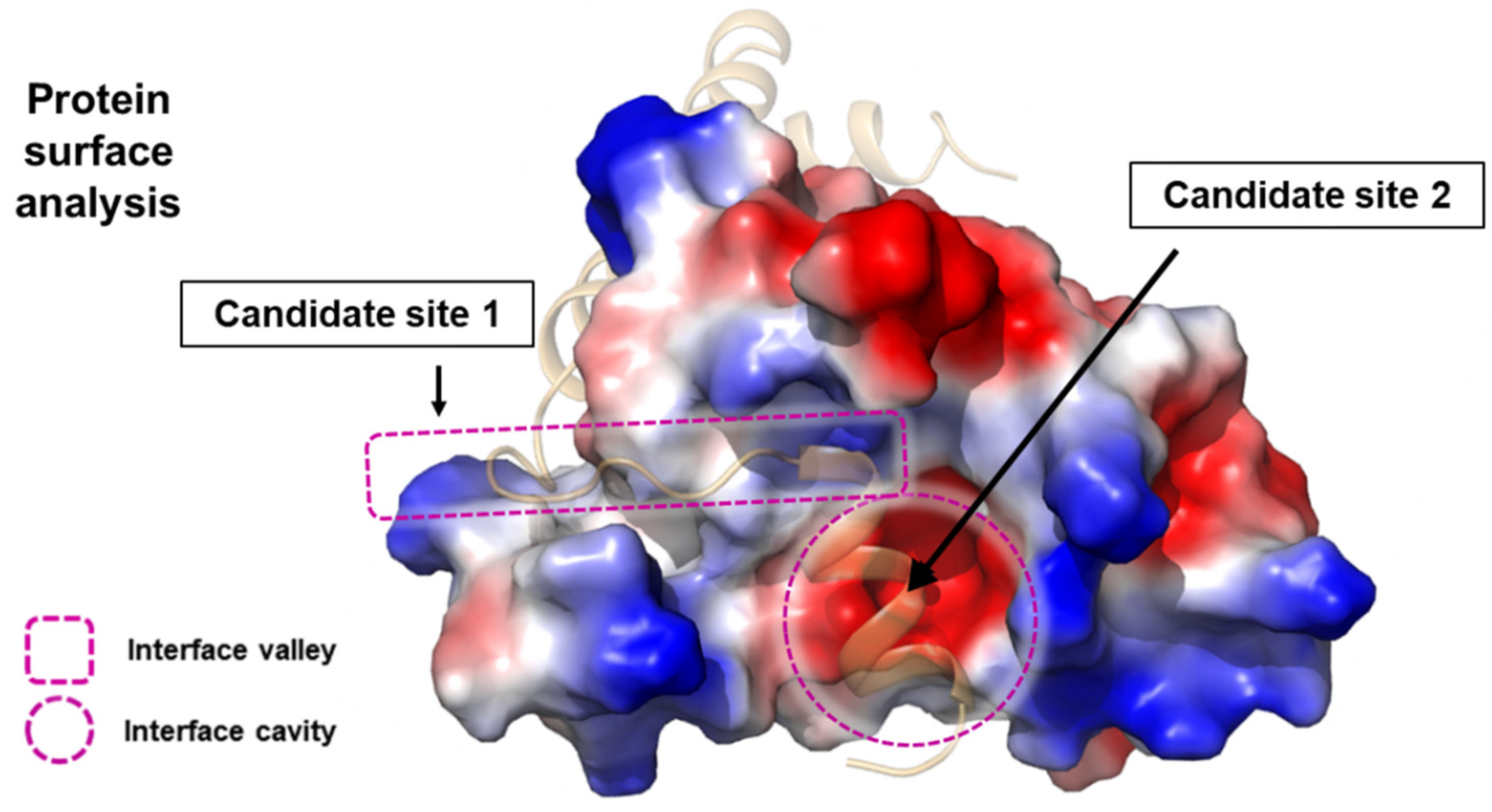
2. Structure-Based Approach
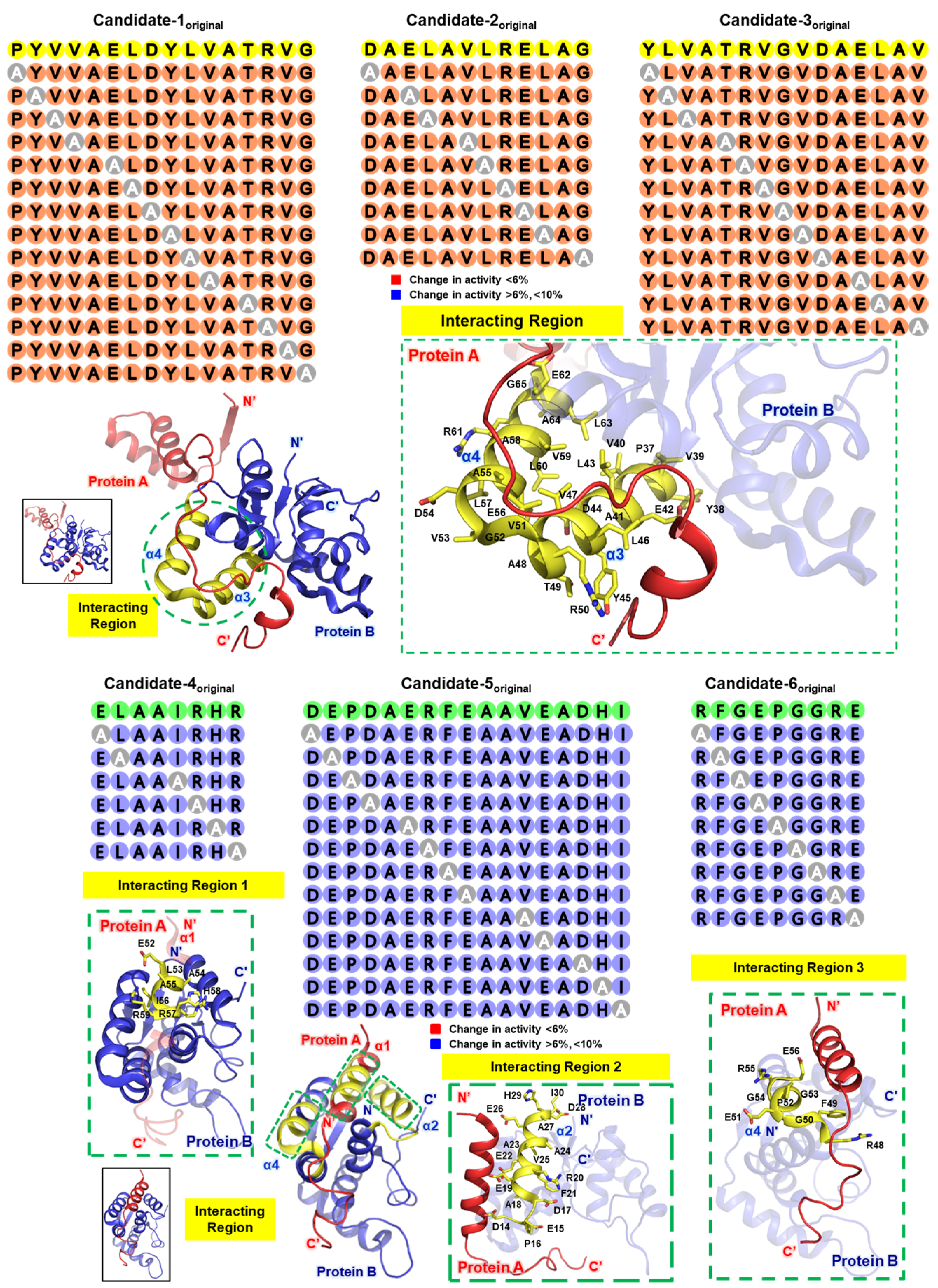
3. Selection of Stapling Residues
4. Alanine Scanning
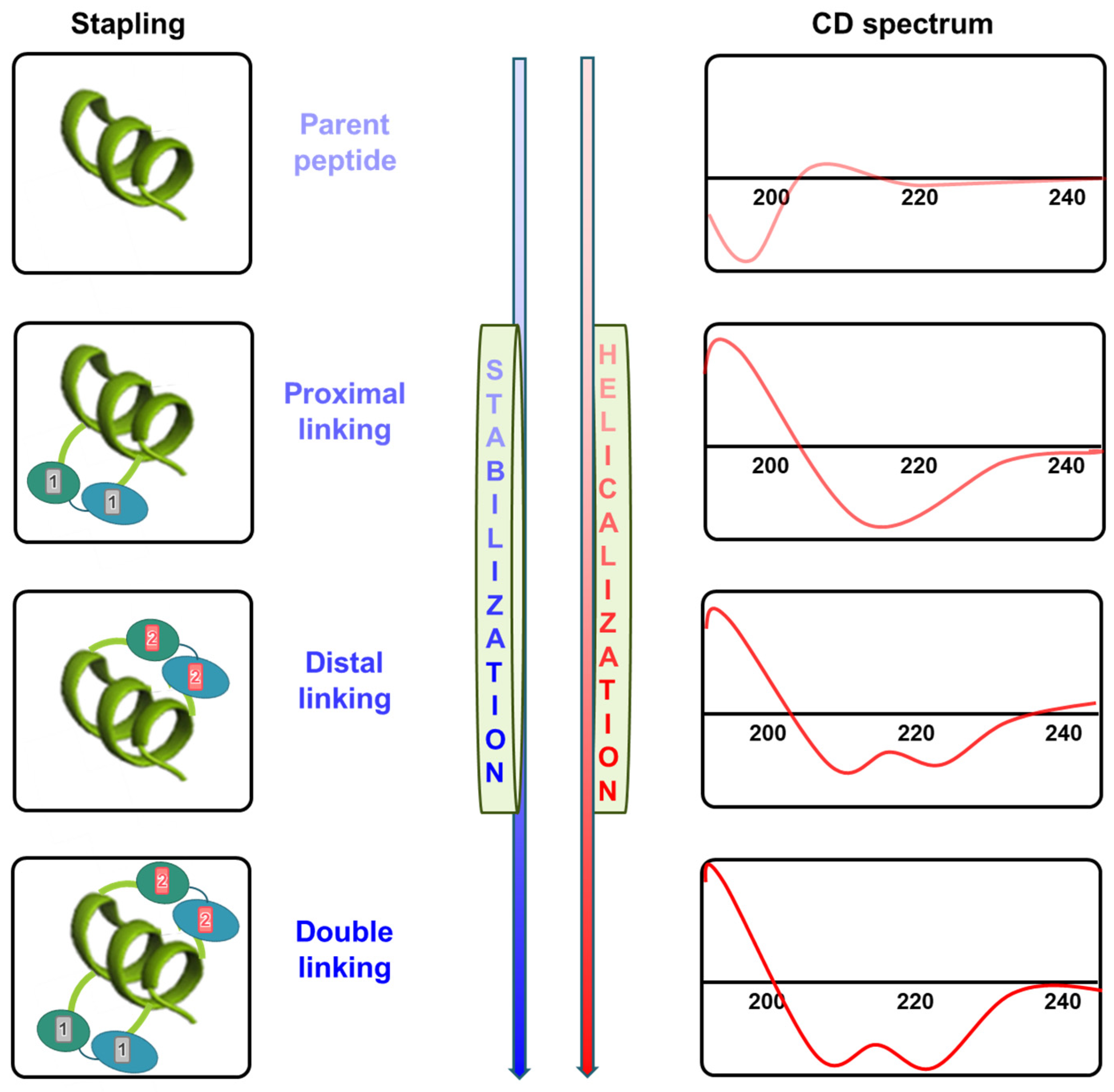
5. CD Spectroscopy
6. Stability Confirmation Post-Synthesis
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sharma, S.; Chauhan, A.; Ranjan, A.; Mathkor, D.M.; Haque, S.; Ramniwas, S.; Tuli, H.S.; Jindal, T.; Yadav, V. Emerging challenges in antimicrobial resistance: Implications for pathogenic microorganisms, novel antibiotics, and their impact on sustainability. Front. Microbiol. 2024, 15, 1403168. [Google Scholar] [CrossRef] [PubMed]
- Belete, M.A.; Gedefie, A.; Alemayehu, E.; Debash, H.; Mohammed, O.; Gebretsadik, D.; Ebrahim, H.; Tilahun, M. The prevalence of vancomycin-resistant Staphylococcus aureus in Ethiopia: A systematic review and meta-analysis. Antimicrob. Resist. Infect. Control 2023, 12, 86. [Google Scholar] [CrossRef] [PubMed]
- Hull, R.C.; Wright, R.C.T.; Sayers, J.R.; Sutton, J.A.F.; Rzaska, J.; Foster, S.J.; Brockhurst, M.A.; Condliffe, A.M. Antibiotics Limit Adaptation of Drug-Resistant Staphylococcus aureus to Hypoxia. Antimicrob. Agents Chemother. 2022, 66, e0092622. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Li, J.; Li, J.; Zhang, J.; Zhao, Y.; Li, W.; Zhang, Y.; Hu, J.; Xie, X.; Zhang, D.; et al. Design, synthesis, and evaluation of novel pleuromutilin aryl acrylate derivatives as promising broad-spectrum antibiotics especially for combatting multi-drug resistant gram-negative bacteria. Eur. J. Med. Chem. 2023, 259, 115653. [Google Scholar] [CrossRef]
- Christensen, S.B. Drugs That Changed Society: History and Current Status of the Early Antibiotics: Salvarsan, Sulfonamides, and beta-Lactams. Molecules 2021, 26, 6057. [Google Scholar] [CrossRef]
- Thakuria, B.; Lahon, K. The Beta Lactam Antibiotics as an Empirical Therapy in a Developing Country: An Update on Their Current Status and Recommendations to Counter the Resistance against Them. J. Clin. Diagn. Res. 2013, 7, 1207–1214. [Google Scholar] [CrossRef]
