1. Introduction
High-dimensional data denote datasets characterized by a large number of features or attributes, wherein the dimensionality typically exceeds the number of observations. However, such data present significant challenges, including the “curse of dimensionality”. This phenomenon adversely impacts the efficacy of conventional data analysis and modeling methodologies, as the distances between data points become increasingly diffuse and sparse in high-dimensional spaces. Consequently, this results in heightened susceptibility to overfitting and elevated computational expenses [
1]. In the analysis of high-dimensional data, feature selection is an essential step to reducing model complexity and computational costs and enhance the model’s generalization capability. By eliminating redundant and irrelevant features, feature selection minimizes noise interference, thereby improving the accuracy and robustness of predictive models. Furthermore, it facilitates a deeper understanding of the underlying data structure, enabling the identification of latent patterns and relationships within high-dimensional datasets. As such, feature selection constitutes a critical component in the preprocessing and modeling of high-dimensional data, contributing significantly to both interpretability and performance [
2].
In fact, feature selection (FS) is a combinatorial optimization problem that aims to optimize two conflicting objectives: maximizing the accuracy of feature classification and minimizing the number of selected features [
3]. Dealing with high-dimensional data remains a challenging task due to the fact that the search space grows exponentially with the number of features. Feature selection methods can broadly be classified into three categories: wrapper methods [
4,
5], filter methods [
6], and embedded methods [
7,
8]. Filter methods evaluate the importance of features regardless of any specific learning algorithm, making them computationally efficient and capable of handling a large number of features. However, they may not capture dependencies between features or their interaction with the learning algorithm. Filter methods are also prone to overfitting, and the selected features may not necessarily lead to the best performance in a specific learning task. Wrapper methods use a specific learning algorithm to evaluate the usefulness of features. They select a subset of features by repeatedly training and testing the learning algorithm with different subsets of features. Wrapper methods are computationally expensive and may overfit the data, but they are often more effective than filter methods in selecting relevant features for a specific learning task. Embedded methods incorporate feature selection into the learning algorithm itself. These methods typically use regularization techniques, such as Lasso or Ridge regression, to penalize the coefficients of irrelevant features and encourage sparsity in the resulting model. Embedded methods are computationally efficient and can produce models with good predictive performance. However, they may require a large amount of data and may not work well with non-linear models.
The wrapper-based approach to feature subset selection constitutes an NP-hard problem. Although exhaustive search strategies that evaluate all possible subsets can theoretically identify the optimal solution, their computational complexity renders them impractical for most real-world applications. Consequently, efficient global search techniques are necessary to navigate the expansive solution space effectively. Meta-heuristic algorithms have emerged as particularly suitable methods for resolving such complex combinatorial optimization challenges. Empirical studies have demonstrated the efficacy of swarm intelligence algorithms—a prominent subclass of meta-heuristics—in addressing feature selection problems [
9]. Notable swarm-intelligence-based optimization techniques applied to this domain include gray wolf optimization (GWO) [
10], particle swarm optimization (PSO) [
11], gravitational search algorithms (GSAs) [
12], and genetic algorithms (GAs) [
13], each offering distinct advantages in feature subset exploration and selection.
In recent years, binary particle swarm optimization (BPSO) has gained significant attention due to its conceptual simplicity, ease of implementation, and fast convergence. It has also been successfully applied to solve feature selection problems [
14]. BPSO was proposed by Kennedy and Eberhart in 1997 [
15]. PSO was converted into BPSO by using a transfer function that maps a continuous search space to a binary one. The updating process is designed to switch the positions of particles between 0 and 1 in binary search spaces. The goal of feature selection is to eliminate as many irrelevant and redundant features as possible. To achieve this goal, existing wrapper-based feature selection methods based on BPSO typically adopt an integrated fitness function that combines maximizing classification performance and minimizing feature subset size [
16]. However, with an increase in data dimension, the search space of the feature selection problem grows exponentially. This leads to a problem that satisfactory feature subsets with some key features may not be found, as a large number of feasible feature subsets pose significant challenges to BPSO. BPSO algorithms often search too slowly to obtain good feature subsets. So, further improvements to the exploration and exploitation capabilities of the BPSO algorithm are needed.
