Advancement in Cancer Vasculogenesis Modeling through 3D Bioprinting Technology
Abstract
1. Introduction
2. Conventional Research Methods for Modeling Cancer Vasculogenesis
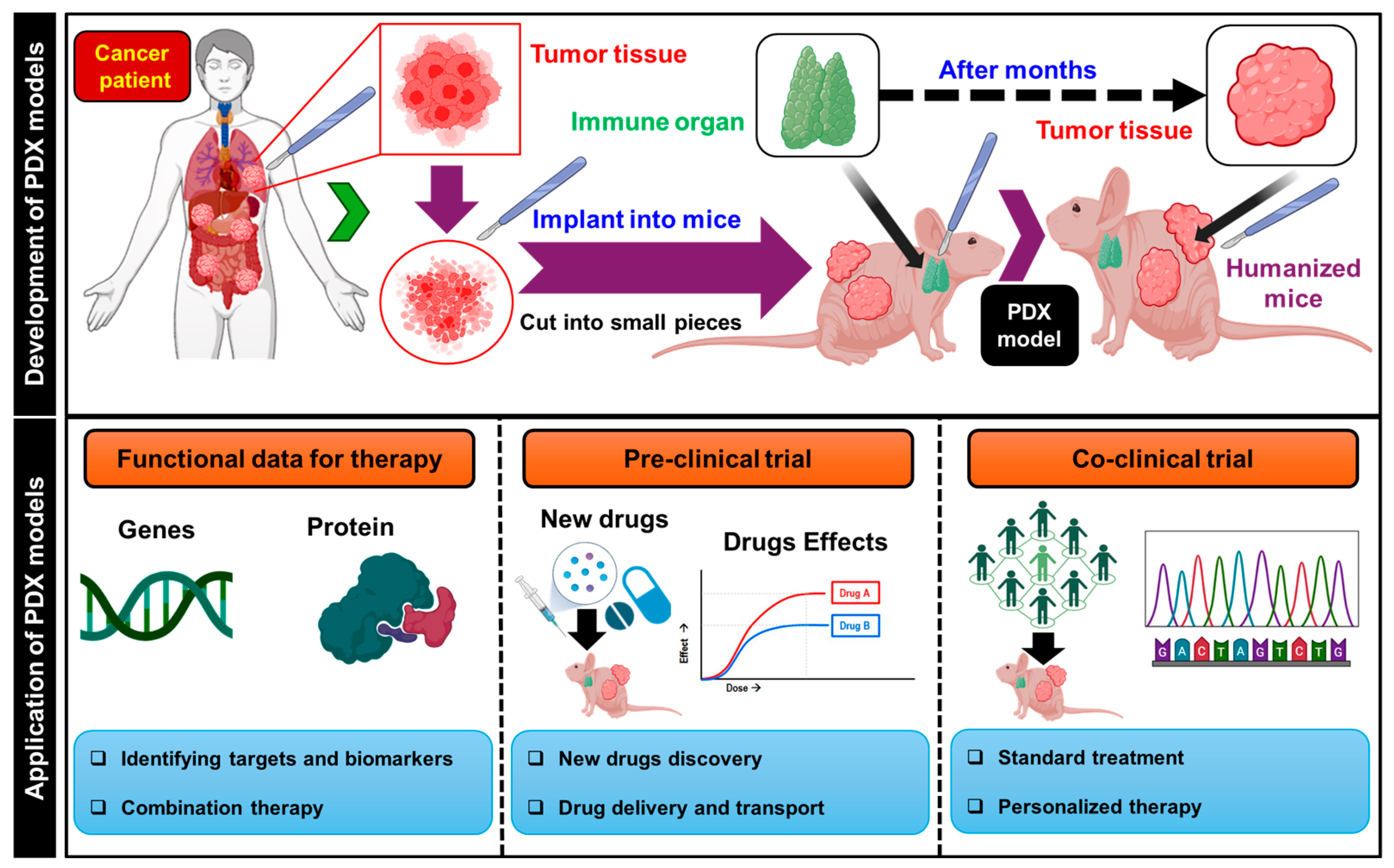
2.1. Study of Cancer Vasculogenesis through Patient-Derived Xenograft (PDX) Models in Conventional Research Methods
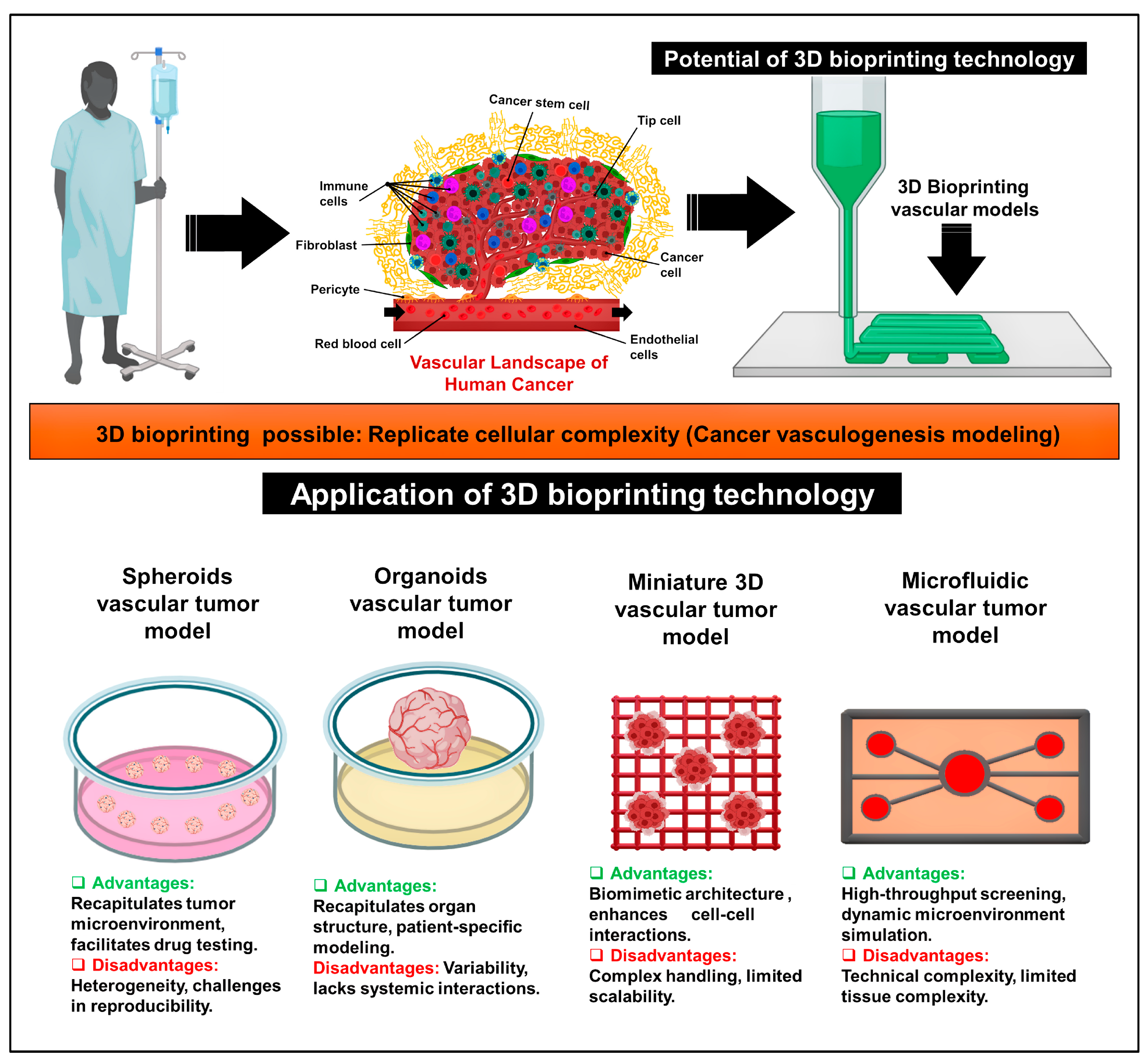
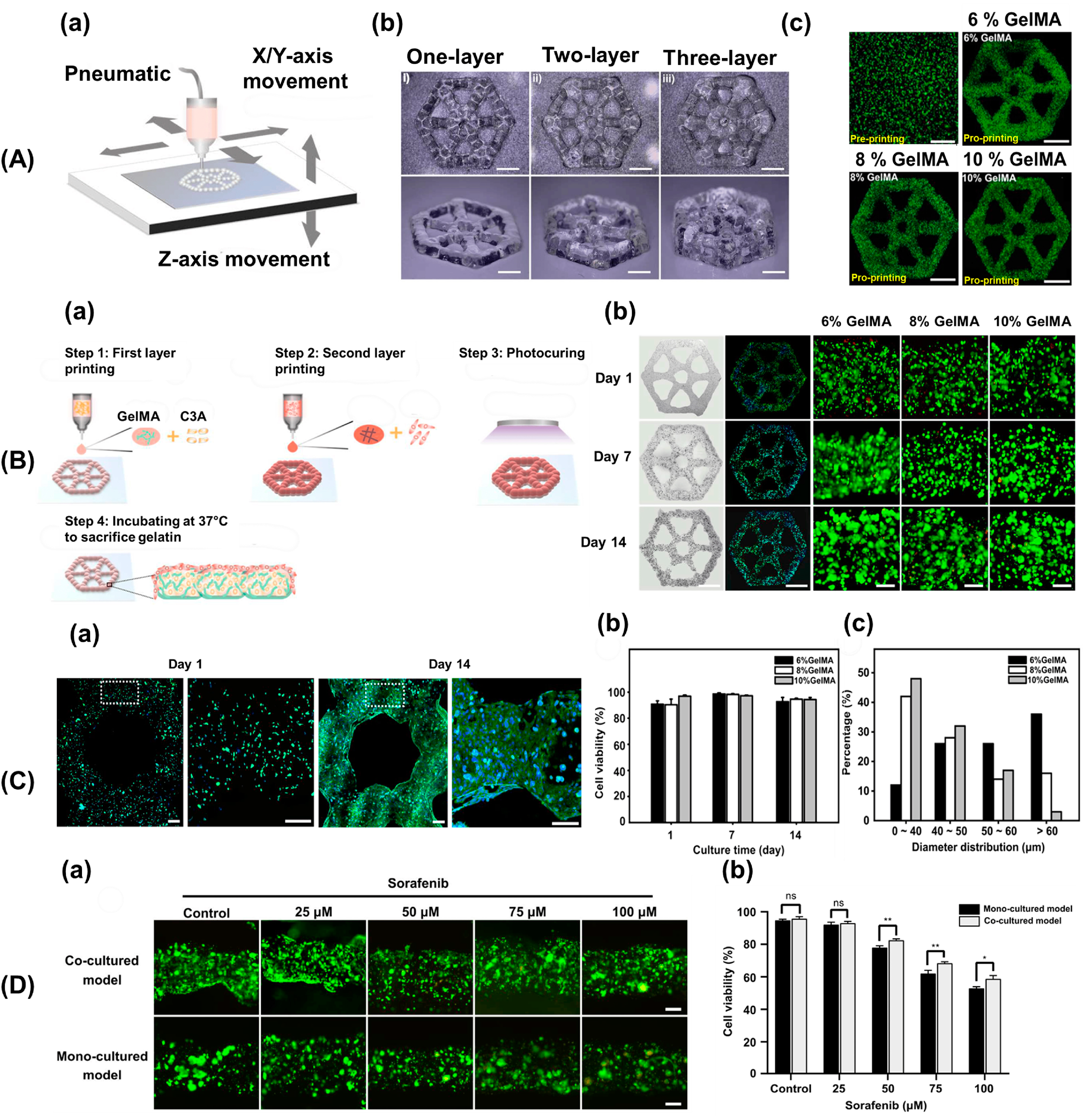
2.2. Application of 3D Bioprinting in the Development of Cancer Vasculogenesis Models
3. Advancements in 3D Bioprinting for Modeling Cancer Vasculogenesis
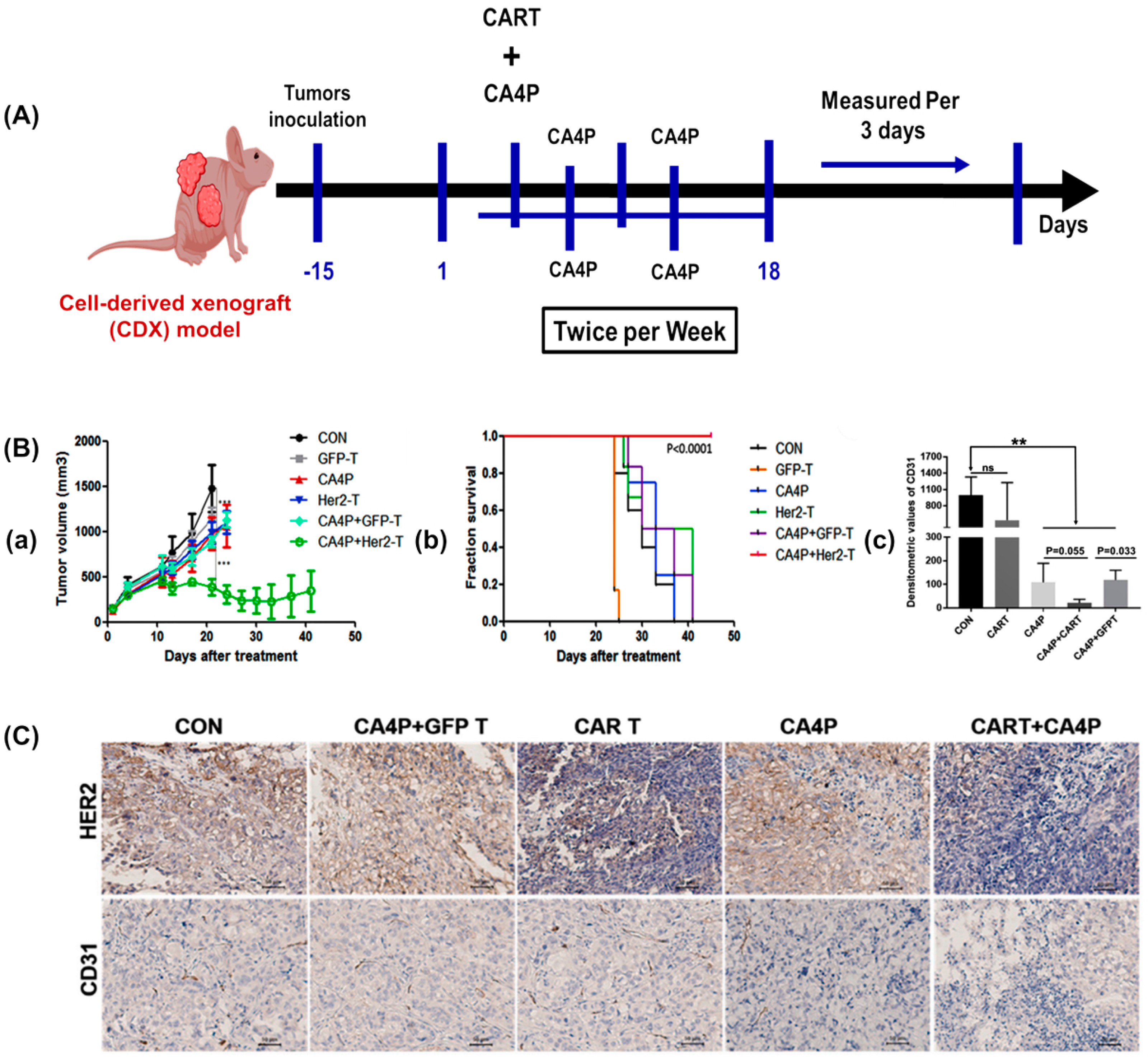
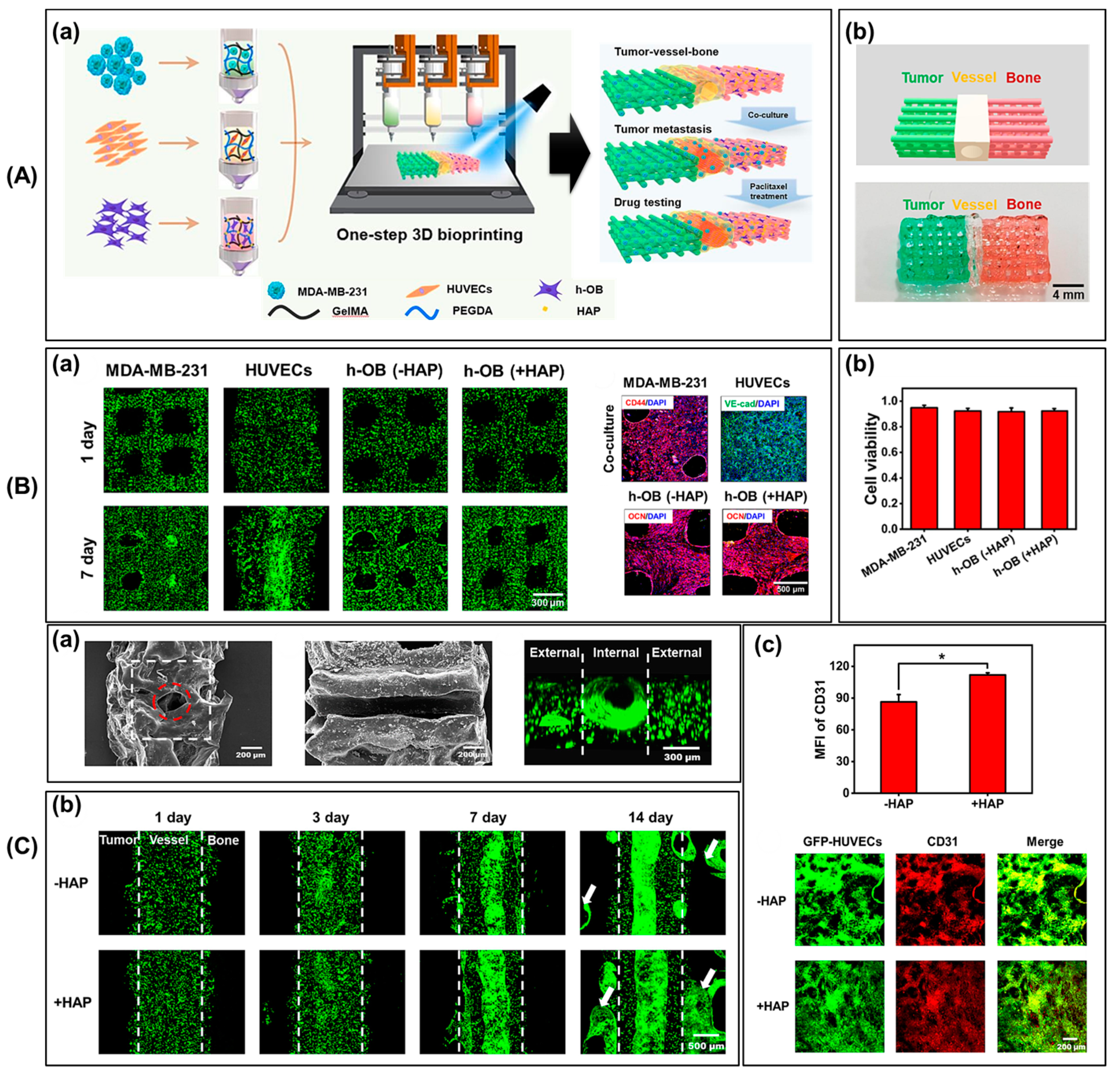
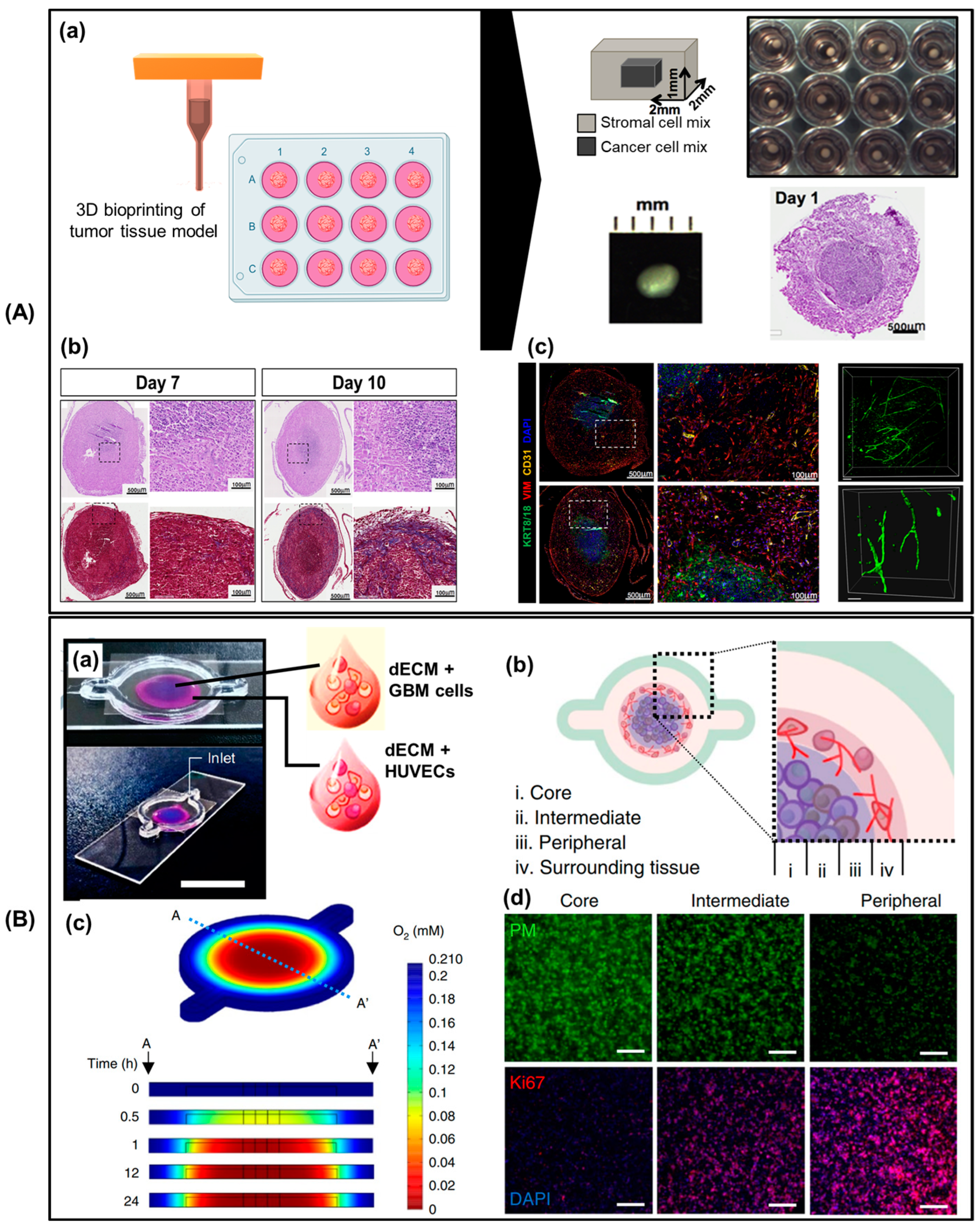
3.1. Pathological Study
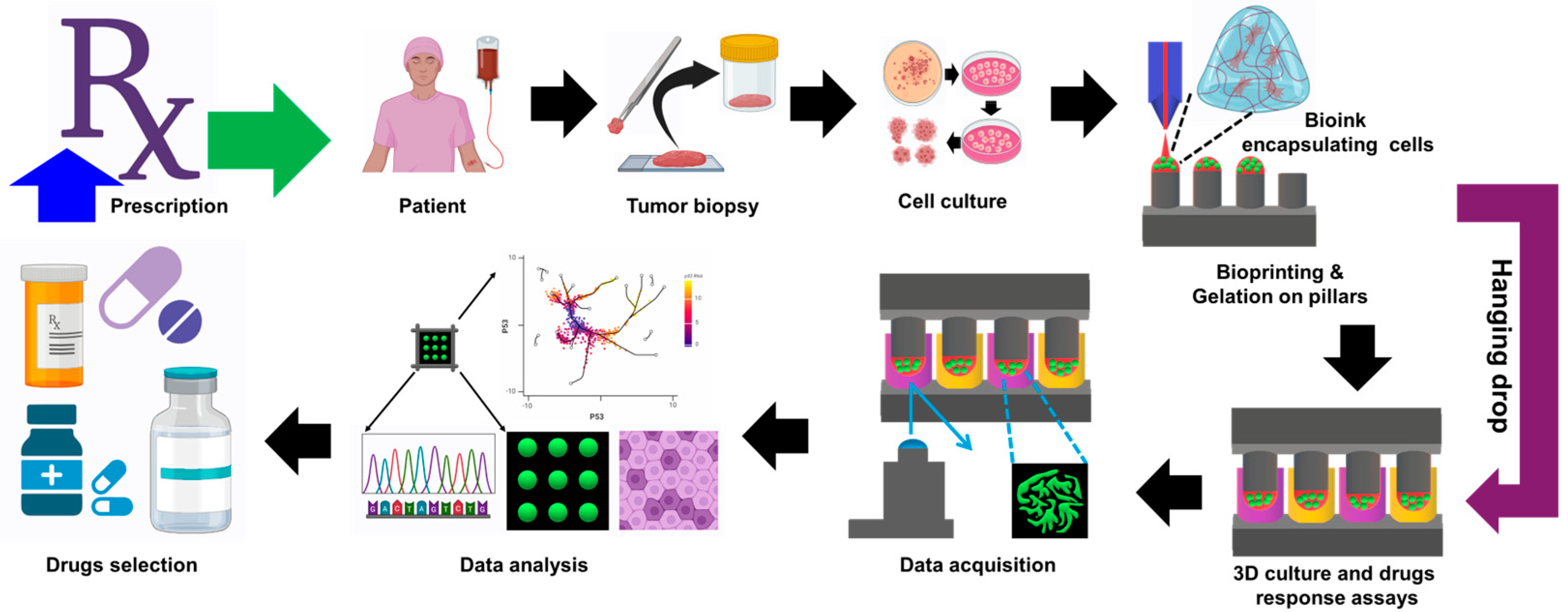
3.2. Preclinical Drug Screening Platform for Personalized Medicine
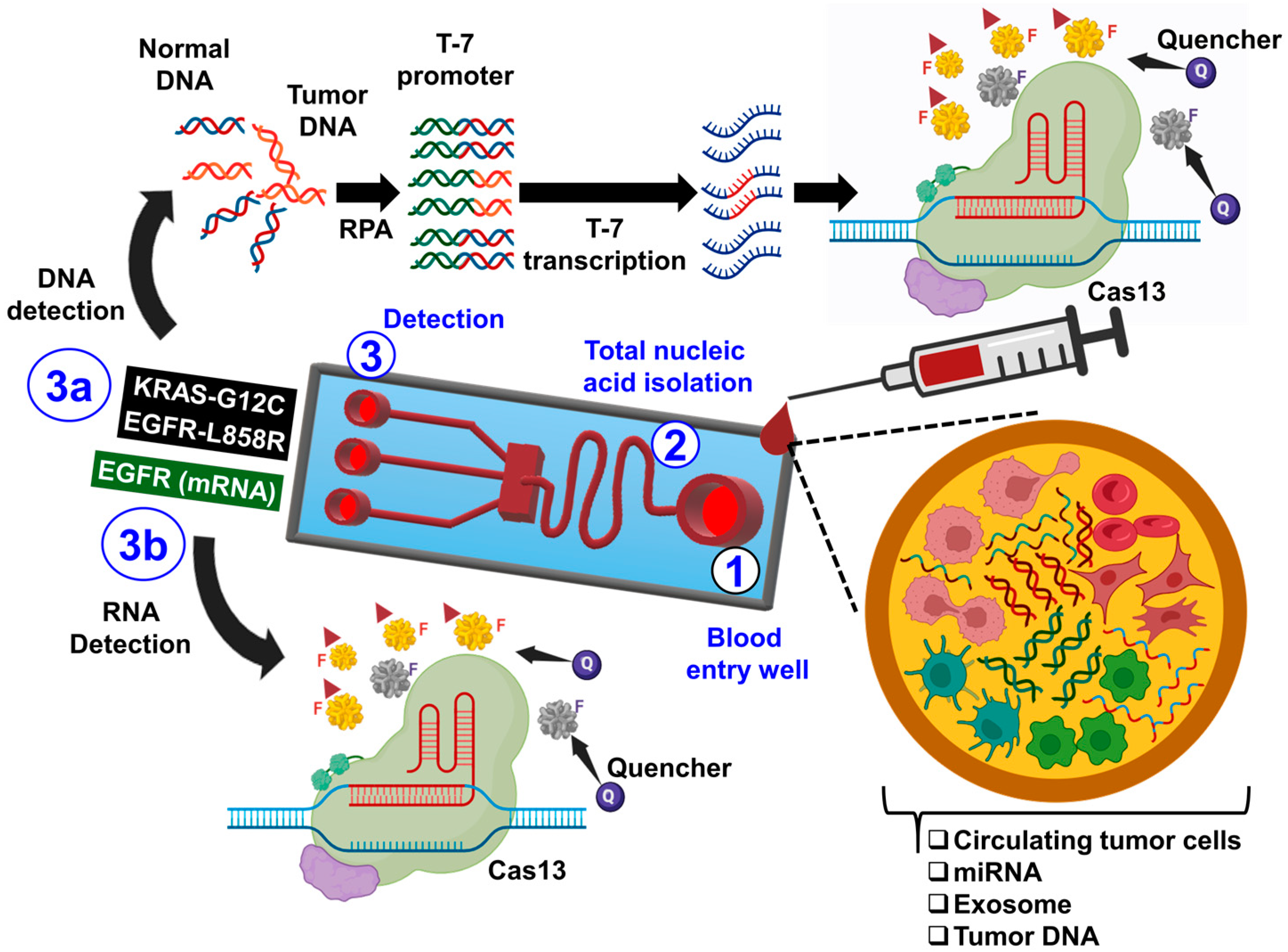
3.3. Cancer Diagnosis
4. Limitation of Cancer Vasculogenesis Models
5. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baiguera, S.; Ribatti, D. Ribatti, Endothelialization approaches for viable engineered tissues. Angiogenesis 2013, 16, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Yin, S.; Zhang, W.; Zhang, Z.; Jiang, X. Recent advances in scaffold design and material for vascularized tissue-engineered bone regeneration. Adv. Healthc. Mater. 2019, 8, 1801433. [Google Scholar] [CrossRef] [PubMed]
- Hantusch, B. Morphological and functional characteristics of blood and lymphatic vessels. In Fundamentals of Vascular Biology; Springer: Berlin/Heidelberg, Germany, 2019; pp. 1–43. [Google Scholar]
- Fleischer, S.; Tavakol, D.N.; Vunjak-Novakovic, G. From arteries to capillaries: Approaches to engineering human vasculature. Adv. Funct. Mater. 2020, 30, 1910811. [Google Scholar] [CrossRef] [PubMed]
- Popel, A.S. Theory of oxygen transport to tissue. Crit. Rev. Biomed. Eng. 1989, 17, 257. [Google Scholar]
- Monahan-Earley, R.; Dvorak, A.M.; Aird, W.C. Evolutionary origins of the blood vascular system and endothelium. J. Thromb. Haemost. 2013, 11, 46–66. [Google Scholar] [CrossRef] [PubMed]
- Pittman, R.N. Regulation of Tissue Oxygenation; Morgan & Claypool Life Sciences: Kentfield, CA, USA, 2016; pp. 3–16. [Google Scholar]
- Zhao, X.; Xu, Z.; Xiao, L.; Shi, T.; Xiao, H.; Wang, Y.; Li, Y.; Xue, F.; Zeng, W. Review on the vascularization of organoids and organoids-on-a-C hip. Front. Bioeng. Biotechnol. 2021, 9, 637048. [Google Scholar] [CrossRef] [PubMed]
- Naderi-Meshkin, H.; Cornelius, V.A.; Eleftheriadou, M.; Potel, K.N.; Setyaningsih, W.A.W.; Margariti, A. Vascular organoids: Unveiling advantages, applications, challenges, and disease modelling strategies. Stem Cell Res. Ther. 2023, 14, 292. [Google Scholar] [CrossRef] [PubMed]
- Devillard, C.D.; Marquette, C.A. Vascular tissue engineering: Challenges and requirements for an ideal large scale blood vessel. Front. Bioeng. Biotechnol. 2021, 9, 721843. [Google Scholar] [CrossRef]
- Laschke, M.W.; Gu, Y.; Menger, M.D. Replacement in angiogenesis research: Studying mechanisms of blood vessel development by animal-free in vitro, in vivo and in silico approaches. Front. Physiol. 2022, 13, 981161. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, N.; Chouhan, D.; Mandal, B.B. Tissue engineered skin and wound healing: Current strategies and future directions. Curr. Pharm. Des. 2017, 23, 3455–3482. [Google Scholar] [CrossRef]
- Choi, J.; Lee, E.J.; Jang, W.B.; Kwon, S.-M. Development of Biocompatible 3D-Printed Artificial Blood Vessels through Multidimensional Approaches. J. Funct. Biomater. 2023, 14, 497. [Google Scholar] [CrossRef] [PubMed]
- Shukla, A.K.; Gao, G.; Kim, B.S. Applications of 3D bioprinting technology in induced pluripotent stem cells-based tissue engineering. Micromachines 2022, 13, 155. [Google Scholar] [CrossRef] [PubMed]
- Schaaf, M.B.; Garg, A.D.; Agostinis, P. Defining the role of the tumor vasculature in antitumor immunity and immunotherapy. Cell Death Dis. 2018, 9, 115. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.S.; Duchamp, M.; Oklu, R.; Ellisen, L.W.; Langer, R.; Khademhosseini, A. Bioprinting the cancer microenvironment. ACS Biomater. Sci. Eng. 2016, 2, 1710–1721. [Google Scholar] [CrossRef] [PubMed]