- Kanannejad, Z.; Pourvali, A.; Esmaeilzadeh, H.; Shokouhi Shoormasti, R.; Reza Fazlollahi, M.; Fallahpour, M.; Zaremehrjardi, F. Diagnosis and selection of alternative antibiotics in beta-lactams hypersensitivity reactions: Current recommendations and challenges. Int. Immunopharmacol. 2023, 122, 110573. [Google Scholar] [CrossRef]
- de Araujo, A.C.J.; Freitas, P.R.; Araujo, I.M.; Siqueira, G.M.; de Oliveira Borges, J.A.; Alves, D.S.; Miranda, G.M.; Dos Santos Nascimento, I.J.; de Araujo-Junior, J.X.; da Silva-Junior, E.F.; et al. Potentiating-antibiotic activity and absorption, distribution, metabolism, excretion and toxicity properties (ADMET) analysis of synthetic thiadiazines against multi-drug resistant (MDR) strains. Fundam. Clin. Pharmacol. 2024, 38, 84–98. [Google Scholar] [CrossRef]
- Carballo, G.M.; Vazquez, K.G.; Garcia-Gonzalez, L.A.; Rio, G.D.; Brizuela, C.A. Embedded-AMP: A Multi-Thread Computational Method for the Systematic Identification of Antimicrobial Peptides Embedded in Proteome Sequences. Antibiotics 2023, 12, 139. [Google Scholar] [CrossRef]
- Morales-Martinez, A.; Bertrand, B.; Hernandez-Meza, J.M.; Garduno-Juarez, R.; Silva-Sanchez, J.; Munoz-Garay, C. Membrane fluidity, composition, and charge affect the activity and selectivity of the AMP ascaphin-8. Biophys. J. 2022, 121, 3034–3048. [Google Scholar] [CrossRef]
- Park, Y.; Hahm, K.S. Novel short AMP: Design and activity study. Protein Pept. Lett. 2012, 19, 652–656. [Google Scholar] [CrossRef] [PubMed]
- Caselli, L.; Kohler, S.; Schirone, D.; Humphreys, B.; Malmsten, M. Conformational control of antimicrobial peptide amphiphilicity: Consequences for boosting membrane interactions and antimicrobial effects of photocatalytic TiO(2) nanoparticles. Phys. Chem. Chem. Phys. 2024, 26, 16529–16539. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Lee, S.; Lee, I.; Nam, H. AMP-BERT: Prediction of antimicrobial peptide function based on a BERT model. Protein Sci. 2023, 32, e4529. [Google Scholar] [CrossRef]
