Different Phenotypes of Pediatric Asthma Show Distinct Bacterial Functional Profiles and Network Relationships
Abstract
1. Introduction
2. Materials and Methods
2.1. Pediatric Asthma Phenotypes
2.2. Bacteriome Analyses
2.2.1. Functional Diversity
2.2.2. Bacterial Networks
3. Results
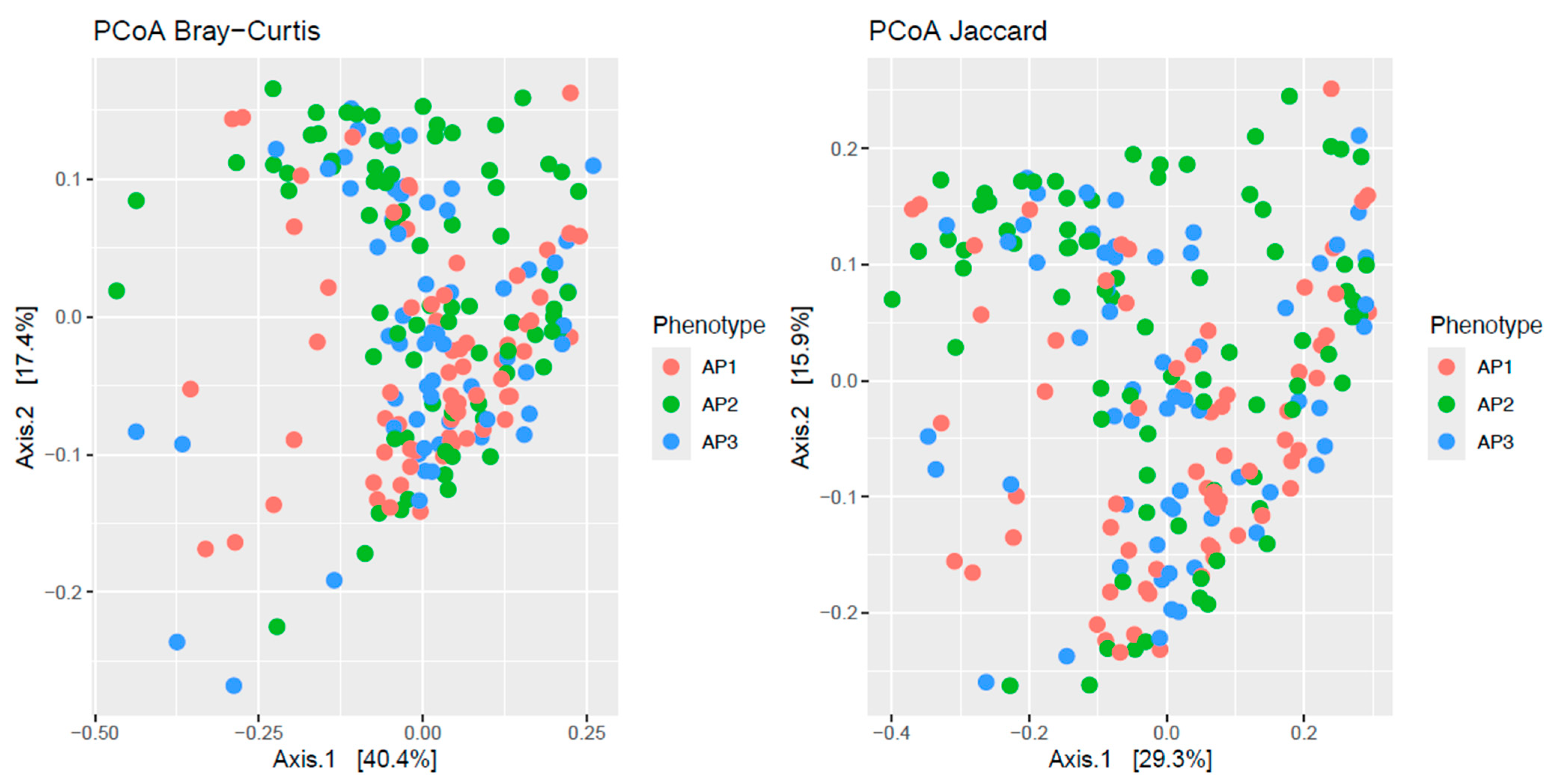
3.1. Functional Diversity
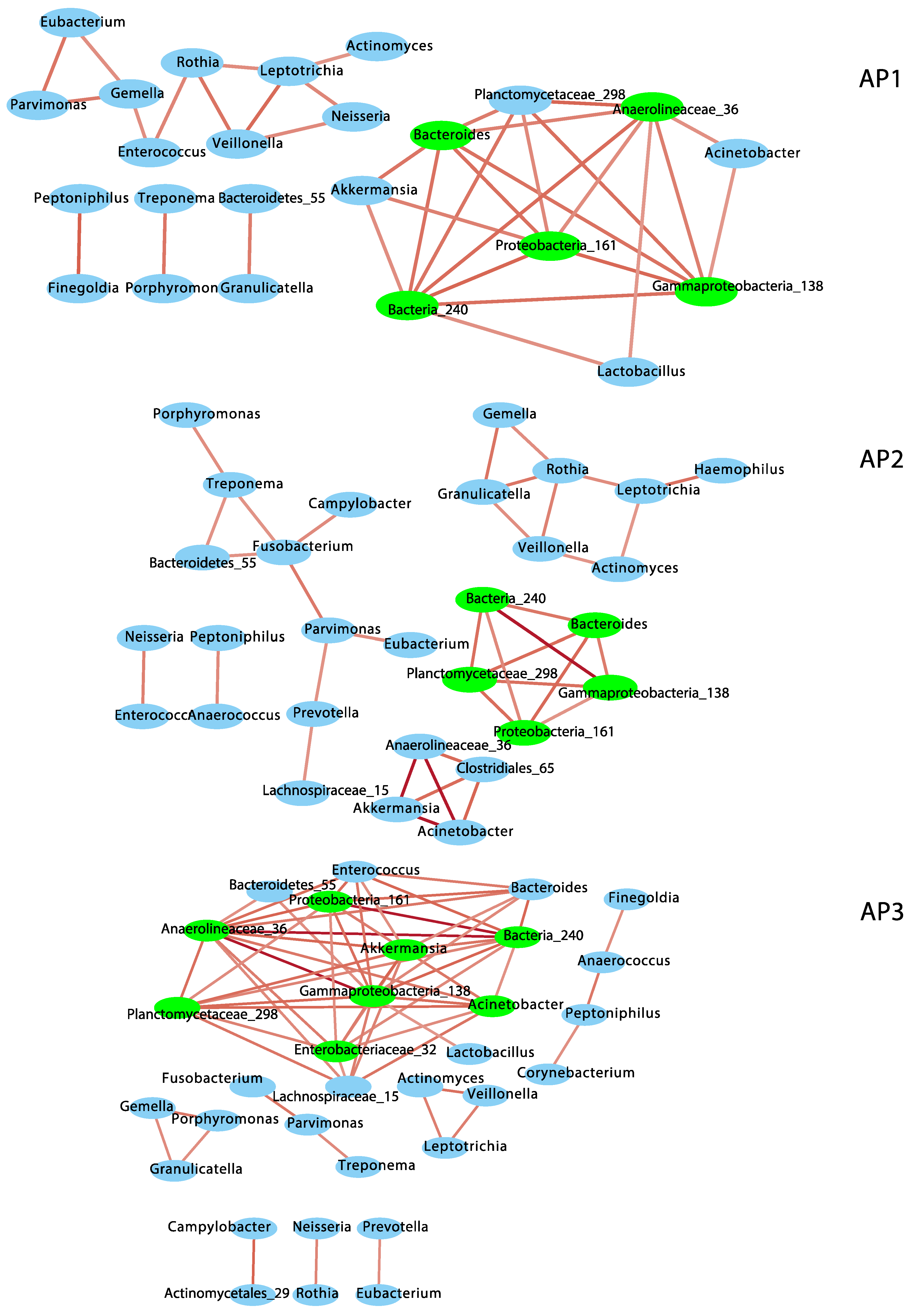
3.2. Network Interactions
4. Discussion
5. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Johnson, C.C.; Chandran, A.; Havstad, S.; Li, X.; McEvoy, C.T.; Ownby, D.R.; Litonjua, A.A.; Karagas, M.R.; Camargo, C.A., Jr.; Gern, J.E.; et al. US Childhood Asthma Incidence Rate Patterns From the ECHO Consortium to Identify High-risk Groups for Primary Prevention. JAMA Pediatr. 2021, 175, 919–927. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention (CDC). Asthma Surveillance in the United States, 2001–2021. 2023. Available online: https://www.cdc.gov/asthma/asthma-prevalence-us-2023-508.pdf (accessed on 12 February 2025).
- American Lung Association. Asthma Trends Brief. 2024. Available online: https://www.lung.org/research/trends-in-lung-disease/asthma-trends-brief (accessed on 12 February 2025).