- Shukla, A.K.; Lee, D.; Yoon, S.; Ahn, M.; Kim, B.S. Vascularization strategies for human skin tissue engineering via 3D bioprinting. Int. J. Bioprinting 2024, 1727. [Google Scholar] [CrossRef]
- Cho, W.; Ahn, M.; Kim, B.S.; Cho, D. Blood-lymphatic integrated system with heterogeneous melanoma spheroids via in-bath three-dimensional bioprinting for modelling of combinational targeted therapy. Adv. Sci. 2022, 9, 2202093. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.S.; Cho, W.W.; Gao, G.; Ahn, M.; Kim, J.; Cho, D.W. Construction of tissue-level cancer-vascular model with high-precision position control via in situ 3D Cell printing. Small Methods 2021, 5, 2100072. [Google Scholar] [CrossRef] [PubMed]
- Ahn, M.; Cho, W.; Lee, H.; Park, W.; Lee, S.; Back, J.W.; Gao, Q.; Gao, G.; Cho, D.; Kim, B.S. Engineering of uniform epidermal layers via sacrificial gelatin bioink-assisted 3D extrusion bioprinting of skin. Adv. Healthc. Mater. 2023, 12, 2301015. [Google Scholar] [CrossRef]
- Liu, X.; Fang, J.; Huang, S.; Wu, X.; Xie, X.; Wang, J.; Liu, F.; Zhang, M.; Peng, Z.; Hu, N. Tumor-on-a-chip: From bioinspired design to biomedical application. Microsyst. Nanoeng. 2021, 7, 50. [Google Scholar] [CrossRef]
- Flores-Torres, S.; Dimitriou, N.M.; Pardo, L.A.; Kort-Mascort, J.; Pal, S.; Peza-Chavez, O.; Kuasne, H.; Berube, J.; Bertos, N.; Park, M.; et al. Bioprinted Multicomponent Hydrogel Co-culture Tumor-Immune Model for Assessing and Simulating Tumor-Infiltrated Lymphocyte Migration and Functional Activation. ACS Appl. Mater. Interfaces 2023, 15, 33250–33262. [Google Scholar] [CrossRef]
- Gao, G.; Park, W.; Kim, B.S.; Ahn, M.; Chae, S.; Cho, W.; Kim, J.; Lee, J.Y.; Jang, J.; Cho, D. Construction of a novel in vitro atherosclerotic model from geometry-tunable artery equivalents engineered via in-bath coaxial cell printing. Adv. Funct. Mater. 2021, 31, 2008878. [Google Scholar] [CrossRef]
- Barrs, R.W.; Jia, J.; Silver, S.E.; Yost, M.; Mei, Y. Biomaterials for bioprinting microvasculature. Chem. Rev. 2020, 120, 10887–10949. [Google Scholar] [CrossRef] [PubMed]
- Seymour, A.J.; Westerfield, A.D.; Cornelius, V.C.; A Skylar-Scott, M.; Heilshorn, S.C. Bioprinted microvasculature: Progressing from structure to function. Biofabrication 2022, 14, 022002. [Google Scholar] [CrossRef] [PubMed]
- Jouybar, M.; de Winde, C.M.; Wolf, K.; Friedl, P.; Mebius, R.E.; Toonder, J.M.D. Cancer-on-chip models for metastasis: Importance of the tumor microenvironment. Trends Biotechnol. 2023, 42, 431–448. [Google Scholar] [CrossRef]
- Benavente, S.; Sánchez-García, A.; Naches, S.; Lleonart, M.E.; Lorente, J. Therapy-induced modulation of the tumor microenvironment: New opportunities for cancer therapies. Front. Oncol. 2020, 10, 582884. [Google Scholar] [CrossRef]
- Fontana, F.; Marzagalli, M.; Sommariva, M.; Gagliano, N.; Limonta, P. In Vitro 3D cultures to model the tumor microenvironment. Cancers 2021, 13, 2970. [Google Scholar] [CrossRef] [PubMed]
- Augustine, R.; Kalva, S.N.; Ahmad, R.; Zahid, A.A.; Hasan, S.; Nayeem, A.; McClements, L.; Hasan, A. 3D Bioprinted cancer models: Revolutionizing personalized cancer therapy. Transl. Oncol. 2021, 14, 101015. [Google Scholar] [CrossRef]
- Wang, X.; Luo, Y.; Ma, Y.; Wang, P.; Yao, R. Converging bioprinting and organoids to better recapitulate the tumor microenvironment. Trends Biotechnol. 2023, 42, 648–663. [Google Scholar] [CrossRef]
- Esmaeili, Y.; Bidram, E.; Bigham, A.; Atari, M.; Azadani, R.N.; Tavakoli, M.; Salehi, S.; Mirhaj, M.; Basiri, A.; Mirzavandi, Z.; et al. Exploring the evolution of tissue engineering strategies over the past decade: From cell-based strategies to gene-activated matrix. Alex. Eng. J. 2023, 81, 137–169. [Google Scholar] [CrossRef]