Researchers have proposed various BPSO algorithms that use different mechanisms to enhance the search process. Mirjalili et al. [
17] introduced six new transfer functions categorized into two types, S-shaped and V-shaped, and significantly enhanced the original BPSO algorithm. Xue et al. [
18] proposed a variable-length representation method for PSO-based feature selection. Although these strategies made PSO obtain better solutions in less time, the computational cost of this method was high. Song et al. [
19] proposed the Mutual Information-based Bare-Bones PSO algorithm (MIBBPSO). This algorithm employed an effective population initialization strategy based on label correlation, which utilized the correlation between features and labels to accelerate population convergence. However, when solving high-dimensional problems, this method required a significant amount of time, and due to the limitations of the initialization strategy, the diversity of particles was constrained, leading to a higher likelihood of the algorithm getting trapped in local optima. Jain et al. [
20] proposed a hybrid method by combining the Correlation-based Feature Selection (CFS) technique with an improved BPSO algorithm. This method also faces the same problem as MIBBPSO. Thaer et al. [
21] proposed a method called Boolean particle swarm optimization with evolutionary population dynamics. Six natural selection mechanisms, including the Best-Based, Tournament, Roulette Wheel, Stochastic Universal Sampling, Linear Rank, and Random-Based mechanisms, were employed to select better solutions. Because of the adoption of multiple selection mechanisms, this algorithm also had high computational costs. Cheng et al. [
22] introduced competitive particle swarm optimization (CSO). CSO enabled particles to learn from better particles randomly selected from the swarm, leading to improved performance. Due to the introduction of a competitive mechanism, the algorithm still incurs significant computational costs when dealing with high-dimensional problems. In summary, the recently proposed algorithms have all improved the performance of BPSO in feature selection. However, their ability to handle high-dimensional datasets is still limited. Therefore, further research is necessary.
In addition to BPSO, researchers have also investigated the application of other non-PSO-based metaheuristic algorithms in feature selection. Genetic algorithms (GAs) have emerged as one of the most popular metaheuristic algorithms used to solve FS problems. Oh et al. [
23] proposed a GA-based FS method that incorporated local search operations and genetic operations, but this method was prone to premature convergence and exhibited low search efficiency. Emary et al. [
24] introduced a feature selection method based on the gray wolf optimization (GWO) algorithm, but this algorithm had poor population diversity and exhibited slow convergence speed in later stages. RYM Nakamura et al. [
25] proposed a binary bat algorithm for feature selection, which combined the exploration ability of the bat algorithm with the Best Path Forest classifier. Majdi et al. [
26] proposed two binary variants of the Whale Optimization Algorithm (WOA). The first variant explored the impact of using Tournament and Roulette Wheel selection mechanisms during the search process, and the second variant incorporated crossover and mutation operators to enhance the exploitation ability of the WOA algorithm. Hichem et al. [
27] introduced a novel binary variant of the Grasshopper Optimization Algorithm (GOA). Their algorithm initialized the positions of grasshoppers with binary values and employed simple operators for position updates. Ibrahim et al. [
28] introduced eight time-varying S-shaped and V-shaped transfer functions into the Binary Dragonfly Algorithm (BDA). These metaheuristic algorithms mentioned above also face similar challenges as the variants of binary particle swarm optimization algorithm. They have high computational costs, the convergence speed is slow, and it is easy to get trapped in local optima when dealing with high-dimensional problems.