- Naja, A.S.; Permaul, P.; Phipatanakul, W. Taming Asthma in School-Aged Children: A Comprehensive Review. J. Allergy Clin. Immunol. Pr. 2018, 6, 726–735. [Google Scholar] [CrossRef] [PubMed]
- Hsu, J.; Qin, X.; Beavers, S.F.; Mirabelli, M.C. Asthma-Related School Absenteeism, Morbidity, and Modifiable Factors. Am. J. Prev. Med. 2016, 51, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Children’s National. IMPACT DC: 2019 Annual Report; Children’s National: Washington, DC, USA, 2019. [Google Scholar]
- Wenzel, S.E. Asthma phenotypes: The evolution from clinical to molecular approaches. Nat. Med. 2012, 18, 716–725. [Google Scholar] [CrossRef]
- Borish, L.; Culp, J.A. Asthma: A syndrome composed of heterogeneous diseases. Ann. Allerg. Asthma Im. 2008, 101, 1–8. [Google Scholar] [CrossRef]
- Martinez, F.D. Definition of pediatric asthma and associated risk factors. Pediatr. Pulm. 1997, 24, 9–12. [Google Scholar] [CrossRef]
- Cowan, K.; Guilbert, T.W. Pediatric asthma phenotypes. Curr. Opin. Pediatr. 2012, 24, 344–351. [Google Scholar] [CrossRef]
- Aujla, S.J.; Ross, K.R.; Chmiel, J.F.; Holguin, F. Airway molecular phenotypes in pediatric asthma. Curr. Opin. Allergy Clin. Immunol. 2011, 11, 122–126. [Google Scholar] [CrossRef]
- Wenzel, S.E. Asthma: Defining of the persistent adult phenotypes. Lancet 2006, 368, 804–813. [Google Scholar] [CrossRef]
- Haldar, P.; Pavord, I.D.; Shaw, D.E.; Berry, M.A.; Thomas, M.; Brightling, C.E.; Wardlaw, A.J.; Green, R.H. Cluster analysis and clinical asthma phenotypes. Am. J. Respir. Crit. Care Med. 2008, 178, 218–224. [Google Scholar] [CrossRef]
- Moore, W.C.; Meyers, D.A.; Wenzel, S.E.; Teague, W.G.; Li, H.; Li, X.; D’Agostino, R., Jr.; Castro, M.; Curran-Everett, D.; Fitzpatrick, A.M.; et al. Identification of asthma phenotypes using cluster analysis in the Severe Asthma Research Program. Am. J. Respir. Crit. Care Med. 2010, 181, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Benton, A.S.; Wang, Z.; Lerner, J.; Foerster, M.; Teach, S.J.; Freishtat, R.J. Overcoming heterogeneity in pediatric asthma: Tobacco smoke and asthma characteristics within phenotypic clusters in an African American cohort. J. Asthma Off. J. Assoc. Care Asthma 2010, 47, 728–734. [Google Scholar] [CrossRef] [PubMed]
- Moore, W.C.; Bleecker, E.R.; Curran-Everett, D.; Erzurum, S.C.; Ameredes, B.T.; Bacharier, L.; Calhoun, W.J.; Castro, M.; Chung, K.F.; Clark, M.P.; et al. Characterization of the severe asthma phenotype by the National Heart, Lung, and Blood Institute’s Severe Asthma Research Program. J. Allergy Clin. Immunol. 2007, 119, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Perez-Losada, M.; Authelet, K.J.; Hoptay, C.E.; Kwak, C.; Crandall, K.A.; Freishtat, R.J. Pediatric asthma comprises different phenotypic clusters with unique nasal microbiotas. Microbiome 2018, 6, 179. [Google Scholar] [CrossRef]
- Pérez-Losada, M.; Castro-Nallar, E.; Bendall, M.L.; Freishtat, R.J.; Crandall, K.A. Dual Transcriptomic Profiling of Host and Microbiota during Health and Disease in Pediatric Asthma. PLoS ONE 2015, 10, e0131819. [Google Scholar] [CrossRef]
- Pérez-Losada, M.; Crandall, K.A.; Freishtat, R.J. Two sampling methods yield distinct microbial signatures in the nasopharynges of asthmatic children. Microbiome 2016, 4, 25. [Google Scholar] [CrossRef]
- Castro-Nallar, E.; Shen, Y.; Freishtat, R.J.; Pérez-Losada, M.; Manimaran, S.; Liu, G.; Spira, A.; Johnson, W.E.; Crandall, K.A. Integrating metagenomics and host gene expression to characterize asthma-associated microbial communities. BMC Med. Genom. 2015, 8, 50. [Google Scholar] [CrossRef]