- Xie, M.; Shi, Y.; Zhang, C.; Ge, M.; Zhang, J.; Chen, Z.; Fu, J.; Xie, Z.; He, Y. In situ 3D bioprinting with bioconcrete bioink. Nat. Commun. 2022, 13, 3597. [Google Scholar] [CrossRef]
- Murphy, S.V.; Atala, A. 3D bioprinting of tissues and organs. Nat. Biotechnol. 2014, 32, 773–785. [Google Scholar] [CrossRef] [PubMed]
- Park, W.; Lee, J.-S.; Gao, G.; Kim, B.S.; Cho, D.-W. 3D bioprinted multilayered cerebrovascular conduits to study cancer extravasation mechanism related with vascular geometry. Nat. Commun. 2023, 14, 7696. [Google Scholar] [CrossRef] [PubMed]
- Ahn, M.; Cho, W.; Kim, B.S.; Cho, D. Engineering densely packed adipose tissue via environmentally controlled in-bath 3D Bioprinting. Adv. Funct. Mater. 2022, 32, 2200203. [Google Scholar] [CrossRef]
- Lin, M.; Tang, M.; Duan, W.; Xia, S.; Liu, W.; Wang, Q. 3D Bioprinting for Tumor Metastasis Research. ACS Biomater. Sci. Eng. 2023, 9, 3116–3133. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, T. Biomaterial-based in vitro 3D modeling of glioblastoma multiforme. Cancer Pathog. Ther. 2023, 1, 177–194. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.S.; Ahn, M.; Cho, W.-W.; Gao, G.; Jang, J.; Cho, D.-W. Engineering of diseased human skin equivalent using 3D cell printing for representing pathophysiological hallmarks of type 2 diabetes in vitro. Biomaterials 2021, 272, 120776. [Google Scholar] [CrossRef] [PubMed]
- Shukla, A.K.; Dey, N.; Nandi, P.; Ranjan, M. Acellular dermis as a dermal matrix of tissue engineered skin substitute for burns treatment. Ann Public Health Res 2015, 2, 1023. [Google Scholar]
- Li, Y.; Liu, J.; Xu, S.; Wang, J. 3D Bioprinting: An Important Tool for Tumor Microenvironment Research. Int. J. Nanomed. 2023, ume 18, 8039–8057. [Google Scholar] [CrossRef]
- Chitty, J.L.; Filipe, E.C.; Lucas, M.C.; Herrmann, D.; Cox, T.R.; Timpson, P. Recent advances in understanding the complexities of metastasis. F1000Research 2018, 7, 1169. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Su, Q.; Zhang, X.; Yang, W.; Ning, J.; Jia, K.; Xin, J.; Li, H.; Yu, L.; Liao, Y.; et al. Recent advances of organ-on-a-chip in cancer modeling research. Biosensors 2022, 12, 1045. [Google Scholar] [CrossRef]
- Parodi, I.; Di Lisa, D.; Pastorino, L.; Scaglione, S.; Fato, M.M. 3D bioprinting as a powerful technique for recreating the tumor microenvironment. Gels 2023, 9, 482. [Google Scholar] [CrossRef]
- Sánchez-Salazar, M.G.; MÁlvarez, M.; Santiago, G.T.-D. Advances in 3D bioprinting for the biofabrication of tumor models. Bioprinting 2021, 21, e00120. [Google Scholar] [CrossRef]
- Xiang, Y.; Miller, K.; Guan, J.; Kiratitanaporn, W.; Tang, M.; Chen, S. 3D bioprinting of complex tissues in vitro: State-of-the-art and future perspectives. Arch. Toxicol. 2022, 96, 691–710. [Google Scholar] [CrossRef]
- Chae, S.; Ha, D.-H.; Lee, H. 3D bioprinting strategy for engineering vascularized tissue models. Int. J. Bioprinting 2023, 9, 15–33. [Google Scholar] [CrossRef] [PubMed]
- Xing, F.; Xu, J.; Yu, P.; Zhou, Y.; Zhe, M.; Luo, R.; Liu, M.; Xiang, Z.; Duan, X.; Ritz, U. Recent advances in biofabrication strategies based on bioprinting for vascularized tissue repair and regeneration. Mater. Des. 2023, 229, 111885. [Google Scholar] [CrossRef]
- You, S.; Xiang, Y.; Hwang, H.H.; Berry, D.B.; Kiratitanaporn, W.; Guan, J.; Yao, E.; Tang, M.; Zhong, Z.; Ma, X.; et al. High cell density and high-resolution 3D bioprinting for fabricating vascularized tissues. Sci. Adv. 2023, 9, eade7923. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.-X.; Wu, Z.; Hou, Y.-Y.; Fang, Z.-X.; Deng, Y.; Wu, H.-T.; Liu, J. Application of three-dimensional (3D) bioprinting in anti-cancer therapy. Heliyon 2023, 9, e20475. [Google Scholar] [CrossRef] [PubMed]
- Cauli, E.; Polidoro, M.A.; Marzorati, S.; Bernardi, C.; Rasponi, M.; Lleo, A. Cancer-on-chip: A 3D model for the study of the tumor microenvironment. J. Biol. Eng. 2023, 17, 53. [Google Scholar] [CrossRef] [PubMed]
- Yeo, M.; Sarkar, A.; Singh, Y.P.; Derman, I.D.; Datta, P.; Ozbolat, I.T. Synergistic coupling between 3D bioprinting and vascularization strategies. Biofabrication 2023, 16, 012003. [Google Scholar] [CrossRef]