Furthermore, numerous hybrid metaheuristic algorithms have been proposed to deal with feature selection problem. For instance, Qasem et al. [
29] presented a binary gray wolf optimization–particle swarm optimization hybrid algorithm, which combining PSO and GWO. Ranya et al. [
30] proposed a binary hybridization of GWO and Harris hawks optimization. This method employed an S-shaped transfer function to convert the continuous search space into a binary search space. Zohre et al. [
31] introduced a three-stage hybrid feature selection method called information gain-based butterfly optimization algorithm (BOA). This method combined information gain technique with the BOA. Lu et al. [
32] proposed a hybrid feature selection technique that integrated mutual information maximization and adaptive genetic algorithm. Ma et al. [
33] proposed a two-stage hybrid ant colony algorithm. This algorithm incorporated an interval strategy to determine the optimal subset size of features searched in the additional stage. Overall, hybrid metaheuristic algorithms are also efficient and effective in finding the best subset of features for classification. However, when applied to high-dimensional feature selection problems, these algorithms still suffer from high computational overhead and low convergence accuracy. Thus, there is still room for further improvement in the convergence speed and accuracy of these algorithms.
Manta ray foraging optimization (MRFO) [
34] is a swarm intelligence algorithm inspired by the foraging behaviors of manta rays. Specifically, the optimization process of MRFO involves three foraging operators: chain foraging, cyclone foraging, and somersault foraging. The chain foraging strategy makes each individual update its position with respect to the one in front of it and the current global best solution. The cyclone foraging strategy makes each individual update its position with respect to both the one in front of it and a reference position, which can either the best position obtained so far or a random position produced in the search space. The choice between the two depends on the value of iteration. The gradual increase in the value of iteration encourages MRFO to smoothly transition from an exploratory search to an exploitative search. With the value of the random number, MRFO can switch between chain foraging and cyclone foraging. Somersault foraging allows individuals to adaptively search in a changing search range.
The MRFO has been widely applied. Abdel-Mawgoud et al. [
35] proposed an improved manta ray foraging optimizer, introducing a simulated annealing operator to enhance the development phase of MRFO and applying it to solve the integration of renewable energy in distribution networks. Supiksha et al. [
36] combined the MRFO with the rider optimization algorithm, proposing general adversarial networks based on a manta ray foraging optimizer for effective glaucoma detection. Ibrahim et al. [
37] proposed a hybrid improved bat foraging optimization algorithm with the slap swarm algorithm to address IoT task scheduling issues in cloud computing. Yuxian et al. [
38] proposed an enhanced elephant herding optimization algorithm and introduced the rolling foraging strategy of bats and Gaussian mutation. Neeraj et al. [
39] utilized the MRFO to optimize multiple locally relevant embedding strength values (MESs), balancing invisibility and robustness, and proposed a novel image adaptive watermarking scheme called MantaRayWmark.
In this paper, we propose a novel algorithm called the BPSO-MRFL, which integrates the MRFO search mechanism into BPSO. BPSO-MRFL involves three search phases—chain learning, cyclone learning, and somersault learning—which enhances the algorithm’s search ability and obtains better performance.