- Abdullah, S.J.; Mu, Y.; Bhattacharjya, S. Structures, Interactions and Activity of the N-Terminal Truncated Variants of Antimicrobial Peptide Thanatin. Antibiotics 2024, 13, 74. [Google Scholar] [CrossRef] [PubMed]
- Sancho-Vaello, E.; Francois, P.; Bonetti, E.J.; Lilie, H.; Finger, S.; Gil-Ortiz, F.; Gil-Carton, D.; Zeth, K. Structural remodeling and oligomerization of human cathelicidin on membranes suggest fibril-like structures as active species. Sci. Rep. 2017, 7, 15371. [Google Scholar] [CrossRef]
- Nielsen, J.E.; Alford, M.A.; Yung, D.B.Y.; Molchanova, N.; Fortkort, J.A.; Lin, J.S.; Diamond, G.; Hancock, R.E.W.; Jenssen, H.; Pletzer, D.; et al. Self-Assembly of Antimicrobial Peptoids Impacts Their Biological Effects on ESKAPE Bacterial Pathogens. ACS Infect. Dis. 2022, 8, 533–545. [Google Scholar] [CrossRef]
- Pirtskhalava, M.; Vishnepolsky, B.; Grigolava, M.; Managadze, G. Physicochemical Features and Peculiarities of Interaction of AMP with the Membrane. Pharmaceuticals 2021, 14, 471. [Google Scholar] [CrossRef]
- Wang, R.; Wang, T.; Zhuo, L.; Wei, J.; Fu, X.; Zou, Q.; Yao, X. Diff-AMP: Tailored designed antimicrobial peptide framework with all-in-one generation, identification, prediction and optimization. Brief. Bioinform. 2024, 25, bbae078. [Google Scholar] [CrossRef]
- Shabir, U.; Ali, S.; Magray, A.R.; Ganai, B.A.; Firdous, P.; Hassan, T.; Nazir, R. Fish antimicrobial peptides (AMP’s) as essential and promising molecular therapeutic agents: A review. Microb. Pathog. 2018, 114, 50–56. [Google Scholar] [CrossRef]
- Monteiro, C.; Fernandes, H.; Oliveira, D.; Vale, N.; Barbosa, M.; Gomes, P.; MC, L.M. AMP-Chitosan Coating with Bactericidal Activity in the Presence of Human Plasma Proteins. Molecules 2020, 25, 3046. [Google Scholar] [CrossRef]