- Swaminathan, S.; Hamid, Q.; Sun, W.; Clyne, A.M. Bioprinting of 3D breast epithelial spheroids for human cancer models. Biofabrication 2019, 11, 025003. [Google Scholar] [CrossRef]
- De Moor, L.; Merovci, I.; Baetens, S.; Verstraeten, J.; Kowalska, P.; Krysko, D.V.; De Vos, W.H.; Declercq, H. High-throughput fabrication of vascularized spheroids for bioprinting. Biofabrication 2018, 10, 035009. [Google Scholar] [CrossRef] [PubMed]
- Rawal, P.; Tripathi, D.M.; Ramakrishna, S.; Kaur, S. Prospects for 3D bioprinting of organoids. Bio-Des. Manuf. 2021, 4, 627–640. [Google Scholar] [CrossRef]
- Choi, Y.-M.; Lee, H.; Ann, M.; Song, M.; Rheey, J.; Jang, J. 3D bioprinted vascularized lung cancer organoid models with underlying disease capable of more precise drug evaluation. Biofabrication 2023, 15, 034104. [Google Scholar] [CrossRef] [PubMed]
- Yigci, D.; Sarabi, M.R.; Ustun, M.; Atceken, N.; Sokullu, E.; Bagci-Onder, T.; Tasoglu, S. 3D bioprinted glioma models. Prog. Biomed. Eng. 2022, 4, 042001. [Google Scholar] [CrossRef]
- Mehrotra, S.; Moses, J.C.; Bandyopadhyay, A.; Mandal, B.B. 3D printing/bioprinting based tailoring of in vitro tissue models: Recent advances and challenges. ACS Appl. Bio Mater. 2019, 2, 1385–1405. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Sun, Q.; Pei, J. Microfluidic-based 3D engineered microvascular networks and their applications in vascularized microtumor models. Micromachines 2018, 9, 493. [Google Scholar] [CrossRef] [PubMed]
- Radhakrishnan, J.; Varadaraj, S.; Dash, S.K.; Sharma, A.; Verma, R.S. Organotypic cancer tissue models for drug screening: 3D constructs, bioprinting and microfluidic chips. Drug Discov. Today 2020, 25, 879–890. [Google Scholar] [CrossRef] [PubMed]
- Germain, N.; Dhayer, M.; Dekiouk, S.; Marchetti, P. Current advances in 3D bioprinting for cancer modeling and personalized medicine. Int. J. Mol. Sci. 2022, 23, 3432. [Google Scholar] [CrossRef] [PubMed]
- Abuwatfa, W.H.; Pitt, W.G.; Husseini, G.A. Scaffold-based 3D cell culture models in cancer research. J. Biomed. Sci. 2024, 31, 7. [Google Scholar] [CrossRef]
- Neufeld, L.; Yeini, E.; Pozzi, S.; Satchi-Fainaro, R. 3D bioprinted cancer models: From basic biology to drug development. Nat. Rev. Cancer 2022, 22, 679–692. [Google Scholar] [CrossRef]
- Gao, G.; Ahn, M.; Cho, W.-W.; Kim, B.-S.; Cho, D.-W. 3D printing of pharmaceutical application: Drug screening and drug delivery. Pharmaceutics 2021, 13, 1373. [Google Scholar] [CrossRef]
- Khalil, A.S.; Jaenisch, R.; Mooney, D.J. Engineered tissues and strategies to overcome challenges in drug development. Adv. Drug Deliv. Rev. 2020, 158, 116–139. [Google Scholar] [CrossRef]
- Fang, Y.; Eglen, R.M. Three-dimensional cell cultures in drug discovery and development. Slas Discov. Adv. Life Sci. R&D 2017, 22, 456–472. [Google Scholar]
- Pognan, F.; Beilmann, M.; Boonen, H.C.M.; Czich, A.; Dear, G.; Hewitt, P.; Mow, T.; Oinonen, T.; Roth, A.; Steger-Hartmann, T.; et al. The evolving role of investigative toxicology in the pharmaceutical industry. Nat. Rev. Drug Discov. 2023, 22, 317–335. [Google Scholar] [CrossRef] [PubMed]
- Sztankovics, D.; Moldvai, D.; Petővári, G.; Gelencsér, R.; Krencz, I.; Raffay, R.; Dankó, T.; Sebestyén, A. 3D bioprinting and the revolution in experimental cancer model systems—A review of developing new models and experiences with in vitro 3D bioprinted breast cancer tissue-mimetic structures. Pathol. Oncol. Res. 2023, 29, 1610996. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Farach-Carson, M.C.; Jia, X. Three-dimensional in vitro tumor models for cancer research and drug evaluation. Biotechnol. Adv. 2014, 32, 1256–1268. [Google Scholar] [CrossRef]
- Santo, V.E.; Rebelo, S.P.; Estrada, M.F.; Alves, P.M.; Boghaert, E.; Brito, C. Drug screening in 3D in vitro tumor models: Overcoming current pitfalls of efficacy read-outs. Biotechnol. J. 2017, 12, 1600505. [Google Scholar] [CrossRef]
