Abstract
Food allergy, particularly peanut allergy (PA), is a growing health concern affecting millions globally. PA can lead to severe reactions, including fatal anaphylaxis. Despite the availability of FDA-approved therapies like Palforzia, a cure remains elusive. Current immunotherapies show promise but lack a definitive cure. This study applies an established computational biology tool to design aptamers targeting Ara h1 and Ara h2. The in silico design aims to streamline the selection process, enabling cost-effective and rapid identification of aptamer candidates. The developed aptamers (AYA22A, including AYA22AR321, AYA22AR211, and AYA22AR524), demonstrated efficacy in inhibiting degranulation of RBL-2H3 cells (rat basophilic leukemia cell line) in vitro. They showed promise in neutralizing peanut allergen-induced immune responses. The selected aptamers inhibited degranulation in RBL-2H3 cells, addressing concerns in raw peanuts. Moreover, these aptamers demonstrated stability and effectiveness in peanut plant seeds and commercial products. Our aptamers exhibited potential in modulating immune responses associated with peanut allergy. They influenced Th1/Th2 balance, indicating a role in cytokine regulation. In vitro studies also showed the aptamers’ impact on immune cell expression and cytokine production, resembling responses observed with established immunotherapies. The findings suggest AYA22A aptamers as a potential therapeutic option for peanut allergy, providing a basis for further in vivo investigations.
1. Introduction
Food allergy is an increasing health issue that affects millions of people in the United States and worldwide. Peanut allergy (PA) is a major adult and pediatric food allergy and a leading cause of fatal anaphylaxis [1]. In the United States, an estimated 1.25 million children (≤17 years old) [2] and 4.6 million adults suffer from peanut allergy [3]. In a study on the burden of peanut allergies, 56.9–59.9% of adolescents and adults reported experiencing at least one PA-related emergency or urgent care visit in the past 12 months [4]. People with PA can experience severe symptoms ranging from urticaria to anaphylaxis. In addition, peanut sensitivity usually appears at an early age and often persists throughout life [5]. While the FDA approved Palforzia (Peanut Allergen powder), the first peanut oral immunotherapy (pOIT), in 2020 to promote desensitization and reduce the risk of peanut allergic reactions in children, it is important to note that Palforzia is not a cure for peanut allergy (PA), meaning it is still necessary for those with PA to avoid peanuts and carry injectable epinephrine. The risk to individuals with peanut sensitivity is on the rise due to several factors. These include the severity of allergic reactions, the expanded utilization of peanuts as protein extenders in processed foods, misleading or inadequate product labeling, contamination from hidden allergens, and the widespread inclusion of peanut products in processed food [6].
PA is caused by peanut allergen-induced lgE-mediated type I hypersensitivity reaction. When patients with PA consume products containing peanuts, peanut allergens pass through mucosal barriers and crosslink with lgE on mast cells and basophils, activating them and leading to degranulation. This release allergic mediators, cytokines, and chemokines, which recruit additional inflammatory cells [7]. A total of 16 peanut allergens have been identified [8]. Among them, Arachis hypogaea 1, 2, and 3 (Ara h1, Ara h2, and Ara h3, respectively) are seed storage proteins and the main allergens in the United States. Ara h2 is a 2S albumin with a 17 KDa molecular weight. It possesses high heat stability and immunogenicity, presenting challenges in removing it from food products during processing or breaking it down in the digestive system [9,10,11]. Ara h2 is a more important peanut allergen in triggering severe allergic reactions and inducing effector cell activation than Ara h1 and Ara h3. Furthermore, Ara h2 is one of the best predictors of peanut allergy in adults and is found in serum lgE of 90% of PA patients [12]. Ten linear lgE-binding epitopes have been identified in Ara h2. Studies showed that the three-dimensional structure of Ara h2 plays a critical role in peanut allergen stability and determines immunodominant lgE-binding epitopes [13].