- Kravchenko, S.V.; Domnin, P.A.; Grishin, S.Y.; Vershinin, N.A.; Gurina, E.V.; Zakharova, A.A.; Azev, V.N.; Mustaeva, L.G.; Gorbunova, E.Y.; Kobyakova, M.I.; et al. Enhancing the Antimicrobial Properties of Peptides through Cell-Penetrating Peptide Conjugation: A Comprehensive Assessment. Int. J. Mol. Sci. 2023, 24, 16723. [Google Scholar] [CrossRef] [PubMed]
- Zhang, O.L.; Niu, J.Y.; Yu, O.Y.; Mei, M.L.; Jakubovics, N.S.; Chu, C.H. Development of a Novel Peptide with Antimicrobial and Mineralising Properties for Caries Management. Pharmaceutics 2023, 15, 2560. [Google Scholar] [CrossRef]
- Soylemez, U.G.; Yousef, M.; Bakir-Gungor, B. Novel Antimicrobial Peptide Design Using Motif Match Score Representation. IEEE/ACM Trans. Comput. Biol. Bioinform. 2024, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Chandarana, C.; Juwarwala, I.; Shetty, S.; Bose, A. Peptide Drugs: Current Status and Treatment for the Treatment of Various Diseases. Curr. Drug Res. Rev. 2024, 16, 381–394. [Google Scholar] [CrossRef] [PubMed]
- Nicze, M.; Borowka, M.; Dec, A.; Niemiec, A.; Buldak, L.; Okopien, B. The Current and Promising Oral Delivery Methods for Protein- and Peptide-Based Drugs. Int. J. Mol. Sci. 2024, 25, 815. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Wang, J.; Wang, Q.; Wang, J.; Qin, S.; Li, W. Advances in peptide-based drug delivery systems. Heliyon 2024, 10, e26009. [Google Scholar] [CrossRef]
- Rizvi, S.F.A.; Zhang, H.; Fang, Q. Engineering peptide drug therapeutics through chemical conjugation and implication in clinics. Med. Res. Rev. 2024. [Google Scholar] [CrossRef]
- Moiola, M.; Memeo, M.G.; Quadrelli, P. Stapled Peptides-A Useful Improvement for Peptide-Based Drugs. Molecules 2019, 24, 3654. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, W.; Xu, L.; Tu, J.; Su, S.; Li, Q.; Zhang, T.; Zheng, L.; Wang, H.; Zhuang, X.; et al. Discovery of a Double-Stapled Short Peptide as a Long-Acting HIV-1 Inactivator with Potential for Oral Bioavailability. J. Med. Chem. 2024, 67, 9991–10004. [Google Scholar] [CrossRef]