- Hickman, J.A.; Graeser, R.; De Hoogt, R.; Vidic, S.; Brito, C.; Gutekunst, M.; Van Der Kuip, H.; IMI PREDECT consortium. Three-dimensional models of cancer for pharmacology and cancer cell biology: Capturing tumor complexity in vitro/ex vivo. Biotechnol. J. 2014, 9, 1115–1128. [Google Scholar] [CrossRef] [PubMed]
- Mao, S.; Pang, Y.; Liu, T.; Shao, Y.; He, J.; Yang, H.; Mao, Y.; Sun, W. Bioprinting of in vitro tumor models for personalized cancer treatment: A review. Biofabrication 2020, 12, 042001. [Google Scholar] [CrossRef]
- Marfe, G.; Mirone, G.; Shukla, A.; Di Stefano, C.; Atta-ur-Rahman. Molecular and therapeutic clues in chronic myeloid leukemia. In Frontiers in Clinical Drug Research: Hematology; Atta-ur-Rahman, Ed.; Bentham Science Publishers: Sharjah, The United Arab Emirates, 2016; pp. 3–80. [Google Scholar]
- Zhang, Y. 3D Printing for Cancer Diagnosis: What Unique Advantages Are Gained? ACS Mater. Au 2023, 3, 620–635. [Google Scholar] [CrossRef]
- McGuckin, C.; Forraz, N.; Milet, C.; Lacroix, M.; Sbirkov, Y.; Sarafian, V.; Ebel, C.; Spindler, A.; Koerper, V.; Balloul, J.-M.; et al. World’s First Long-Term Colorectal Cancer Model by 3D Bioprinting as a Mechanism for Screening Oncolytic Viruses. Cancers 2023, 15, 4724. [Google Scholar] [CrossRef] [PubMed]
- Shukla, A.K. Genome Editing and Personalized Medicine Is a Next Generation of Solid Cancer Treatment for Targeting the Cancer Epigenome. In Proceedings of the Multidisciplinary International Conference on Green Earth: A Panoromic View, Maharashtra, India, 12–13 January 2018; pp. 236–239. [Google Scholar]
- Hwang, D.G.; Choi, Y.-M.; Jang, J. 3D bioprinting-based vascularized tissue models mimicking tissue-specific architecture and pathophysiology for in vitro studies. Front. Bioeng. Biotechnol. 2021, 9, 685507. [Google Scholar] [CrossRef] [PubMed]
- Song, K.; Zu, X.; Du, Z.; Hu, Z.; Wang, J.; Li, J. Diversity models and applications of 3d breast tumor-on-a-chip. Micromachines 2021, 12, 814. [Google Scholar] [CrossRef] [PubMed]
- Edri, S.; Frisch, A.N.; Safina, D.; Machour, M.; Zavin, J.; Landsman, L.; Pierreux, C.E.; Spagnoli, F.M.; Levenberg, S. 3D Bioprinting of Multicellular Stem Cell-Derived Constructs to Model Pancreatic Cell Differentiation. Adv. Funct. Mater. 2024, 2315488. [Google Scholar] [CrossRef]
- Tomás-Bort, E.; Kieler, M.; Sharma, S.; Candido, J.B.; Loessner, D. 3D approaches to model the tumor microenvironment of pancreatic cancer. Theranostics 2020, 10, 5074. [Google Scholar] [CrossRef]
- Pacheco, C.; Martins, C.; Monteiro, J.; Baltazar, F.; Costa, B.M.; Sarmento, B. Glioblastoma vasculature: From its critical role in tumor survival to relevant in vitro modelling. Front. Drug Deliv. 2022, 2, 823412. [Google Scholar] [CrossRef]
- Baghban, R.; Roshangar, L.; Jahanban-Esfahlan, R.; Seidi, K.; Ebrahimi-Kalan, A.; Jaymand, M.; Kolahian, S.; Javaheri, T.; Zare, P. Tumor microenvironment complexity and therapeutic implications at a glance. Cell Commun. Signal. 2020, 18, 59. [Google Scholar] [CrossRef] [PubMed]
- Niu, G.; Chen, X. Vascular endothelial growth factor as an anti-angiogenic target for cancer therapy. Curr. Drug Targets 2010, 11, 1000–1017. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.-L.; Chen, H.-H.; Zheng, L.-L.; Sun, L.-P.; Shi, L. Angiogenic signaling pathways and anti-angiogenic therapy for cancer. Signal Transduct. Target. Ther. 2023, 8, 198. [Google Scholar] [CrossRef]
- Huskin, G.; Chen, J.; Davis, T.; Jun, H.-W. Tissue-engineered 3D in vitro disease models for high-throughput drug screening. Tissue Eng. Regen. Med. 2023, 20, 523–538. [Google Scholar] [CrossRef]
- Ahn, M.; Cho, W.-W.; Park, W.; Lee, J.-S.; Choi, M.-J.; Gao, Q.; Gao, G.; Cho, D.-W.; Kim, B.S. 3D biofabrication of diseased human skin models in vitro. Biomater. Res. 2023, 27, 80. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Koo, I.S.; Hwang, H.J.; Lee, D.W. In Vitro three-dimensional (3D) cell culture tools for spheroid and organoid models. SLAS Discov. 2023, in press.