Due to the IgE-mediated nature of food allergy, several efforts have been made to develop immunotherapy methods that are both safe and effective. Currently reported immunotherapy approaches encompass both antigen-specific and non-specific methods. Antigen-specific approaches consist of oral, sublingual, and epicutaneous immunotherapy. A recent study found that peanut oral immunotherapy can induce desensitization [14]. Another study by Kim et al. [15] also demonstrated that peanut-specific sublingual immunotherapy can induce significant desensitization after 12 months of therapy. Nonspecific approaches include anti-IgE antibodies and a Chinese herbal formulation [16]. Although the new approaches show promising results [17], no curative immunotherapy is available for the treatment of peanut allergy [18].
Aptamers are short, single stands of DNA or RNA that bind to target molecules with high specificity and affinity. They are typically generated through a laboratory process known as systematic evolution of ligands by exponential enrichment (SELEX), in which a library of randomized sequences is incubated with its target molecule and selected for its ability to bind [19,20,21]. Aptamers have been explored as a potential strategy for preventing allergic reactions. One approach is to use aptamers to sequester IgE in the bloodstream, preventing it from binding to allergens and triggering an allergic response [22]. Another approach is to use aptamers to block the binding of IgE to its receptor on immune cells, thereby preventing the activation of the immune response. The aptamer was shown to inhibit the binding of IgE to FcRI in vitro and to reduce allergic responses in a mouse model of anaphylaxis [23,24]. These studies proposed that an aptamer could be developed as a potential therapeutic agent for the treatment of IgE-mediated allergic reactions.
In this study, we applied an established computational biology tool to design aptamers, demonstrating its effectiveness in identifying novel aptamer candidates targeting a diverse range of molecules. Our results highlight the tool’s robustness and versatility, building on our previous findings [25] to further validate its application in aptamer design and predict aptamers that bind to specified target molecules with high affinity. This approach reduces the time and cost associated with traditional SELEX methods and can effectively identify novel aptamer candidates targeting a diverse range of molecules, as demonstrated in a prior study [25]. Using an in silico approach, we developed an aptamer that binds to peanut allergens Ara h2 and Ara h1 and neutralizes the immune response induced by them. Our in vitro data show that AYA22A aptamers were able to inhibit the degranulation of RBL-2H3 cells (rat basophilic leukemia cell line) [26,27,28]. The presence of allergenic proteins such as Ara h1 and Ara h2 in raw peanuts can pose a risk of allergic reaction. Our research indicates that post processing of peanut products using aptamers may provide a solution to this problem, as they have the potential to effectively mask the allergenic agent and ensure the safety of the product for consumption. Overall, while aptamers are still in the early stages of development as therapeutics for allergic reactions, their potential as a new class of allergy treatments is an active area of research.
2. Materials and Methods
2.1. Initial Aptamer Generation
Fifty aptamer sequences, each consisting of 40 nucleotides, were produced using a random sequence generator to establish a diverse population of candidates for further analysis. The nucleotide composition of these sequences maintained an equal ratio of 1:1:1:1 of adenine (A), thymine (T), guanine (G), and cytosine (C), respectively. This ratio is crucial, as it reduces bias towards specific nucleotide motifs and shows a balanced representation of the sequence space.
2.2. Iterative Selection and Mutation
An iterative selection process was then carried out to refine and improve the aptamer sequences. The top 15 aptamers with the lowest docking scores, as determined by structure prediction and docking simulation, were selected for the next round of evolution. To introduce diversity into the aptamer pool, two sites within each selected aptamer sequence were randomly chosen, and a random nucleotide mutation was introduced at each site. The mutation involved replacing the original nucleotide with another nucleotide (A, T, G, or C) with equal probability. A total of 40 out of the 50 new aptamers were generated based on the sequences from the previous round, ensuring a degree of continuity and cumulative improvement. The remaining 10 aptamers were randomly generated following the same protocol as in the initial generation step. This process was repeated for a total of 10 rounds of selection. This iterative approach aimed to gradually optimize the aptamer sequences for enhanced binding affinity and specificity towards the target Ara h1 and Ara h2 proteins. After the 10th round of iterative selection, the top 23 aptamers were identified based on their docking scores and RMSD scores. These aptamers exhibited the most favorable binding interactions with the target Ara h1 and Ara h2 proteins. The selection of these top aptamers marked the completion of the computational phase and paved the way for subsequent experimental validation studies.