- Teo, S.M.; Mok, D.; Pham, K.; Kusel, M.; Serralha, M.; Troy, N.; Holt, B.J.; Hales, B.J.; Walker, M.L.; Hollams, E.; et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe 2015, 17, 704–715. [Google Scholar] [CrossRef]
- Bogaert, D.; Keijser, B.; Huse, S.; Rossen, J.; Veenhoven, R.; van Gils, E.; Bruin, J.; Montijn, R.; Bonten, M.; Sanders, E. Variability and diversity of nasopharyngeal microbiota in children: A metagenomic analysis. PLoS ONE 2011, 6, e17035. [Google Scholar] [CrossRef]
- Pérez-Losada, M.; Alamri, L.; Crandall, K.A.; Freishtat, R.J. Nasopharyngeal Microbiome Diversity Changes over Time in Children with Asthma. PLoS ONE 2017, 12, e0170543. [Google Scholar] [CrossRef]
- Esposito, S.; Principi, N. Impact of nasopharyngeal microbiota on the development of respiratory tract diseases. Eur. J. Clin. Microbiol. Infect. Dis. Off. Publ. Eur. Soc. Clin. Microbiol. 2018, 37, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.J. Nasopharyngeal Microbiota: Gatekeepers or Fortune Tellers of Susceptibility to Respiratory Tract Infections? Am. J. Respir. Crit. Care Med. 2017, 196, 1504–1505. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.L.; Leong, L.E.X.; Choo, J.M.; Wesselingh, S.; Yang, I.A.; Upham, J.W.; Reynolds, P.N.; Hodge, S.; James, A.L.; Jenkins, C.; et al. Inflammatory phenotypes in patients with severe asthma are associated with distinct airway microbiology. J. Allergy Clin. Immunol. 2018, 141, 94–103.e15. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Cox, M.; Liang, Z.; Brinkmann, F.; Cardenas, P.A.; Duff, R.; Bhavsar, P.; Cookson, W.; Moffatt, M.; Chung, K.F. Airway Microbiota in Severe Asthma and Relationship to Asthma Severity and Phenotypes. PLoS ONE 2016, 11, e0152724. [Google Scholar] [CrossRef]
- Chung, K.F. Potential Role of the Lung Microbiome in Shaping Asthma Phenotypes. Ann. Am. Thorac. Soc. 2017, 14, S326–S331. [Google Scholar] [CrossRef]
- Kozik, A.J.; Huang, Y.J. The microbiome in asthma: Role in pathogenesis, phenotype, and response to treatment. Ann. Allergy Asthma Immunol. Off. Publ. Am. Coll. Allergy Asthma Immunol. 2019, 122, 270–275. [Google Scholar] [CrossRef]
- Liu, C.; Makrinioti, H.; Saglani, S.; Bowman, M.; Lin, L.L.; Camargo, C.A., Jr.; Hasegawa, K.; Zhu, Z. Microbial dysbiosis and childhood asthma development: Integrated role of the airway and gut microbiome, environmental exposures, and host metabolic and immune response. Front. Immunol. 2022, 13, 1028209. [Google Scholar] [CrossRef]
- van Beveren, G.J.; Said, H.; van Houten, M.A.; Bogaert, D. The respiratory microbiome in childhood asthma. J. Allergy Clin. Immunol. 2023, 152, 1352–1367. [Google Scholar] [CrossRef]
- Pijnenburg, M.W.; Frey, U.; De Jongste, J.C.; Saglani, S. Childhood asthma: Pathogenesis and phenotypes. Eur. Respir. J. 2022, 59, 2100731. [Google Scholar] [CrossRef]
- Shibata, R.; Zhu, Z.; Ooka, T.; Freishtat, R.J.; Mansbach, J.M.; Perez-Losada, M.; Ramos-Tapia, I.; Teach, S.; Camargo, C.A., Jr.; Hasegawa, K. Immunoglobulin E-virus phenotypes of infant bronchiolitis and risk of childhood asthma. Front. Immunol. 2023, 14, 1187065. [Google Scholar] [CrossRef]
- Tsai, M.H.; Shih, H.J.; Su, K.W.; Liao, S.L.; Hua, M.C.; Yao, T.C.; Lai, S.H.; Yeh, K.W.; Chen, L.C.; Huang, J.L.; et al. Nasopharyngeal microbial profiles associated with the risk of airway allergies in early childhood. J. Microbiol. Immunol. Infect. 2022, 55, 777–785. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Aziz, M.I.; Thorsen, J.; Hashimoto, S.; Vijverberg, S.J.H.; Neerincx, A.H.; Brinkman, P.; van Aalderen, W.; Stokholm, J.; Rasmussen, M.A.; Roggenbuck-Wedemeyer, M.; et al. Oropharyngeal Microbiota Clusters in Children with Asthma or Wheeze Associate with Allergy, Blood Transcriptomic Immune Pathways, and Exacerbation Risk. Am. J. Respir. Crit. Care Med. 2023, 208, 142–154. [Google Scholar] [CrossRef] [PubMed]