- Suarez, A.C.; JHammel, H.; Munson, J.M. Modeling lymphangiogenesis: Pairing in vitro and in vivo metrics. Microcirculation 2023, 30, e12802. [Google Scholar] [CrossRef]
- Rahman, H.S.; Tan, B.L.; Othman, H.H.; Chartrand, M.S.; Pathak, Y.; Mohan, S.; Abdullah, R.; Alitheen, N.B. An overview of in vitro, in vivo, and computational techniques for cancer-associated angiogenesis studies. BioMed Res. Int. 2020, 2020, 8857428. [Google Scholar] [CrossRef] [PubMed]
- Vilanova, G.; Colominas, I.; Gomez, H. Computational modeling of tumor-induced angiogenesis. Arch. Comput. Methods Eng. 2017, 24, 1071–1102. [Google Scholar] [CrossRef]
- Subramanian, A.; Zakeri, P.; Mousa, M.; Alnaqbi, H.; Alshamsi, F.Y.; Bettoni, L.; Damiani, E.; Alsafar, H.; Saeys, Y.; Carmeliet, P. Angiogenesis goes computational–The future way forward to discover new angiogenic targets? Comput. Struct. Biotechnol. J. 2022, 20, 5235–5255. [Google Scholar] [CrossRef] [PubMed]
- Vitale, C.; Marzagalli, M.; Scaglione, S.; Dondero, A.; Bottino, C.; Castriconi, R. Tumor microenvironment and hydrogel-based 3D cancer models for in vitro testing immunotherapies. Cancers 2022, 14, 1013. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Calar, K.; de la Puente, P. Mimicking tumor hypoxia and tumor-immune interactions employing three-dimensional in vitro models. J. Exp. Clin. Cancer Res. 2020, 39, 75. [Google Scholar] [CrossRef] [PubMed]
- Butler, K.S.; Brinker, C.J.; Leong, H.S. Bridging the in vitro to in vivo gap: Using the chick embryo model to accelerate nanoparticle validation and qualification for in vivo studies. ACS Nano 2022, 16, 19626–19650. [Google Scholar] [CrossRef]
- Hicklin, D.J.; Ellis, L.M. Role of the vascular endothelial growth factor pathway in tumor growth and angiogenesis. J. Clin. Oncol. 2005, 23, 1011–1027. [Google Scholar] [CrossRef]
- Ghalehbandi, S.; Yuzugulen, J.; Pranjol, Z.I.; Pourgholami, M.H. The role of VEGF in cancer-induced angiogenesis and research progress of drugs targeting VEGF. Eur. J. Pharmacol. 2023, 949, 175586. [Google Scholar] [CrossRef]
- Deng, C.; Zhao, J.; Zhou, S.; Dong, J.; Cao, J.; Gao, J.; Bai, Y.; Deng, H. The vascular disrupting agent CA4P improves the antitumor efficacy of CAR-T cells in preclinical models of solid human tumors. Mol. Ther. 2020, 28, 75–88. [Google Scholar] [CrossRef]
- He, J.; Zhang, C.; Ozkan, A.; Feng, T.; Duan, P.; Wang, S.; Yang, X.; Xie, J.; Liu, X. Patient-derived tumor models and their distinctive applications in personalized drug therapy. Mechanobiol. Med. 2023, 1, 100014. [Google Scholar] [CrossRef]
- Teleanu, R.I.; Chircov, C.; Grumezescu, A.M.; Teleanu, D.M. Tumor angiogenesis and anti-angiogenic strategies for cancer treatment. J. Clin. Med. 2019, 9, 84. [Google Scholar] [CrossRef] [PubMed]
- Genta, S.; Coburn, B.; Cescon, D.W.; Spreafico, A. Patient-derived cancer models: Valuable platforms for anticancer drug testing. Front. Oncol. 2022, 12, 976065. [Google Scholar] [CrossRef] [PubMed]
- Jubelin, C.; Muñoz-Garcia, J.; Griscom, L.; Cochonneau, D.; Ollivier, E.; Heymann, M.-F.; Vallette, F.M.; Oliver, L.; Heymann, D. Three-dimensional in vitro culture models in oncology research. Cell Biosci. 2022, 12, 155. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Wang, Y.; Yang, H. Revolutionizing preclinical research for pancreatic cancer: The potential of 3D bioprinting technology for personalized therapy. Hepatobiliary Surg. Nutr. 2023, 12, 616. [Google Scholar] [CrossRef]
- Datta, P.; Dey, M.; Ataie, Z.; Unutmaz, D.; Ozbolat, I.T. 3D bioprinting for reconstituting the cancer microenvironment. NPJ Precis. Oncol. 2020, 4, 18. [Google Scholar] [CrossRef]
- Fedi, A.; Vitale, C.; Ponschin, G.; Ayehunie, S.; Fato, M.; Scaglione, S. In vitro models replicating the human intestinal epithelium for absorption and metabolism studies: A systematic review. J. Control. Release 2021, 335, 247–268. [Google Scholar] [CrossRef]
- Kong, Z.; Wang, X. Bioprinting technologies and bioinks for vascular model establishment. Int. J. Mol. Sci. 2023, 24, 891. [Google Scholar] [CrossRef]
- Ozbolat, I.T.; Hospodiuk, M. Current advances and future perspectives in extrusion-based bioprinting. Biomaterials 2016, 76, 321–343. [Google Scholar] [CrossRef] [PubMed]
- Gudapati, H.; Dey, M.; Ozbolat, I. A comprehensive review on droplet-based bioprinting: Past, present and future. Biomaterials 2016, 102, 20–42. [Google Scholar] [CrossRef] [PubMed]
- Datta, P.; Ayan, B.; Ozbolat, I.T. Bioprinting for vascular and vascularized tissue biofabrication. Acta Biomater. 2017, 51, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Boland, T.; D’Lima, D.; Lotz, M.K. Thermal inkjet printing in tissue engineering and regenerative medicine. Recent Pat. Drug Deliv. Formul. 2012, 6, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Perez, M.R.; da Silva, V.A.; Thomsen, J.; Bhardwaj, L.; Andrade, T.A.M.; Alhussan, A.; Willerth, S.M. 3D bioprinting complex models of cancer. Biomater. Sci. 2023, 11, 3414–3430. [Google Scholar] [CrossRef]
- Hölzl, K.; Lin, S.; Tytgat, L.; Van Vlierberghe, S.; Gu, L.; Ovsianikov, A. Bioink properties before, during and after 3D bioprinting. Biofabrication 2016, 8, 032002. [Google Scholar] [CrossRef] [PubMed]
- Dababneh, A.B.; Ozbolat, I.T. Bioprinting technology: A current state-of-the-art review. J. Manuf. Sci. Eng. 2014, 136, 061016. [Google Scholar] [CrossRef]
- Guillotin, B.; Souquet, A.; Catros, S.; Duocastella, M.; Pippenger, B.; Bellance, S.; Bareille, R.; Rémy, M.; Bordenave, L.; Amédée, J.; et al. Laser assisted bioprinting of engineered tissue with high cell density and microscale organization. Biomaterials 2010, 31, 7250–7256. [Google Scholar] [CrossRef]
- Billiet, T.; Gevaert, E.; De Schryver, T.; Cornelissen, M.; Dubruel, P. The 3D printing of gelatin methacrylamide cell-laden tissue-engineered constructs with high cell viability. Biomaterials 2014, 35, 49–62. [Google Scholar] [CrossRef]
- Chimene, D.; Lennox, K.K.; Kaunas, R.R.; Gaharwar, A.K. Advanced bioinks for 3D printing: A materials science perspective. Ann. Biomed. Eng. 2016, 44, 2090–2102. [Google Scholar] [CrossRef]
- Rutz, A.L.; Hyland, K.E.; Jakus, A.E.; Burghardt, W.R.; Shah, R.N. A multi-material bioink method for 3D printing tunable, cell-compatible hydrogels. Adv. Mater. 2015, 27, 1607. [Google Scholar] [CrossRef]
- Xing, F.; Xiang, Z.; Rommens, P.M.; Ritz, U. 3D bioprinting for vascularized tissue-engineered bone fabrication. Materials 2020, 13, 2278. [Google Scholar] [CrossRef]
- Wang, Z.; Abdulla, R.; Parker, B.; Samanipour, R.; Ghosh, S.; Kim, K. A simple and high-resolution stereolithography-based 3D bioprinting system using visible light crosslinkable bioinks. Biofabrication 2015, 7, 045009. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, L.; Yao, R.; Zhao, Y.; Sun, W. Effect of bioink properties on printability and cell viability for 3D bioplotting of embryonic stem cells. Biofabrication 2016, 8, 035020. [Google Scholar] [CrossRef]
- Pati, F.; Ha, D.-H.; Jang, J.; Han, H.H.; Rhie, J.-W.; Cho, D.-W. Biomimetic 3D tissue printing for soft tissue regeneration. Biomaterials 2015, 62, 164–175. [Google Scholar] [CrossRef] [PubMed]
- Gao, G.; Yonezawa, T.; Hubbell, K.; Dai, G.; Cui, X. Inkjet-bioprinted acrylated peptides and PEG hydrogel with human mesenchymal stem cells promote robust bone and cartilage formation with minimal printhead clogging. Biotechnol. J. 2015, 10, 1568–1577. [Google Scholar] [CrossRef]
- Koch, L.; Deiwick, A.; Franke, A.; Schwanke, K.; Haverich, A.; Zweigerdt, R.; Chichkov, B. Laser bioprinting of human induced pluripotent stem cells—The effect of printing and biomaterials on cell survival, pluripotency, and differentiation. Biofabrication 2018, 10, 035005. [Google Scholar] [CrossRef] [PubMed]
- Guillemot, F.; Souquet, A.; Catros, S.; Guillotin, B.; Lopez, J.; Faucon, M.; Pippenger, B.; Bareille, R.; Rémy, M.; Bellance, S.; et al. High-throughput laser printing of cells and biomaterials for tissue engineering. Acta Biomater. 2010, 6, 2494–2500. [Google Scholar] [CrossRef]
- Ovsianikov, A.; Deiwick, A.; Van Vlierberghe, S.; Dubruel, P.; Möller, L.; Dräger, G.; Chichkov, B. Laser fabrication of three-dimensional CAD scaffolds from photosensitive gelatin for applications in tissue engineering. Biomacromolecules 2011, 12, 851–858. [Google Scholar] [CrossRef]
- Gruene, M.; Pflaum, M.; Deiwick, A.; Koch, L.; Schlie, S.; Unger, C.; Wilhelmi, M.; Haverich, A.; Chichkov, B.N. Adipogenic differentiation of laser-printed 3D tissue grafts consisting of human adipose-derived stem cells. Biofabrication 2011, 3, 015005. [Google Scholar] [CrossRef]
- Jiang, H.; Li, X.; Chen, T.; Liu, Y.; Wang, Q.; Wang, Z.; Jia, J. Bioprinted vascular tissue: Assessing functions from cellular, tissue to organ levels. Mater. Today Bio 2023, 23, 100846. [Google Scholar] [CrossRef] [PubMed]
- Ze, Y.; Li, Y.; Huang, L.; Shi, Y.; Li, P.; Gong, P.; Lin, J.; Yao, Y. Biodegradable inks in indirect three-dimensional bioprinting for tissue vascularization. Front. Bioeng. Biotechnol. 2022, 10, 856398. [Google Scholar] [CrossRef] [PubMed]
- Echeverria Molina, M.I.; Malollari, K.G.; Komvopoulos, K. Design challenges in polymeric scaffolds for tissue engineering. Front. Bioeng. Biotechnol. 2021, 9, 617141. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Xiong, Z.; Wang, X.; Yan, Y.; Liu, H.; Zhang, R. Direct fabrication of a hybrid cell/hydrogel construct by a double-nozzle assembling technology. J. Bioact. Compat. Polym. 2009, 24, 249–265. [Google Scholar]
- Skardal, A.; Zhang, J.; McCoard, L.; Xu, X.; Oottamasathien, S.; Prestwich, G.D. Photocrosslinkable hyaluronan-gelatin hydrogels for two-step bioprinting. Tissue Engineering Part A 2010, 16, 2675–2685. [Google Scholar] [CrossRef] [PubMed]