2.3. Secondary and Tertiary Structure Prediction
In each round of selection, a comprehensive computational predication approach was utilized to predict the secondary and tertiary structures of the generated aptamers. The DNA fold service [29] is a well-established bioinformatic tool that uses advanced algorithms and thermodynamic calculations to predict secondary structures and calculates the energy associated with different potential folding patterns for each aptamer sequence. The sequences with the lowest energy were selected for further investigation. To determine the three-dimensional (3D) structure of the selected aptamer sequences, we first utilized the RNAcomposer tool [30,31], which is known for its accuracy and reliability in predicting 3D RNA structures. Each aptamer sequence, paired with its corresponding secondary structure, was provided as input to RNAcomposer. Energy minimization procedures were then applied to refine and optimize the 3D structures of the aptamers, enhancing their stability and accuracy.
2.4. Docking Simulation
To investigate the potential interactions between the aptamers and Ara h1 and Ara h2 proteins, docking simulations were performed using HADDOCK 2.4 [32,33]. The predicted 3D protein structure was obtained from the AlphaFold protein structure database [34]. PyMOL (The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC, New York, NY, USA) was used to visualize the proteins and prep them for docking. The aptamer structures generated during the iterative selection were docked onto these epitopes using appropriate software and algorithms. Docking scores were recorded as a measure of the binding affinity between each aptamer and the target. The sequences for the selected AYA22A aptamers are listed in the Supplementary Materials, Table S1.
2.5. Cell Lines
Rat basophilic leukemia cell line RBL-2H3 were purchased from the American Type Culture Collection (ATCC, #CRL-2256). Cells were cultured with Eagle’s Minimum Essential Medium (EMEM) (ATCC, #30-2003) supplemented with 10% HyClone Standard Fetal Bovine Serum (FBS) (Cytiva life Sciences, Burlington, NJ, USA SH30088.03HI) and 50 U/mL penicillin/streptomycin (Gibco, New York, NY, USA). Cells were cultured in a humidified incubator at 37 °C in 5% CO2 as previously described [35]. Materials and key resources used in the methods are mentioned in detail in Supplementary Materials, Table S2.
2.6. Characterization of Aptamer Binding to Ara h2 Peanut Protein by ELISA
Overnight, a MaxiSorp plate was coated with 2 µg/mL of recombinant Ara h2 peanut protein (Inbio, VA, USA) prepared in a 50 mM carbonate-bicarbonate solution. Following incubation, the plate underwent blocking for 1 h using 2% BSA in 1 mM MgCl2 supplemented with 1XPBS/Tween solution. Subsequently, after washing twice with 1XPBS/Tween solution, biotinylated AYA22A aptamers (the selected AYA22A aptamers; Supplementary Table S1) were added at a concentration of 100 nM in duplicate and incubated at room temperature for 1 h. For controls, non-specific aptamers (control aptamers 1–6; Supplementary Table S1), and a no-aptamer condition were included. The wells were washed three times with 1XPBS/Tween (0.05%) solution, and HRP-conjugated streptavidin (Thermofisher Scientific, Plaquemine, LA, USA) was employed, along with TMB, for colorimetric detection. The reaction was halted using 1 M H2SO4, and absorbance was measured using a spectrophotometer at 450 nm.
2.7. Assessment of the Specificity of AYA22A Aptamers to Ara h2 Protein by Competition ELISA
A competition assay was conducted to ascertain the specificity of AYA22A aptamers to Ara h2 protein. Overnight, a MaxiSorp plate was coated with 2 µg/mL of recombinant Ara h2 peanut protein (Inbio, VA, USA) prepared in a 50 mM carbonate–bicarbonate solution. Subsequently, the plate was blocked for 1 h with 2% BSA in 1 mM MgCl2 supplemented with 1XPBS/Tween solution. Following extensive washing using 1XPBS/Tween (0.05%) solution, biotinylated aptamers were added to the wells in the presence of 100× non-biotinylated aptamers at various concentrations (500 nM, 200 nM, 100 nM, and 50 nM). The plate was then incubated at room temperature for 1 h. Direct binding of biotinylated aptamers to Ara h2 protein served as the positive control compared to wells containing the 100× non-biotinylated aptamers. The binding of the aptamers was detected using HRP-conjugated streptavidin (Thermofisher Scientific, Plaquemine, LA, USA) with incubation for 1 h. After the addition of TMB and incubation for up to 10 min for color development, the reaction was terminated using 1 M H2SO4, and absorbance was measured using a spectrophotometer at 450 nm.