The novelty of this work is as follows:
The algorithm introduces the chain learning of the manta ray foraging optimization algorithm into the binary particle swarm optimization algorithm, which effectively improves the exploration ability of the algorithm;
The algorithm introduces the rolling learning of the manta ray foraging optimization algorithm into the binary particle swarm optimization algorithm, which effectively improves the development ability of the algorithm;
The algorithm introduces the whirlwind learning of the manta ray foraging optimization algorithm into the binary particle swarm optimization algorithm to balance the development and exploration ability of the algorithm;
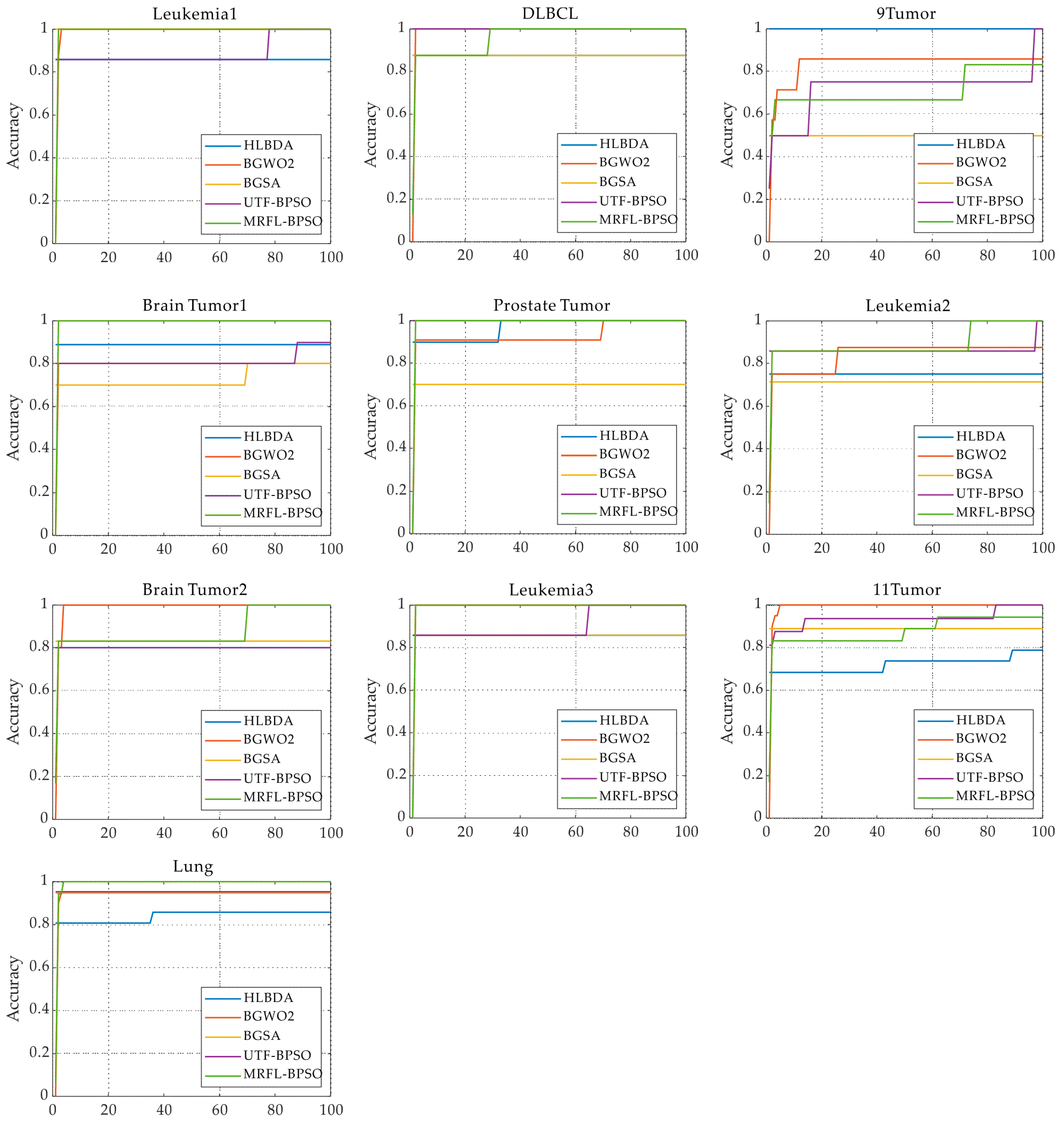