- Dongrui, Z.; Miyamoto, M.; Yokoo, H.; Demizu, Y. Innovative peptide architectures: Advancements in foldamers and stapled peptides for drug discovery. Expert. Opin. Drug Discov. 2024, 19, 699–723. [Google Scholar] [CrossRef]
- Holdbrook, D.A.; Marzinek, J.K.; Boncel, S.; Boags, A.; Tan, Y.S.; Huber, R.G.; Verma, C.S.; Bond, P.J. The nanotube express: Delivering a stapled peptide to the cell surface. J. Colloid. Interface Sci. 2021, 604, 670–679. [Google Scholar] [CrossRef] [PubMed]
- Anananuchatkul, T.; Chang, I.V.; Miki, T.; Tsutsumi, H.; Mihara, H. Construction of a Stapled alpha-Helix Peptide Library Displayed on Phage for the Screening of Galectin-3-Binding Peptide Ligands. ACS Omega 2020, 5, 5666–5674. [Google Scholar] [CrossRef]
- Cornillie, S.P.; Bruno, B.J.; Lim, C.S.; Cheatham, T.E., 3rd. Computational Modeling of Stapled Peptides toward a Treatment Strategy for CML and Broader Implications in the Design of Lengthy Peptide Therapeutics. J. Phys. Chem. B 2018, 122, 3864–3875. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, E.E.; Menon, A.; Chmiel, A.F.; Deprey, K.; Kritzer, J.A.; Garner, A.L. A cell-penetrant lactam-stapled peptide for targeting eIF4E protein-protein interactions. Eur. J. Med. Chem. 2020, 205, 112655. [Google Scholar] [CrossRef]
- Ma, B.; Liu, D.; Zheng, M.; Wang, Z.; Zhang, D.; Jian, Y.; Ma, J.; Fan, Y.; Chen, Y.; Gao, Y.; et al. Development of a Double-Stapled Peptide Stabilizing Both alpha-Helix and beta-Sheet Structures for Degrading Transcription Factor AR-V7. JACS Au 2024, 4, 816–827. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Yin, F.; Li, Z. Helical Stabilization of Peptide Macrocycles by Stapled Architectures. Methods Mol. Biol. 2022, 2371, 391–409. [Google Scholar] [CrossRef]
- Zhang, J.; Dong, S. In-Bridge Stereochemistry: A Determinant of Stapled Peptide Conformation and Activity. Chembiochem 2024, 25, e202300747. [Google Scholar] [CrossRef]