2.8. Aptamer Specificity to Ara Proteins (Ara h1, Ara h2, and Ara h6) Determined by ELISA
Overnight, a MaxiSorp plate was coated with 2 µg/mL of natural Ara h1 (Inbio, VA, USA), recombinant Ara h2 (Inbio, VA, USA) and recombinant Ara h6 (Biosynth, Compton, UK) peanut proteins prepared in a 50 mM carbonate–bicarbonate solution. Subsequently, the plate was blocked for 1 h using 2% BSA in 1 mM MgCl2 supplemented with 1XPBS/Tween (0.05%) solution. After washing twice with 1XPBS/Tween (0.05%) solution, biotinylated AYA22A aptamers (AYA22AR211, AYA22AR321, and AYA22AR524) were added at various concentrations (500 nM, 100 nM, and 50 nM) in duplicate and incubated at room temperature for 1 h. The wells were washed three times with 1XPBS/Tween (0.05%) solution, and HRP-conjugated streptavidin (Thermofisher Scientific, Plaquemine, LA, USA) was utilized in conjunction with TMB for colorimetric detection. The reaction was terminated using 1 M H2SO4, and absorbance was measured using a spectrophotometer at 450 nm.
2.9. Extraction of Crude Peanut Protein
Crude peanut protein was extracted as previously described [36], with slight modifications. Briefly, peanuts were purchased from a local shop and ground in the laboratory. Then, 2 mg of peanut flour was suspended in 2 mL of EMEM media (pH 7.0 to 7.4), and the mixture was vortexed at high speed for 20 min at room temperature. Subsequently, the mixture was centrifuged at 500× g at room temperature for 5 min, and the supernatant was collected. The collected supernatant then underwent an additional vortexing step at high speed for 20 min. The crude mixture was then centrifuged at 16,000× g for 5 min. The resulting supernatant, referred to as crude protein extract, was collected for experiments, and the protein concentration was measured using a bicinchoninic acid assay (BCA assay).
2.10. Extraction of Crude Protein from Peanut Butter
Peanut butter is primarily prepared by roasting, a process that induces biophysical mechanisms leading to enhanced allergenic properties of major peanut allergens Ara h1 and Ara h2. Additionally, roasted Ara h2 has been observed to shield Ara h1 from trypsin digestion. Thus, thermal processing has been demonstrated to modify both the structure and allergenicity of peanuts [37]. Consequently, we assessed the aptamers’ efficacy in mitigating the anaphylactic potential of Ara h2 in peanut butter through a degranulation assay using RBL-2H3 cells. Briefly, peanut butter extract was prepared using the same protocol utilized for crude peanut extract. This involved suspending 2 mg of peanut butter in 2 mL of EMEM media, following the instructions outlined in Section 2.9. The resulting supernatant, comprising the crude protein extract of peanut butter, was collected for experiments, and the protein concentration was measured using a BCA assay.
2.11. RBL-2H3 Cells Degranulation and -Hexosaminidase Assay
RBL-2H3 cells (1 × 106 cells/well) were cultured in EMEM media without FBS. The cells were sensitized with 1 µg/mL of IgE dansyl (clone 27–74) for 30 min at 37 °C in 5% CO2. Sensitized cells were then stimulated with or without peanut protein at different serial dilutions in the presence or absence of Ara h2-specific aptamers (5 µM) and non-specific aptamers (5 µM) for 1 h at 37 °C in 5% CO2. Degranulation was detected spectroscopically by measuring the activity of the granule-stored enzyme -hexosaminidase secreted into the supernatant. As a positive control, to detect the total -hexosaminidase in the cytoplasm, the cells were lysed with 1% Triton X-100. After 1 h, 25 µL of the cell culture supernatants and cell lysates were incubated with 1 mM p-nitrophenyl-N-acetyl--d-glucosamine in 0.05 M citrate buffer (pH 4.5) for 1 h at 37 °C, followed by stopping solution [200 µL of 0.05 M sodium carbonate buffer (pH 10.0)]. The -hexosaminidase activity was determined by measuring the difference in absorbance at 405 nm and 630 nm [38], and the percentage of degranulation was calculated as follows [39]: percent (%) degranulation = (experimental -hexosaminidase release − vehicle control -hexosaminidase release)/(Triton-X-100 -hexosaminidase release − vehicle control -hexosaminidase release) × 100. All the degranulation assays were repeated in triplicate, at least.