- McCauley, K.E.; Durack, J.; Lynch, K.V.; Fadrosh, D.W.; Fujimura, K.E.; Vundla, F.; Ozcam, M.; LeBeau, P.; Caltroni, A.; Burns, P.; et al. Early-life nasal microbiota dynamics relate to longitudinal respiratory phenotypes in urban children. J. Allergy Clin. Immunol. 2024, 153, 1563–1573. [Google Scholar] [CrossRef] [PubMed]
- Aldriwesh, M.G.; Al-Mutairi, A.M.; Alharbi, A.S.; Aljohani, H.Y.; Alzahrani, N.A.; Ajina, R.; Alanazi, A.M. Paediatric Asthma and the Microbiome: A Systematic Review. Microorganisms 2023, 11, 939. [Google Scholar] [CrossRef]
- Logon, K.; Swirkosz, G.; Nowak, M.; Wrzesniewska, M.; Szczygiel, A.; Gomulka, K. The Role of the Microbiome in the Pathogenesis and Treatment of Asthma. Biomedicines 2023, 11, 1618. [Google Scholar] [CrossRef]
- Valverde-Molina, J.; Garcia-Marcos, L. Microbiome and Asthma: Microbial Dysbiosis and the Origins, Phenotypes, Persistence, and Severity of Asthma. Nutrients 2023, 15, 486. [Google Scholar] [CrossRef]
- Santacroce, L.; Charitos, I.A.; Ballini, A.; Inchingolo, F.; Luperto, P.; De Nitto, E.; Topi, S. The Human Respiratory System and its Microbiome at a Glimpse. Biology 2020, 9, 318. [Google Scholar] [CrossRef]
- Liu, X.F.; Shao, J.H.; Liao, Y.T.; Wang, L.N.; Jia, Y.; Dong, P.J.; Liu, Z.Z.; He, D.D.; Li, C.; Zhang, X. Regulation of short-chain fatty acids in the immune system. Front. Immunol. 2023, 14, 1186892. [Google Scholar] [CrossRef]
- Liu, T.; Sun, Z.; Yang, Z.; Qiao, X. Microbiota-derived short-chain fatty acids and modulation of host-derived peptides formation: Focused on host defense peptides. Biomed. Pharmacother. 2023, 162, 114586. [Google Scholar] [CrossRef]
- Qian, G.; Jiang, W.; Zou, B.; Feng, J.; Cheng, X.; Gu, J.; Chu, T.; Niu, C.; He, R.; Chu, Y.; et al. LPS inactivation by a host lipase allows lung epithelial cell sensitization for allergic asthma. J. Exp. Med. 2018, 215, 2397–2412. [Google Scholar] [CrossRef]
- Dipak, G.V.; Nami, S.P.; Harissios, V. Protease-activated receptor-2: Role in asthma pathogenesis and utility as a biomarker of disease severity. Front. Med. 2022, 9, 954990. [Google Scholar]
- Gulliver, E.L.; Young, R.B.; Chonwerawong, M.; D’Adamo, G.L.; Thomason, T.; Widdop, J.T.; Rutten, E.L.; Rossetto Marcelino, V.; Bryant, R.V.; Costello, S.P.; et al. Review article: The future of microbiome-based therapeutics. Aliment. Pharmacol. Ther. 2022, 56, 192–208. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, M.; Azcarate-Peril, M.A.; Barnard, A.; Benoit, V.; Grimaldi, R.; Guyonnet, D.; Holscher, H.D.; Hunter, K.; Manurung, S.; Obis, D.; et al. Shaping the Future of Probiotics and Prebiotics. Trends Microbiol. 2021, 29, 667–685. [Google Scholar] [CrossRef] [PubMed]
- Logotheti, M.; Agioutantis, P.; Katsaounou, P.; Loutrari, H. Microbiome Research and Multi-Omics Integration for Personalized Medicine in Asthma. J. Pers. Med. 2021, 11, 1299. [Google Scholar] [CrossRef]
- Campbell, C.D.; Gleeson, M.; Sulaiman, I. The role of the respiratory microbiome in asthma. Front. Allergy 2023, 4, 1120999. [Google Scholar] [CrossRef]
- Zhao, H.; Liu, J.; Zhu, J.; Yang, F.; Wu, H.; Ba, Y.; Cui, L.; Chen, R.; Chen, S. Bacterial composition and community structure of the oropharynx of adults with asthma are associated with environmental factors. Microb. Pathog. 2020, 149, 104505. [Google Scholar] [CrossRef]