- Fathi, F.; Alizadeh, B.; Tabarzad, M.V.; Tabarzad, M. Important structural features of antimicrobial peptides towards specific activity: Trends in the development of efficient therapeutics. Bioorg. Chem. 2024, 149, 107524. [Google Scholar] [CrossRef]
- Dougherty, P.G.; Wen, J.; Pan, X.; Koley, A.; Ren, J.G.; Sahni, A.; Basu, R.; Salim, H.; Appiah Kubi, G.; Qian, Z.; et al. Enhancing the Cell Permeability of Stapled Peptides with a Cyclic Cell-Penetrating Peptide. J. Med. Chem. 2019, 62, 10098–10107. [Google Scholar] [CrossRef]
- Ito, T.; Hashimoto, W.; Ohoka, N.; Misawa, T.; Inoue, T.; Kawano, R.; Demizu, Y. Structure-Activity Relationship Study of Helix-Stabilized Antimicrobial Peptides Containing Nonproteinogenic Amino Acids. ACS Biomater. Sci. Eng. 2023, 9, 4654–4661. [Google Scholar] [CrossRef]
- Feurstein, C.; Meyer, V.; Jung, S. Structure-Activity Predictions From Computational Mining of Protein Databases to Assist Modular Design of Antimicrobial Peptides. Front. Microbiol. 2022, 13, 812903. [Google Scholar] [CrossRef]
- Hillman, R.A.; Nadraws, J.W.; Bertucci, M.A. The Hydrocarbon Staple & Beyond: Recent Advances Towards Stapled Peptide Therapeutics that Target Protein-Protein Interactions. Curr. Top. Med. Chem. 2018, 18, 611–624. [Google Scholar] [CrossRef]
- Tan, Y.S.; Lane, D.P.; Verma, C.S. Stapled peptide design: Principles and roles of computation. Drug Discov. Today 2016, 21, 1642–1653. [Google Scholar] [CrossRef] [PubMed]
- Zhen, B.; Geng, C.; Yang, Y.; Liang, H.; Jiang, Y.; Li, X.; Ye, G. Systematic alanine and stapling mutational analysis of antimicrobial peptide Chem-KVL. Bioorg. Med. Chem. Lett. 2024, 107, 129794. [Google Scholar] [CrossRef] [PubMed]
- You, Y.; Liu, H.; Zhu, Y.; Zheng, H. Rational design of stapled antimicrobial peptides. Amino Acids 2023, 55, 421–442. [Google Scholar] [CrossRef] [PubMed]
- Hirano, M.; Saito, C.; Goto, C.; Yokoo, H.; Kawano, R.; Misawa, T.; Demizu, Y. Rational Design of Helix-Stabilized Antimicrobial Peptide Foldamers Containing alpha,alpha-Disubstituted Amino Acids or Side-Chain Stapling. Chempluschem 2020, 85, 2731–2736. [Google Scholar] [CrossRef]