2.12. Detection of Aptamer Uptake by Peanut Plant by Real-Time PCR Assay
This study examined the uptake of the AYA2012004_L aptamer (control aptamer 1) in peanut plants. Peanut plants were watered daily for 4 days with a 5 µM solution of the AYA2012004_L aptamer, which is 86 nucleotides long and contains primer binding sites (Supplementary Table S1). A separate set of plants was watered without any aptamer as a mock control. After the treatment period, the roots, stems, leaves, and seeds were collected from the peanut plants and processed for nucleic acid extraction. The presence of the AYA2012004_L aptamer in the plant tissues was detected using a real-time PCR assay, following the previously described method for detecting this aptamer [40]. In addition, peanut seeds were planted and watered with either 5 µM AYA22A aptamers or control aptamers (Supplementary Table S1) daily from sprouting until the plants reached the fruiting stage (approximately 100 days). Laboratory-developed aptamer-treated seeds from RBL-2H3 cells were utilized for the degranulation-induced study and for ELISA to detect the aptamers in different parts of the plant body.
2.13. Enzyme-Linked Immunosorbent Assay (ELISA)-Based Based Detection of AYA22AR321 in Peanut Crude Samples
Crude peanut samples with protein concentrations of 500 µg/mL and 250 µg/mL were prepared in a 50 mM carbonate–bicarbonate solution and added to MaxiSorp plates for overnight incubation. The wells were then spiked with 5 µM of the AYA22AR321 aptamer. Following incubation, the wells were washed twice and subsequently blocked for 60 min with 2% BSA in a solution containing 1 mM MgCl2 supplemented with 1XPBS/Tween. After washing, the presence of the AYA22AR321 aptamer was detected using biotinylated 3′ and 5′ complementary DNA at a concentration of 5 µM. Colorimetric detection was achieved using HRP-conjugated streptavidin in conjunction with TMB. The reaction was terminated with 1M H2SO4, and absorbance was measured using a spectrophotometer at 450 nm.
2.14. Aptamer Detection in AYA22AR321-Watered Peanut Plants Using ELISA
Overnight, a MaxiSorp plate was coated with preparations of AYA22AR321-watered peanut plant crude extract prepared from seeds, leaves, roots, and stem at a protein concentration of 0.25 mg/mL. Details regarding the preparation of these peanut plant sections are provided in the Supplementary Material. Following the overnight coating, the plate underwent extensive washing with a 1XPBS/Tween solution. Subsequently, the wells were blocked with 2% BSA in a solution containing 1mM MgCl2 supplemented with 1XPBS/Tween for 60 min at room temperature. After washing twice, AYA22AR321 was detected using biotinylated 3′ complementary DNA at a concentration of 2 µM. HRP-conjugated streptavidin (Thermo Fisher Scientific, Plaquemine, LA, USA) was employed in conjunction with TMB for colorimetric detection. The reaction was terminated using 1 M H2SO4, and absorbance was measured using a spectrophotometer at 450 nm.