- Tan, E.Y.S.; Yeong, W.Y. Concentric bioprinting of alginate-based tubular constructs using multi-nozzle extrusion-based technique. Int. J. Bioprinting 2015, 1, 49–56. [Google Scholar] [CrossRef]
- Sasmal, P.; Datta, P.; Wu, Y.; Ozbolat, I.T. 3D bioprinting for modelling vasculature. Microphysiol. Syst. 2018, 2, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Hinton, T.J.; Jallerat, Q.; Palchesko, R.N.; Park, J.H.; Grodzicki, M.S.; Shue, H.-J.; Ramadan, M.H.; Hudson, A.R.; Feinberg, A.W. Three-dimensional printing of complex biological structures by freeform reversible embedding of suspended hydrogels. Sci. Adv. 2015, 1, e1500758. [Google Scholar] [CrossRef] [PubMed]
- Pinnock, C.B.; Meier, E.M.; Joshi, N.N.; Wu, B.; Lam, M.T. Customizable engineered blood vessels using 3D printed inserts. Methods 2016, 99, 20–27. [Google Scholar] [CrossRef]
- Jia, W.; Gungor-Ozkerim, P.S.; Zhang, Y.S.; Yue, K.; Zhu, K.; Liu, W.; Pi, Q.; Byambaa, B.; Dokmeci, M.R.; Shin, S.R.; et al. Direct 3D bioprinting of perfusable vascular constructs using a blend bioink. Biomaterials 2016, 106, 58–68. [Google Scholar] [CrossRef]
- Selvaganapathy, P.R.; Attalla, R. Microfluidic vascular channels in gels using commercial 3D printers. In Microfluidics, BioMEMS, and Medical Microsystems XIV; SPIE: San Francisco, USA, 2016. [Google Scholar]
- Cheng, S.; Li, Y.; Yu, C.; Deng, Z.; Huang, J.; Zhang, Z. 3D bioprinted tumor-vessel-bone co-culture scaffold for breast cancer bone metastasis modeling and drug testing. Chem. Eng. J. 2023, 476, 146685. [Google Scholar] [CrossRef]
- Lee, G.; Kim, S.J.; Park, J.-K. Fabrication of a self-assembled and vascularized tumor array via bioprinting on a microfluidic chip. Lab A Chip 2023, 23, 4079–4091. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Datta, P.; Shanmughapriya, S.; Ozbolat, I.T. 3D bioprinting of tumor models for cancer research. ACS Appl. Bio Mater. 2020, 3, 5552–5573. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.P.; Toksoy, Z.; Davis, B.A.; Geibel, J.P. 3D Bioprinting of Vascularized Tissues for in vitro and in vivo Applications. Front. Bioeng. Biotechnol. 2021, 9, 664188. [Google Scholar] [CrossRef]
- Dudley, A.C.; Griffioen, A.W. Pathological angiogenesis: Mechanisms and therapeutic strategies. Angiogenesis 2023, 26, 313–347. [Google Scholar] [CrossRef] [PubMed]
- Haas, G.; Fan, S.; Ghadimi, M.; De Oliveira, T.; Conradi, L.-C. Different forms of tumor vascularization and their clinical implications focusing on vessel co-option in colorectal cancer liver metastases. Front. Cell Dev. Biol. 2021, 9, 612774. [Google Scholar] [CrossRef] [PubMed]
- Bouchalova, P.; Bouchal, P. Current methods for studying metastatic potential of tumor cells. Cancer Cell Int. 2022, 22, 394. [Google Scholar] [CrossRef] [PubMed]
- Al-Ostoot, F.H.; Salah, S.; Khamees, H.A.; Khanum, S.A. Tumor angiogenesis: Current challenges and therapeutic opportunities. Cancer Treat. Res. Commun. 2021, 28, 100422. [Google Scholar] [CrossRef] [PubMed]
- Yi, H.-G.; Jeong, Y.H.; Kim, Y.; Choi, Y.-J.; Moon, H.E.; Park, S.H.; Kang, K.S.; Bae, M.; Jang, J.; Youn, H.; et al. A bioprinted human-glioblastoma-on-a-chip for the identification of patient-specific responses to chemoradiotherapy. Nat. Biomed. Eng. 2019, 3, 509–519. [Google Scholar] [CrossRef]
- Noor, N.; Shapira, A.; Edri, R.; Gal, I.; Wertheim, L.; Dvir, T. 3D printing of personalized thick and perfusable cardiac patches and hearts. Adv. Sci. 2019, 6, 1900344. [Google Scholar] [CrossRef]
- Gallego-Perez, D.; Higuita-Castro, N.; Denning, L.; DeJesus, J.; Dahl, K.; Sarkar, A.; Hansford, D.J. Microfabricated mimics of in vivo structural cues for the study of guided tumor cell migration. Lab A Chip 2012, 12, 4424–4432. [Google Scholar] [CrossRef] [PubMed]
- Langer, E.M.; Allen-Petersen, B.L.; King, S.M.; Kendsersky, N.D.; Turnidge, M.A.; Kuziel, G.M.; Riggers, R.; Samatham, R.; Amery, T.S.; Jacques, S.L.; et al. Modeling tumor phenotypes in vitro with three-dimensional bioprinting. Cell Rep. 2019, 26, 608–623.e6. [Google Scholar] [CrossRef] [PubMed]
- Roy, V.; Magne, B.; Vaillancourt-Audet, M.; Blais, M.; Chabaud, S.; Grammond, E.; Piquet, L.; Fradette, J.; Laverdière, I.; Moulin, V.J.; et al. Human organ-specific 3D cancer models produced by the stromal self-assembly method of tissue engineering for the study of solid tumors. BioMed Res. Int. 2020, 2020, 6051210. [Google Scholar] [CrossRef] [PubMed]
- Dey, M.; Kim, M.H.; Nagamine, M.; Dogan, M.; Kozhaya, L.; Unutmaz, D.; Ozbolat, I.T. 3D Bioprinted perfusable and vascularized breast tumor model for dynamic screening of chemotherapeutics and CAR-T cells. BioRxiv 2022. [Google Scholar] [CrossRef]
- González-Callejo, P.; Vázquez-Aristizabal, P.; García-Astrain, C.; de Aberasturi, D.J.; Henriksen-Lacey, M.; Izeta, A.; Liz-Marzán, L.M. 3D bioprinted breast tumor-stroma models for pre-clinical drug testing. Mater. Today Biol. 2023, 23, 100826. [Google Scholar] [CrossRef] [PubMed]
- de Andrés, J.L.; Ruiz-Toranzo, M.; Antich, C.; Chocarro-Wrona, C.; López-Ruíz, E.; Jiménez, G.; Marchal, J.A. Biofabrication of a tri-layered 3D-bioprinted CSC-based malignant melanoma model for personalized cancer treatment. Biofabrication 2023, 15, 035016. [Google Scholar] [CrossRef] [PubMed]
- Lam, E.H.Y.; Yu, F.; Zhu, S.; Wang, Z. 3D bioprinting for next-generation personalized medicine. Int. J. Mol. Sci. 2023, 24, 6357. [Google Scholar] [CrossRef] [PubMed]
- Yi, H.-G.; Kim, H.; Kwon, J.; Choi, Y.-J.; Jang, J.; Cho, D.-W. Application of 3D bioprinting in the prevention and the therapy for human diseases. Signal Transduct. Target. Ther. 2021, 6, 177. [Google Scholar] [CrossRef] [PubMed]
- Molander, D.; Sbirkov, Y.; Sarafian, V. 3D bioprinting as an emerging standard for cancer modeling and drug testing. Folia Medica 2022, 64, 559–565. [Google Scholar] [CrossRef]
- Dankó, T.; Petővári, G.; Raffay, R.; Sztankovics, D.; Moldvai, D.; Vetlényi, E.; Krencz, I.; Rókusz, A.; Sipos, K.; Visnovitz, T.; et al. Characterisation of 3D bioprinted human breast cancer model for in vitro drug and metabolic targeting. Int. J. Mol. Sci. 2022, 23, 7444. [Google Scholar] [CrossRef]
- Fan, Z.; Wei, X.; Chen, K.; Wang, L.; Xu, M. 3D bioprinting of an endothelialized liver lobule-like construct as a tumor-scale drug screening platform. Micromachines 2023, 14, 878. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Kim, S.; Chen, Z.; Shin, H.K.; Lee, S.-Y.; Moon, H.E.; Paek, S.H.; Park, S. 3D bioprinted vascularized tumour for drug testing. Int. J. Mol. Sci. 2020, 21, 2993. [Google Scholar] [CrossRef] [PubMed]
- Proietto, M.; Crippa, M.; Damiani, C.; Pasquale, V.; Sacco, E.; Vanoni, M.; Gilardi, M. Tumor heterogeneity: Preclinical models, emerging technologies, and future applications. Front. Oncol. 2023, 13, 1164535. [Google Scholar] [CrossRef] [PubMed]
- Pries, A.R.; Cornelissen, A.J.M.; Sloot, A.A.; Hinkeldey, M.; Dreher, M.R.; Höpfner, M.; Dewhirst, M.W.; Secomb, T.W. Structural adaptation and heterogeneity of normal and tumor microvascular networks. PLoS Comput. Biol. 2009, 5, e1000394. [Google Scholar] [CrossRef] [PubMed]
- Palaz, F.; Kalkan, A.K.; Can, Ö.; Demir, A.N.; Tozluyurt, A.; Özcan, A.; Ozsoz, M. CRISPR-Cas13 system as a promising and versatile tool for cancer diagnosis, therapy, and research. ACS Synth. Biol. 2021, 10, 1245–1267. [Google Scholar] [CrossRef] [PubMed]
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar] [CrossRef] [PubMed]
- Nussenzweig, P.M.; Marraffini, L.A. Molecular mechanisms of CRISPR-Cas immunity in bacteria. Annu. Rev. Genet. 2020, 54, 93–120. [Google Scholar] [CrossRef] [PubMed]
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A.; et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442. [Google Scholar] [CrossRef] [PubMed]
- Kadimisetty, K.; Song, J.; Doto, A.M.; Hwang, Y.; Peng, J.; Mauk, M.G.; Bushman, F.D.; Gross, R.; Jarvis, J.N.; Liu, C. Fully 3D printed integrated reactor array for point-of-care molecular diagnostics. Biosens. Bioelectron. 2018, 109, 156–163. [Google Scholar] [CrossRef]
- Liu, P.; Griffiths, S.; Veljanoski, D.; Vaughn-Beaucaire, P.; Speirs, V.; Brüning-Richardson, A. Preclinical models of glioblastoma: Limitations of current models and the promise of new developments. Expert Rev. Mol. Med. 2021, 23, e20. [Google Scholar] [CrossRef]
- Marusyk, A.; Polyak, K. Tumor heterogeneity: Causes and consequences. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2010, 1805, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.; Maswikiti, E.P.; Yu, Y.; Gao, L.; Ma, C.; Ma, H.; Deng, X.; Wang, N.; Wang, B.; Chen, H. Advances in the application of preclinical models in photodynamic therapy for tumor: A narrative review. Pharmaceutics 2023, 15, 197. [Google Scholar] [CrossRef] [PubMed]
- Pennes, H.H. Analysis of tissue and arterial blood temperatures in the resting human forearm. J. Appl. Physiol. 1948, 1, 93–122. [Google Scholar] [CrossRef] [PubMed]
- Krishna, S.M.; Omer, S.M.; Golledge, J. Evaluation of the clinical relevance and limitations of current pre-clinical models of peripheral artery disease. Clin. Sci. 2016, 130, 127–150. [Google Scholar] [CrossRef] [PubMed]
- Bentley, K.; Chakravartula, S. The temporal basis of angiogenesis. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20150522. [Google Scholar] [CrossRef]
- Cox, M.C. Modeling the Heterogeneous Brain Tumor Microenvironment to Analyze Mechanisms of Vascular Development and Chemoresistance. 2018. Available online: http://hdl.handle.net/10919/95947 (accessed on 30 April 2024).