- Valiente, P.A.; Becerra, D.; Kim, P.M. A Method to Calculate the Relative Binding Free Energy Differences of alpha-Helical Stapled Peptides. J. Org. Chem. 2020, 85, 1644–1651. [Google Scholar] [CrossRef]
- Bittrich, S.; Segura, J.; Duarte, J.M.; Burley, S.K.; Rose, Y. RCSB protein Data Bank: Exploring protein 3D similarities via comprehensive structural alignments. Bioinformatics 2024, 40, btae370. [Google Scholar] [CrossRef]
- Kang, S.M. Focused Overview of Mycobacterium tuberculosis VapBC Toxin-Antitoxin Systems Regarding Their Structural and Functional Aspects: Including Insights on Biomimetic Peptides. Biomimetics 2023, 8, 412. [Google Scholar] [CrossRef]
- Tu, L.; Wang, D.; Li, Z. Design and Synthetic Strategies for Helical Peptides. Methods Mol. Biol. 2019, 2001, 107–131. [Google Scholar] [CrossRef]
- Makura, Y.; Ueda, A.; Kato, T.; Iyoshi, A.; Higuchi, M.; Doi, M.; Tanaka, M. X-ray Crystallographic Structure of alpha-Helical Peptide Stabilized by Hydrocarbon Stapling at i,i + 1 Positions. Int. J. Mol. Sci. 2021, 22, 5364. [Google Scholar] [CrossRef] [PubMed]
- McWhinnie, F.S.; Sepp, K.; Wilson, C.; Kunath, T.; Hupp, T.R.; Baker, T.S.; Houston, D.R.; Hulme, A.N. Mono-Substituted Hydrocarbon Diastereomer Combinations Reveal Stapled Peptides with High Structural Fidelity. Chemistry 2018, 24, 2094–2097. [Google Scholar] [CrossRef] [PubMed]
- Lighezan, L.; Georgieva, R.; Neagu, A. The secondary structure and the thermal unfolding parameters of the S-layer protein from Lactobacillus salivarius. Eur. Biophys. J. 2016, 45, 491–509. [Google Scholar] [CrossRef] [PubMed]
- Glibowicka, M.; He, S.; Deber, C.M. Enhanced proteolytic resistance of cationic antimicrobial peptides through lysine side chain analogs and cyclization. Biochem. Biophys. Res. Commun. 2022, 612, 105–109. [Google Scholar] [CrossRef]