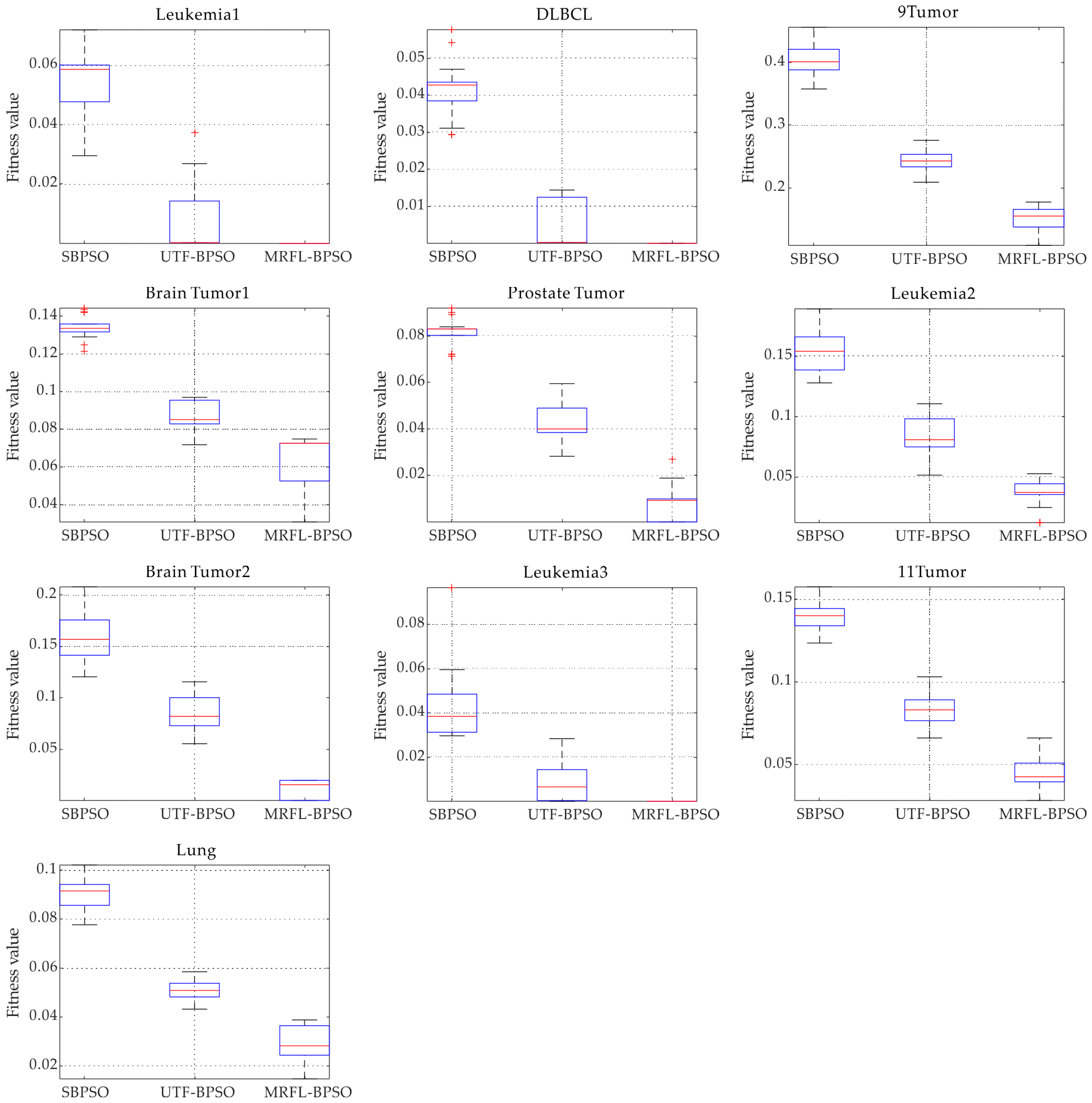
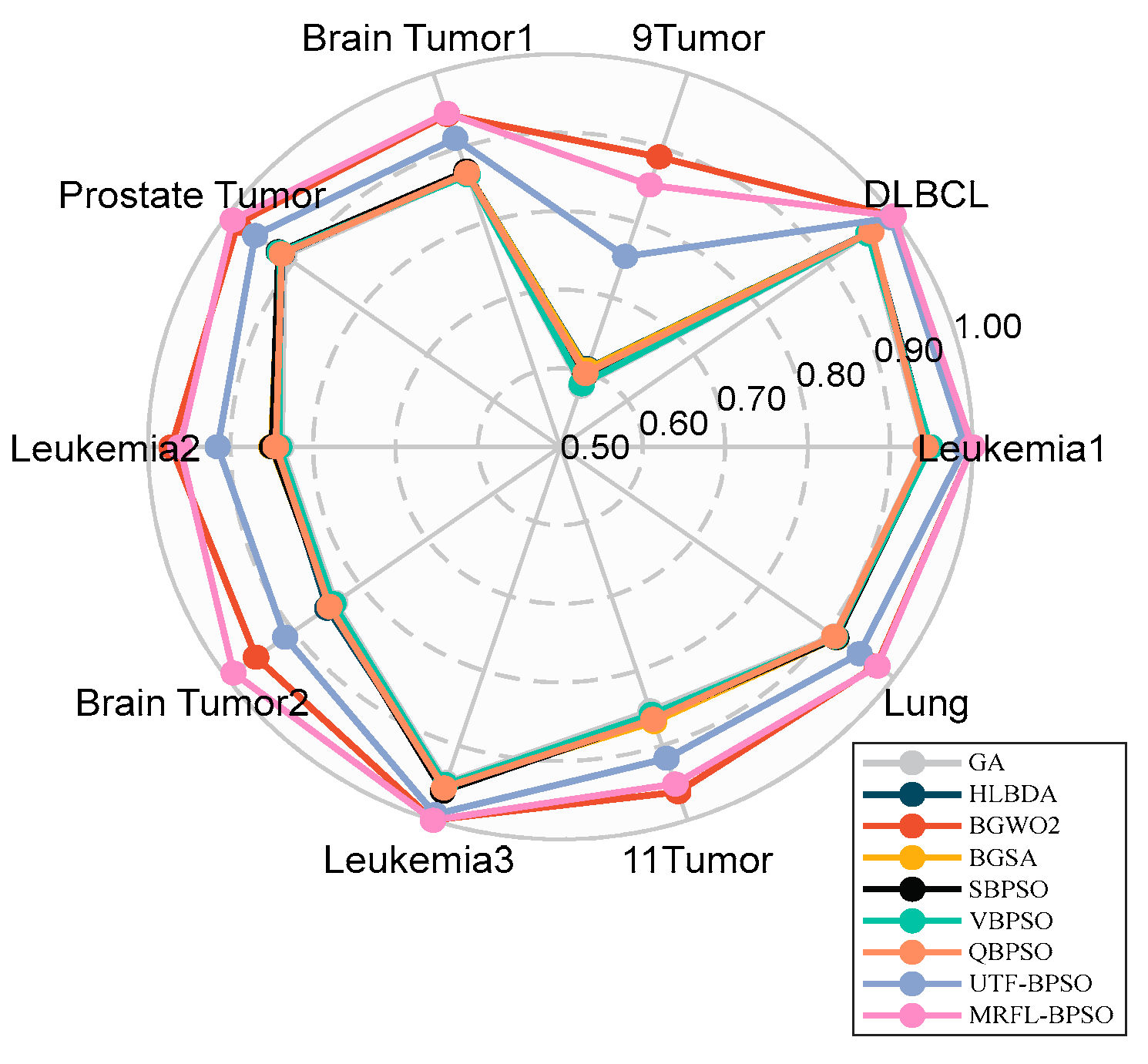
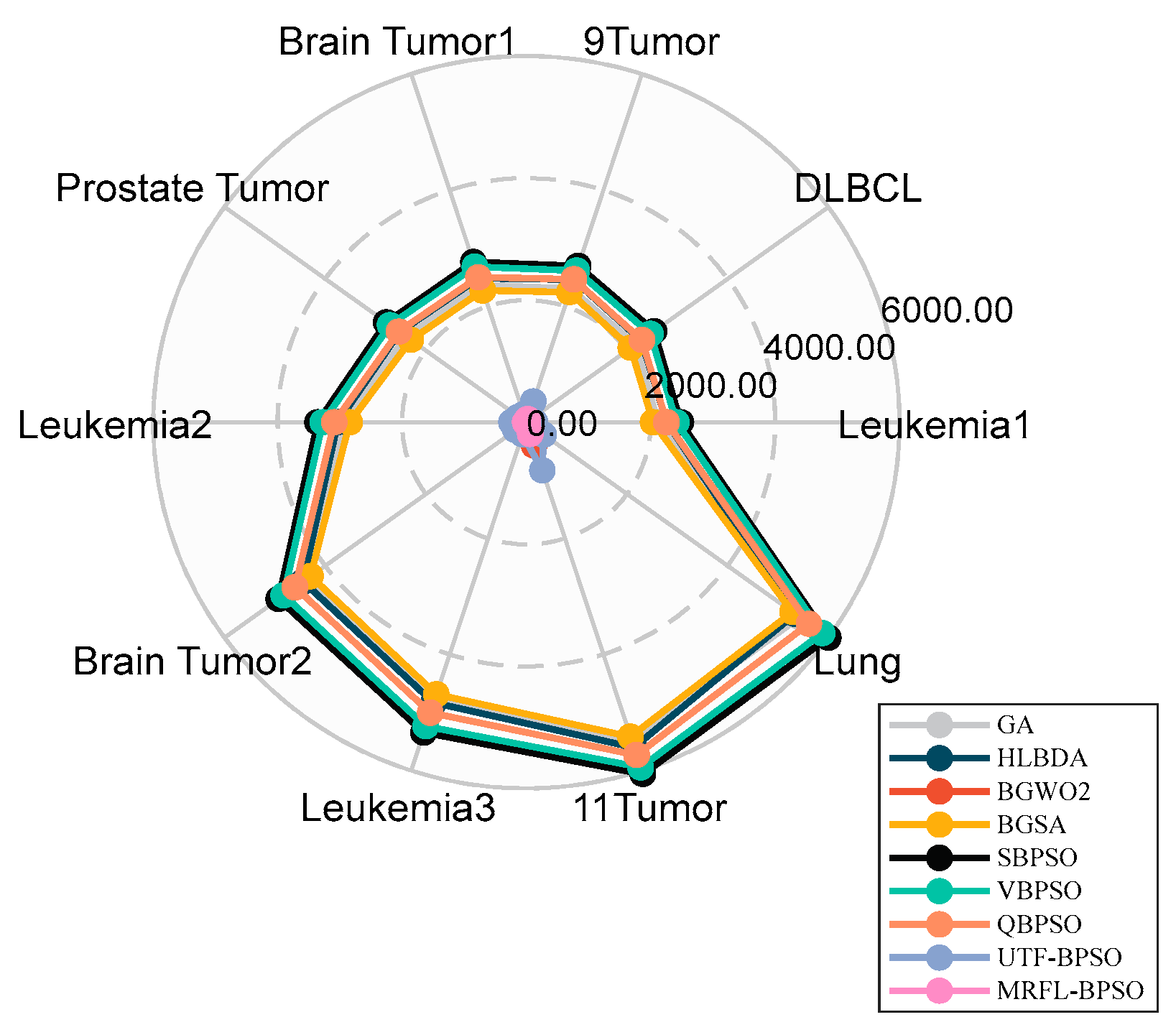
We conduct experiments on 10 gene expression datasets with thousands of features that are publicly available to evaluate the performance of the proposed algorithms;
The remainder of this paper is organized as follows.
Section 2 presents the background and related work.
Section 3 introduces the proposing BPSO-MRFL algorithm.
Section 4 presents the experimental datasets, the evaluating metrics of experimental results, and the parameter settings of the comparing algorithms.
Section 5 provides the experimental results and comparisons. Finally,
Section 6 presents the conclusions of this paper.