- Rossant, J.; Howard, L. Signaling pathways in vascular development. Annu. Rev. Cell Dev. Biol. 2002, 18, 541–573. [Google Scholar] [CrossRef]
- Asghar, W.; El Assal, R.; Shafiee, H.; Pitteri, S.; Paulmurugan, R.; Demirci, U. Engineering cancer microenvironments for in vitro 3-D tumor models. Mater. Today 2015, 18, 539–553. [Google Scholar] [CrossRef]
- Tiwari, A.P.; Thorat, N.D.; Pricl, S.; Patil, R.M.; Rohiwal, S.; Townley, H. Bioink: A 3D-bioprinting tool for anticancer drug discovery and cancer management. Drug Discov. Today 2021, 26, 1574–1590. [Google Scholar] [CrossRef]
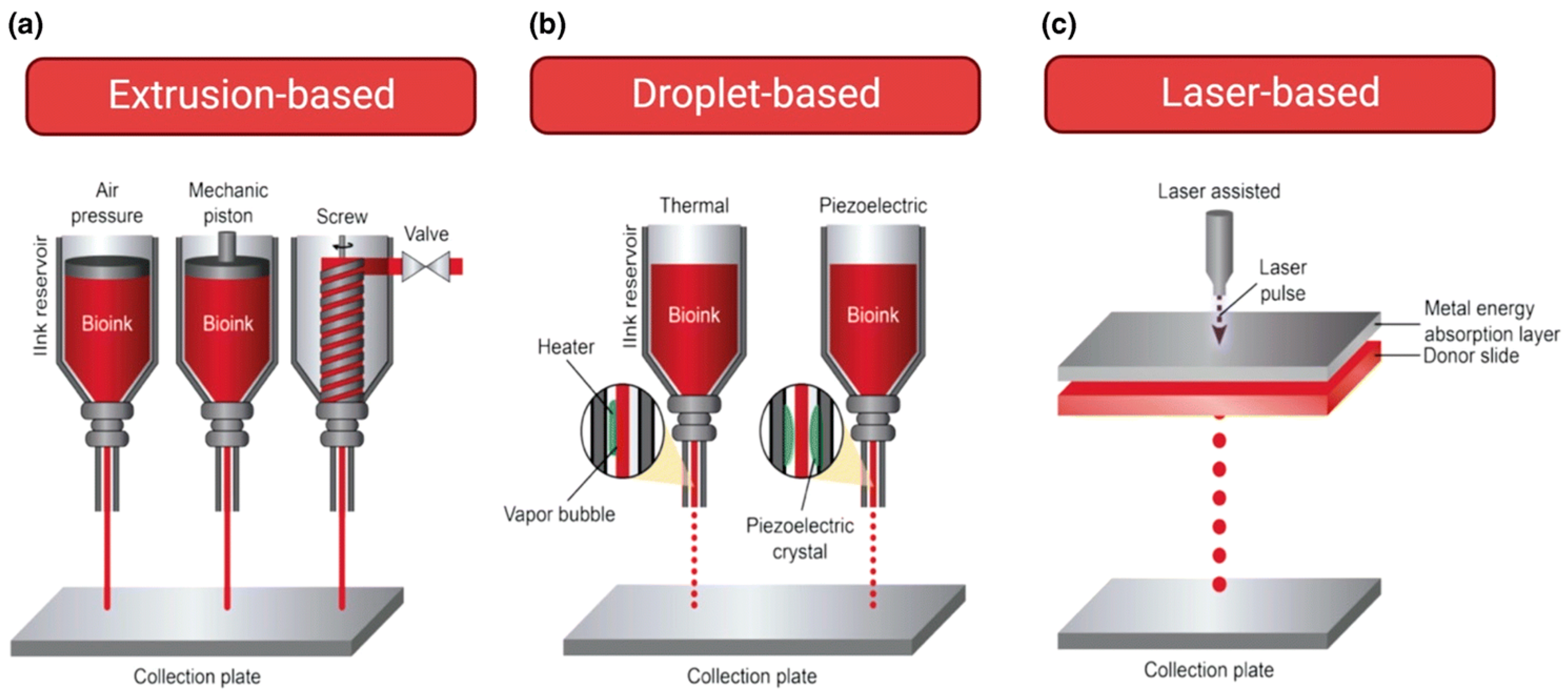

| Method | Advantages | Limitations |
|---|---|---|
| In vitro tube formation assay |
|
|
| In vivo chick CAM assay |
|
|
| Xenograft mouse models |
|
|
| Patient-derived xenograft models |
|
|
| Biomaterial | Advantage | Disadvantage | Application |
|---|---|---|---|
| Gelatin | Excellent biocompatibility, good cell adhesion, physical crosslinking properties | Low shape fidelity, especially unstable at temperatures suitable for cell growth, and low mechanical strength | Modification such as methacryloyl anhydride, or cross-linking, enhances its mechanical strength and printing resolution |
| PU | Excellent histocompatibility, super mechanical strength | Cells cannot be encapsulated directly | 3D printing vascular networks, bioartificial liver manufacturing |
| PLGA | Poor biocompatibility, middle mechanical properties | Cells cannot be encapsulated directly | 3D printing vascular networks, bioartificial liver manufacturing |
| Alginate | Shear thinning properties, very short time polymerizable, porous properties | Poor biocompatibility, low cell adhesion properties | Often mixed with gelatin, hyaluronic acid, etc. for printing; as a sacrificial material for vascular stents |
| Fibrinogen | Excellent biocompatibility, good cell adhesion | Low mechanical strength, fast degradation rate | Commonly used for thrombin cross-linking, blending or double cross-linking with gelatin, sodium alginate, etc. |
| Hyaluronic Acid | High water absorption, excellent biocompatibility, low molecular weight has the ability to promote cell proliferation | Low mechanical strength and poor formability | Modification such as methacryloyl anhydride, or compounded with other materials |
| dECM | Promotes cell adhesion, proliferation and functionalization, especially has a certain antithrombotic effect | Low mechanical strength, slow gelation, complicated preparation process | Often used with fast cross-linking materials such as sodium alginate |
| Pluronic® F127 | High resolution printing, special temperature sensitive properties | Low mechanical strength, fast degradation rate | As a sacrificial material for vascular stents |
| Bioprinting Techniques | Advantages | Disadvantages | Outcomes | References |
|---|---|---|---|---|
| 1. Extrusion-Based |
|
|
| [105,110,111,112,113,114,115,116] |
| 2. Droplet-Based |
|
|
| [106,117,118,119,120] |
| 3. Laser-Based (LBB) |
|
|
| [112,121,122,123,124] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shukla, A.K.; Yoon, S.; Oh, S.-O.; Lee, D.; Ahn, M.; Kim, B.S. Advancement in Cancer Vasculogenesis Modeling through 3D Bioprinting Technology. Biomimetics 2024, 9, 306. https://doi.org/10.3390/biomimetics9050306
Shukla AK, Yoon S, Oh S-O, Lee D, Ahn M, Kim BS. Advancement in Cancer Vasculogenesis Modeling through 3D Bioprinting Technology. Biomimetics. 2024; 9(5):306. https://doi.org/10.3390/biomimetics9050306
Chicago/Turabian StyleShukla, Arvind Kumar, Sik Yoon, Sae-Ock Oh, Dongjun Lee, Minjun Ahn, and Byoung Soo Kim. 2024. "Advancement in Cancer Vasculogenesis Modeling through 3D Bioprinting Technology" Biomimetics 9, no. 5: 306. https://doi.org/10.3390/biomimetics9050306
APA StyleShukla, A. K., Yoon, S., Oh, S.-O., Lee, D., Ahn, M., & Kim, B. S. (2024). Advancement in Cancer Vasculogenesis Modeling through 3D Bioprinting Technology. Biomimetics, 9(5), 306. https://doi.org/10.3390/biomimetics9050306