2.15. PBMC Isolation and Generation of Human Monocyte-Derived Dendritic Cells
Blood samples were obtained from healthy donors’ buffy coats (Carter BloodCare), and peripheral blood mononuclear cells (PBMCs) were isolated as described previously [25]. Monocytes were then enriched from the PBMCs using an EasySep™ Human Monocyte Enrichment Kit (#19059) and cultured in complete RPMI-1640 (cRPMI) media. The cRPMI media consisted of RPMI-1640 media (Gibco, 11875-093) supplemented with 10% Fetal Bovine Serum (FBS) (Cytiva Life Sciences HyClone, SH30088.03HI), penicillin/streptomycin (Gibco, 15140-122), MEM non-essential amino acids (Gibco, 11140-050), 1 mM sodium pyruvate (Gibco, 11360-070), and 25 mM HEPES (Cytiva, SH40003.01). To induce differentiation of monocytes into dendritic cells (DCs), the monocytes were cultured in media supplemented with 20 ng/mL IL-4 and 40 ng/mL granulocyte–macrophage colony-stimulating factor (GM-CSF) for 6 days in a 37 °C incubator with 5% CO2 [41]. Media were refreshed with the aforementioned cytokines every other day. Following the differentiation period, the cultured DCs were harvested and stimulated with 10 ng/mL of IL-1, 10 ng/mL IL-6, 10 ng/mL TNF-, and 1 µg/mL prostaglandin E2 (PGE2). The stimulation of the DCs was confirmed through the expression analysis of various markers by staining with BV510-CD80, PE-CD86, PE-Dazzel-PD-L1, AF700-CD14, FITC-CD11c, Kro-HLA-DR, and live–dead dyes, followed by fixation using flow cytometry (Supplementary Table S2). Data were analyzed by FlowJoTM v10.8.1 software (Becton Dickinson Life Sciences, Franklin Lakes, NJ, USA), and results were expressed as geometric mean fluorescence intensity (gMFI).
2.16. AYA22A Aptamer Attenuates Cross-Presentation of Peanut Allergens by Mo-DCs
Mo-DCs were then pulsed with or without a cell stimulation cocktail (IL-1b, IL-6, TNF, and PGE2) in the presence and absence of peanut crude protein at dilutions of 1:4 (2.75 mg/mL), 1:8 (1.36 mg/mL), 1:16 (0.69 mg/mL), and 1:32 (0.34 mg/mL); rAra h2 (0.016 mg/mL) with or without 5 µM of AYA22AR321 and control aptamer for 48 h at 37 °C incubator with 5% CO2. Cells were then stained with biotinylated anti-Ara h2 antibody (0.5 µL/well; Absolute antibody, #AB01494-23.0), followed by streptavidin APC (1.0 µL/50 µL cell staining buffer/well) and live–dead dye (1:1000 dilution with cell-staining buffer). The stained cells were analyzed using a NaviosTM EX Flow Cytometer 2.0 (Beckman Coulter Inc., Brea, CA, USA). Flow cytometry data were subsequently analyzed using FlowJoTM v10.8.1 software (Becton Dickinson Life Sciences), and results were expressed as geometric mean fluorescence intensity (gMFI).
2.17. In Vitro Co-Culture Model of Mo-DCs and T Cells to Evaluate the Effect of AYA22A Aptamers on Th2 Cytokines Triggered by Peanut Allergens
A mixed lymphocyte reaction (MLR) was undertaken to assess the capacity of Mo-DCs to prime T cells. Total T cells isolated using an EasySepTM Human T Cell Isolation Kit (Catalog #17951) from autologous PBMCs were labeled with 10 µM carboxyfluorescein succinimidyl ester (CFSE; Invitrogen, Carlsbad, CA, USA) for 20 min at 37 °C. Mo-DCs were exposed to 10 ng/mL of IL-1, 10 ng/mL IL-6, 10 ng/mL TNF-, and 1 µg/mL prostaglandin E2 (PGE2) in the presence and absence of peanut crude extract at a dilution of 1:4 (2.75 mg/mL) and/or recombinant Ara h2 (rAra h2, 0.08 mg/mL) alone or in combination with AYA22AR321 (5 µM) and/or AYA22AR211 (5 µM) for 48 h, then co-cultured with CFSE-stained autologous enriched T cells for 48 h. protein transport inhibitors containing brefeldin A and monesin were added as described previously [42]. Th2 cell cytokine expression in proliferated, non-proliferated, and total CD4+ T cells was assessed by intracellular cytokine staining using AF700 CD3, PerCP/Cy5.5 CD8, APC/Cy7 CD4, APC IL-5, PE/Cy7 IL-13, and PE/Dazzle 594 IL-4 (Supplementary Table S2). Th2 cell cytokine expression in proliferated, non-proliferated, and total CD4+ T cells was assessed by intracellular cytokine staining.