- Li, Y.; Zou, C.; Li, J.; Wang, W.; Wang, F.; Guo, Y. Airway Microbiome Composition and Co-Occurrence Network Are Associated with Inflammatory Phenotypes of Asthma. Int. Arch. Allergy Immunol. 2023, 184, 1254–1263. [Google Scholar] [CrossRef]
- Huang, C.; Yu, Y.; Du, W.; Liu, Y.; Dai, R.; Tang, W.; Wang, P.; Zhang, C.; Shi, G. Fungal and bacterial microbiome dysbiosis and imbalance of trans-kingdom network in asthma. Clin. Transl. Allergy 2020, 10, 42. [Google Scholar] [CrossRef]
- Earl, C.S.; An, S.Q.; Ryan, R.P. The changing face of asthma and its relation with microbes. Trends Microbiol. 2015, 23, 408–418. [Google Scholar] [CrossRef]
- Littman, D.R.; Pamer, E.G. Role of the commensal microbiota in normal and pathogenic host immune responses. Cell Host Microbe 2011, 10, 311–323. [Google Scholar] [CrossRef]
- Perez-Losada, M.; Castro-Nallar, E.; Laerte Boechat, J.; Delgado, L.; Azenha Rama, T.; Berrios-Farias, V.; Oliveira, M. The oral bacteriomes of patients with allergic rhinitis and asthma differ from that of healthy controls. Front. Microbiol. 2023, 14, 1197135. [Google Scholar] [CrossRef] [PubMed]
- Perez-Losada, M.; Castro-Nallar, E.; Laerte Boechat, J.; Delgado, L.; Azenha Rama, T.; Berrios-Farias, V.; Oliveira, M. Nasal Bacteriomes of Patients with Asthma and Allergic Rhinitis Show Unique Composition, Structure, Function and Interactions. Microorganisms 2023, 11, 683. [Google Scholar] [CrossRef] [PubMed]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microb. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef]
- Chen, I.A.; Chu, K.; Palaniappan, K.; Ratner, A.; Huang, J.; Huntemann, M.; Hajek, P.; Ritter, S.; Varghese, N.; Seshadri, R.; et al. The IMG/M data management and analysis system v.6.0: New tools and advanced capabilities. Nucleic Acids Res. 2021, 49, D751–D763. [Google Scholar] [CrossRef]
- Caspi, R.; Billington, R.; Fulcher, C.A.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Latendresse, M.; Midford, P.E.; Ong, Q.; Ong, W.K.; et al. The MetaCyc database of metabolic pathways and enzymes. Nucleic Acids Res. 2018, 46, D633–D639. [Google Scholar] [CrossRef]
- Caspi, R.; Billington, R.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Midford, P.E.; Ong, W.K.; Paley, S.; Subhraveti, P.; Karp, P.D. The MetaCyc database of metabolic pathways and enzymes—A 2019 update. Nucleic Acids Res. 2020, 48, D445–D453. [Google Scholar] [CrossRef]
- Yang, C.; Mai, J.; Cao, X.; Burberry, A.; Cominelli, F.; Zhang, L. ggpicrust2: An R package for PICRUSt2 predicted functional profile analysis and visualization. Bioinformatics 2023, 39, btad470. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Statist. Soc. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Cook, R.D. Detection of influential observation in linear regression. Technometrics 1977, 19, 15–18. [Google Scholar] [CrossRef]
- Dixon, P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2003, 14, 927–930. [Google Scholar] [CrossRef]
- Schwager, E.; Weingart, G.; Bielski, C.; Huttenhower, C. CCREPE: Compositionality Corrected by Permutation and Renormalization. 2024. Available online: https://huttenhower.sph.harvard.edu/ccrepe/ (accessed on 12 February 2025).
- Doncheva, N.T.; Morris, J.H.; Holze, H.; Kirsch, R.; Nastou, K.C.; Cuesta-Astroz, Y.; Rattei, T.; Szklarczyk, D.; von Mering, C.; Jensen, L.J. Cytoscape stringApp 2.0: Analysis and Visualization of Heterogeneous Biological Networks. J. Proteome Res. 2023, 22, 637–646. [Google Scholar] [CrossRef] [PubMed]
- Csárdi, G.; Nepusz, T.; Traag, V.; Horvát, S.; Zanini, F.; Noom, D.; Müller, K. igraph: Network Analysis and Visualization in R, 2.1.4. 2025. Available online: https://igraph.r-universe.dev/igraph (accessed on 12 February 2025).