- Grison, C.M.; Burslem, G.M.; Miles, J.A.; Pilsl, L.K.A.; Yeo, D.J.; Imani, Z.; Warriner, S.L.; Webb, M.E.; Wilson, A.J. Double quick, double click reversible peptide “stapling”. Chem. Sci. 2017, 8, 5166–5171. [Google Scholar] [CrossRef]
- Nero, T.L.; Parker, M.W.; Morton, C.J. Protein structure and computational drug discovery. Biochem. Soc. Trans. 2018, 46, 1367–1379. [Google Scholar] [CrossRef]
- Winter, A.; Higueruelo, A.P.; Marsh, M.; Sigurdardottir, A.; Pitt, W.R.; Blundell, T.L. Biophysical and computational fragment-based approaches to targeting protein-protein interactions: Applications in structure-guided drug discovery. Q. Rev. Biophys. 2012, 45, 383–426. [Google Scholar] [CrossRef]
- Zhang, Y.; Guo, J.; Cheng, J.; Zhang, Z.; Kang, F.; Wu, X.; Chu, Q. High-Throughput Screening of Stapled Helical Peptides in Drug Discovery. J. Med. Chem. 2023, 66, 95–106. [Google Scholar] [CrossRef]
- Al Musaimi, O.; Lombardi, L.; Williams, D.R.; Albericio, F. Strategies for Improving Peptide Stability and Delivery. Pharmaceuticals 2022, 15, 1283. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, D.-H.; Kang, S.-M. Stapled Peptides: An Innovative and Ultimate Future Drug Offering a Highly Powerful and Potent Therapeutic Alternative. Biomimetics 2024, 9, 537. https://doi.org/10.3390/biomimetics9090537
Kim D-H, Kang S-M. Stapled Peptides: An Innovative and Ultimate Future Drug Offering a Highly Powerful and Potent Therapeutic Alternative. Biomimetics. 2024; 9(9):537. https://doi.org/10.3390/biomimetics9090537
Chicago/Turabian StyleKim, Do-Hee, and Sung-Min Kang. 2024. "Stapled Peptides: An Innovative and Ultimate Future Drug Offering a Highly Powerful and Potent Therapeutic Alternative" Biomimetics 9, no. 9: 537. https://doi.org/10.3390/biomimetics9090537
APA StyleKim, D.-H., & Kang, S.-M. (2024). Stapled Peptides: An Innovative and Ultimate Future Drug Offering a Highly Powerful and Potent Therapeutic Alternative. Biomimetics, 9(9), 537. https://doi.org/10.3390/biomimetics9090537