2.18. Mechanistic Evaluation of AYA22A Aptamers in Inducing T-Cell Cytokines: Co-Culture Study with Autologous PBMCs
We implemented a mechanistic approach to determine if AYA22A aptamers can induce Th1 cytokines in the co-culture of autologous PBMCs. The PBMCS were spiked with or without peanut crude extract at a dilution of 1:64 (0.17 mg/mL) and incubated for 24 h at 37 °C in a 5% CO2 incubator. After 24 h incubation, cells were collected and washed by centrifugation at 300× g with cRPMI media and further co-cultured with autologous human PBMCs (0.5 × 106) stained with CFSE at a 1:1 ratio in the presence or absence of 5 µM AYA22A-aptamers (AYA22AR321, AYA22AR211, and AYA22AR524) and control aptamer in 200 µL of cRPMI media for 72 h (3 days) at 37 °C in a 5% CO2 incubator. After incubation, the supernatant and cells were collected. T-cell cytokines were assessed by legendplexTM and intracellular cytokine staining.
2.19. Mechanistic Evaluation of AYA22A Aptamers in Regulating CD23 and CD63 Expression on B Cells: Co-Culture Study with Autologous PBMCs
It has been demonstrated that the density of CD23 on the surface of B cells correlates with IgE levels and plays a crucial role in facilitating allergen uptake by IgE and activating allergen-specific T cells [43]. Additionally, the significance of CD23 has been underscored in a non-inflammatory pathway involving IgE–allergen complexes [44]. In this study, we employed a mechanistic approach to investigate whether peanut allergen-specific aptamers can induce the expression of CD23 and CD63 in co-cultures of autologous PBMCs. PBMCs were exposed or not to peanut crude extract at dilutions of 1:4 (2.75 mg/mL), 1:8 (1.38 mg/mL), 1:128 (0.86 mg/mL), and 1:256 (0.43 mg/mL) (for assessment of CD23 expression). PBMCs were also exposed or not to peanut crude extract at a dilution of 1:256 (for evaluation of CD63 expression). Exposed PBMCs were incubated for 24 h at 37 °C in a 5% CO2 incubator. Subsequently, the exposed PBMCs were collected, washed, and co-cultured with autologous human PBMCs stained with CFSE at a 1:1 ratio in the presence or absence of 5 µM AYA22A-aptamers (AYA22AR321, AYA22AR211, and AYA22AR524), along with a control aptamer, in 200 µL of cRPMI media for 72 h at 37 °C in a 5% CO2 incubator. After incubation, cells were stained with AF700 CD3, PerCP FcRI, APC/Cy7 CD23, PE-CF594 CD19, PE CD63, BV510 CD45, and live–dead dyes. Subsequently, they were fixed and analyzed using flow cytometry. Data analysis was conducted using FlowJoTM v10.8.1 software (Becton Dickinson Life Sciences).
2.20. Preventative and Therapeutic Basophil Activation Test
To assess the preventative and therapeutic effects of the AYA22AR321 aptamer on the reduction of the expression of degranulation markers on basophils triggered by peanut allergens, we conducted in vitro experiments using a basophil activation test (BAT) [45] with human peripheral blood mononuclear cells (PBMCs) obtained from healthy donors (N = 2). PBMCs were treated with 5 µM of AYA22AR321 aptamer for the preventative study. For the therapeutic study, PBMCs were exposed to peanut crude samples at a dilution 1:64 (0.17 mg/mL) and/or recombinant Ara h2 (rAra h2) (0.08 mg/mL). In the induced study, PBMCs were untreated. All treatments were incubated at 37 °C in a 5% CO2 incubator for 24 h. Subsequently, the cells were washed once with media and further incubated for 15 min under various conditions, For the induced study, conditions included IgE (human Anti-Ara h2/Ara h3 [PA12P3F10], 1 mg/mL), IgE (1 mg/mL) + peanut crude (0.17 mg/mL), IgE (1 mg/mL) + peanut crude (0.17 mg/mL) + AYA22AR321 (5 µM), and no exposure. For the preventative and therapeutic studies, conditions included IgE (1 mg/mL), IgE (1 mg/mL) + peanut crude (0.17 mg/mL) / rAra h2 (0.08 mg/mL), IgE (1 mg/mL) + peanut crude (0.17 mg/mL) / rAra h2 (0.08 mg/mL) + AYA22AR321 (5 µM), and no exposure. After incubation, cells were stained with CD3-AF700, CD123-APC, CD63-PE, HLA-DR-BV510, and live–dead dyes, followed by fixation using flow cytometry. Data were analyzed by FlowJoTM v10.8.1 software (Becton Dickinson Life Sciences), and results were expressed the gated percent frequency of CD3−CD123+HLA-DR−CD63+ cells.