- Hoffman, D.J.; Campos-Ponce, M.; Taddei, C.R.; Doak, C.M. Microbiome, growth retardation and metabolism: Are they related? Ann. Hum. Biol. 2017, 44, 201–207. [Google Scholar] [CrossRef]
- Ta, L.D.H.; Yap, G.C.; Tay, C.J.X.; Lim, A.S.M.; Huang, C.H.; Chu, C.W.; De Sessions, P.F.; Shek, L.P.; Goh, A.; Van Bever, H.P.S.; et al. Establishment of the nasal microbiota in the first 18 months of life: Correlation with early-onset rhinitis and wheezing. J. Allergy Clin. Immunol. 2018, 142, 86–95. [Google Scholar] [CrossRef]
- Chiang, T.Y.; Yang, Y.R.; Zhuo, M.Y.; Yang, F.; Zhang, Y.F.; Fu, C.H.; Lee, T.J.; Chung, W.H.; Chen, L.; Chang, C.J. Microbiome profiling of nasal extracellular vesicles in patients with allergic rhinitis. World Allergy Organ. J. 2022, 15, 100674. [Google Scholar] [CrossRef]
- Losol, P.; Choi, J.P.; Kim, S.H.; Chang, Y.S. The Role of Upper Airway Microbiome in the Development of Adult Asthma. Immune Netw. 2021, 21, e19. [Google Scholar] [CrossRef]
- Lynch, S.V. The Lung Microbiome and Airway Disease. Ann. Am. Thorac. Soc. 2016, 13 (Suppl. S2), S462–S465. [Google Scholar] [CrossRef]
- Huffnagle, G.B.; Dickson, R.P.; Lukacs, N.W. The respiratory tract microbiome and lung inflammation: A two-way street. Mucosal Immunol. 2017, 10, 299–306. [Google Scholar] [CrossRef]
- Lira-Lucio, J.A.; Falfan-Valencia, R.; Ramirez-Venegas, A.; Buendia-Roldan, I.; Rojas-Serrano, J.; Mejia, M.; Perez-Rubio, G. Lung Microbiome Participation in Local Immune Response Regulation in Respiratory Diseases. Microorganisms 2020, 8, 1059. [Google Scholar] [CrossRef]
- Paudel, K.R.; Dharwal, V.; Patel, V.K.; Galvao, I.; Wadhwa, R.; Malyla, V.; Shen, S.S.; Budden, K.F.; Hansbro, N.G.; Vaughan, A.; et al. Role of Lung Microbiome in Innate Immune Response Associated With Chronic Lung Diseases. Front. Med. 2020, 7, 554. [Google Scholar] [CrossRef] [PubMed]
- Costello, E.K.; Stagaman, K.; Dethlefsen, L.; Bohannan, B.J.; Relman, D.A. The application of ecological theory toward an understanding of the human microbiome. Science 2012, 336, 1255–1262. [Google Scholar] [CrossRef] [PubMed]
- Chiu, C.Y.; Cheng, M.L.; Chiang, M.H.; Wang, C.J.; Tsai, M.H.; Lin, G. Integrated metabolic and microbial analysis reveals host-microbial interactions in IgE-mediated childhood asthma. Sci. Rep. 2021, 11, 23407. [Google Scholar] [CrossRef] [PubMed]
- Georas, S.N.; Khurana, S. Update on asthma biology. J. Allergy Clin. Immunol. 2024, 153, 1215–1228. [Google Scholar] [CrossRef]
- Hu, Q.; Jin, L.; Zeng, J.; Wang, J.; Zhong, S.; Fan, W.; Liao, W. Tryptophan metabolite-regulated Treg responses contribute to attenuation of airway inflammation during specific immunotherapy in a mouse asthma model. Hum. Vaccin. Immunother. 2020, 16, 1891–1899. [Google Scholar] [CrossRef]
- Chiu, C.Y.; Cheng, M.L.; Chiang, M.H.; Wang, C.J.; Tsai, M.H.; Lin, G. Metabolomic Analysis Reveals Distinct Profiles in the Plasma and Urine Associated with IgE Reactions in Childhood Asthma. J. Clin. Med. 2020, 9, 887. [Google Scholar] [CrossRef]
- Samra, M.S.; Lim, D.H.; Han, M.Y.; Jee, H.M.; Kim, Y.K.; Kim, J.H. Bacterial Microbiota-derived Extracellular Vesicles in Children With Allergic Airway Diseases: Compositional and Functional Features. Allergy Asthma Immunol. Res. 2021, 13, 56–74. [Google Scholar] [CrossRef]
- Li, K.J.; Chen, Z.L.; Huang, Y.; Zhang, R.; Luan, X.Q.; Lei, T.T.; Chen, L. Dysbiosis of lower respiratory tract microbiome are associated with inflammation and microbial function variety. Respir. Res. 2019, 20, 272. [Google Scholar] [CrossRef]
- Al Bataineh, M.T.; Hamoudi, R.A.; Dash, N.R.; Ramakrishnan, R.K.; Almasalmeh, M.A.; Sharif, H.A.; Al-Hajjaj, M.S.; Hamid, Q. Altered respiratory microbiota composition and functionality associated with asthma early in life. BMC Infect. Dis. 2020, 20, 697. [Google Scholar] [CrossRef]
- Perez-Losada, M.; Castro-Nallar, E.; Garcia-Huidobro, J.; Boechat, J.L.; Delgado, L.; Rama, T.A.; Oliveira, M. Characterization of the oral mycobiome of Portuguese with allergic rhinitis and asthma. Curr. Res. Microb. Sci. 2024, 7, 100300. [Google Scholar] [CrossRef]
- Perez-Losada, M.; Castro-Nallar, E.; Garcia-Huidobro, J.; Boechat, J.L.; Delgado, L.; Rama, T.A.; Oliveira, M. The nasal mycobiome of individuals with allergic rhinitis and asthma differs from that of healthy controls in composition, structure and function. Front. Microbiol. 2024, 15, 1464257. [Google Scholar] [CrossRef] [PubMed]
- Faust, K.; Raes, J. Microbial interactions: From networks to models. Nat. Res. Microbiol. 2012, 10, 538–550. [Google Scholar] [CrossRef] [PubMed]
- Faust, K.; Sathirapongsasuti, J.F.; Izard, J.; Segata, N.; Gevers, D.; Raes, J.; Huttenhower, C. Microbial co-occurrence relationships in the human microbiome. PLoS Comput. Biol. 2012, 8, e1002606. [Google Scholar] [CrossRef] [PubMed]
- Sedghi, L.; DiMassa, V.; Harrington, A.; Lynch, S.V.; Kapila, Y.L. The oral microbiome: Role of key organisms and complex networks in oral health and disease. Periodontol 2000 2021, 87, 107–131. [Google Scholar] [CrossRef]
- Dickson, R.P.; Erb-Downward, J.R.; Huffnagle, G.B. Towards an ecology of the lung: New conceptual models of pulmonary microbiology and pneumonia pathogenesis. Lancet. Respir. Med. 2014, 2, 238–246. [Google Scholar] [CrossRef]
- Hooks, K.B.; O’Malley, M.A. Dysbiosis and Its Discontents. mBio 2017, 8, e01492–e01517. [Google Scholar] [CrossRef]
- Lozupone, C.A.; Stombaugh, J.I.; Gordon, J.I.; Jansson, J.K.; Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 2012, 489, 220–230. [Google Scholar] [CrossRef]
- Lynch, M.D.; Neufeld, J.D. Ecology and exploration of the rare biosphere. Nat. Rev. Microbiol. 2015, 13, 217–229. [Google Scholar] [CrossRef]
- Litchman, E.; Villeger, S.; Zinger, L.; Auguet, J.C.; Thuiller, W.; Munoz, F.; Kraft, N.J.B.; Philippot, L.; Violle, C. Refocusing the microbial rare biosphere concept through a functional lens. Trends Ecol. Evol. 2024, 39, 923–936. [Google Scholar] [CrossRef]
- Reyman, M.; Clerc, M.; van Houten, M.A.; Arp, K.; Chu, M.; Hasrat, R.; Sanders, E.A.M.; Bogaert, D. Microbial community networks across body sites are associated with susceptibility to respiratory infections in infants. Commun. Biol. 2021, 4, 1233. [Google Scholar] [CrossRef]
- Hilton, S.K.; Castro-Nallar, E.; Perez-Losada, M.; Toma, I.; McCaffrey, T.A.; Hoffman, E.P.; Siegel, M.O.; Simon, G.L.; Johnson, W.E.; Crandall, K.A. Metataxonomic and Metagenomic Approaches vs. Culture-Based Techniques for Clinical Pathology. Front. Microbiol. 2016, 7, 484. [Google Scholar] [CrossRef]
- Perez-Losada, M.; Narayanan, D.B.; Kolbe, A.R.; Ramos-Tapia, I.; Castro-Nallar, E.; Crandall, K.A.; Dominguez, J. Comparative Analysis of Metagenomics and Metataxonomics for the Characterization of Vermicompost Microbiomes. Front. Microbiol. 2022, 13, 854423. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pérez-Losada, M. Different Phenotypes of Pediatric Asthma Show Distinct Bacterial Functional Profiles and Network Relationships. Allergies 2025, 5, 14. https://doi.org/10.3390/allergies5020014
Pérez-Losada M. Different Phenotypes of Pediatric Asthma Show Distinct Bacterial Functional Profiles and Network Relationships. Allergies. 2025; 5(2):14. https://doi.org/10.3390/allergies5020014
Chicago/Turabian StylePérez-Losada, Marcos. 2025. "Different Phenotypes of Pediatric Asthma Show Distinct Bacterial Functional Profiles and Network Relationships" Allergies 5, no. 2: 14. https://doi.org/10.3390/allergies5020014
APA StylePérez-Losada, M. (2025). Different Phenotypes of Pediatric Asthma Show Distinct Bacterial Functional Profiles and Network Relationships. Allergies, 5(2), 14. https://doi.org/10.3390/allergies5020014