2.21. Statistical Analysis
Data are presented as mean ± standard deviation(SD). GraphPad Software 5.0 was applied to perform Student’s unpaired t tests. p < 0.05 was considered to be statistically significant.
3. Results
3.1. Design and Molecular Docking Analysis of Aptamers Targeting Ara h1 and Ara h2 Proteins Using In Silico Approaches
We utilized an in silico (computational) methodology to rationally design DNA aptamers—short single-stranded oligonucleotides—with specific binding affinity towards the Ara h1 and Ara h2 proteins. Figure 1(a1,b1,c1) visualize the molecular docking complexes and interactions between the Ara h1 protein and the aptamer in illustrations generated using the HADDOCK protein-DNA docking software suite (versions 2.4/2.6) [32,33]. These figures provide detailed representations of the structural complex formed between Ara h1 and the aptamer. In contrast, Figure 1(a2,b2,c2) illustrate analogous docking analysis outcomes between the aptamer and the Ara h2 protein. Overall, these computational docking results predict and characterize successful in silico binding between the engineered aptamers and their target proteins. Lower (negative) docking scores typically correlate with stronger theoretical binding affinities and the formation of more structurally and energetically favorable complexes, providing a quantitative assessment. The comprehensive structural analysis and biophysical characterization of these docking poses and interfaces (Figure 1) offer valuable mechanistic insights into potential binding modes, molecular recognition, and key contacts governing aptamer/protein interactions. This computational framework establishes a foundation for subsequent empirical binding validation and further optimization of these aptamers. For example, having validated binding and determined favorable aptamer/protein poses, directed evolution and mutagenesis studies can now rationally modify the aptamer sequence to improve affinity and specificity. Specific aptamer nucleotides making key contacts at the binding interface can be mutated in silico and re-docked to evaluate changes in binding.
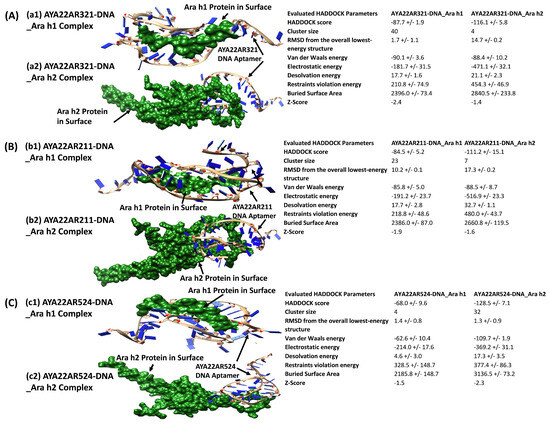
Figure 1.
In silico binding of the aptamer with Ara h1 and Ara h2 proteins. (A) The docking complex of Ara h1 protein and the AYA22AR321 aptamer obtained from HADDOCK Docking Software [33]. The docking simulation produced a docking score of −87.7 ± 1.9 (a1). The docking complex of Ara h2 and the AYA22AR321 aptamer obtained from HADDOCK Docking Software [33]. The docking simulation produced a docking score of −116.1 ± 5.8 (a2). (B) The docking complex of Ara h1 protein and the AYA22AR211 aptamer obtained from HADDOCK Docking Software [33]. The docking simulation produced a docking score of −84.5 ± 5.2 (b1). The docking complex of Ara h2 and the AYA22AR211 aptamer obtained from HADDOCK Docking Software [33]. The docking simulation produced a docking score of −111.2 ± 15.1 (b2). (C) The docking complex of Ara h1 protein and the AYA22AR524 aptamer obtained from HADDOCK Docking Software [33]. The docking simulation produced a docking score of −68.0 ± 9.6 (c1). The docking complex of Ara h2 and the AYA22AR524 aptamer obtained from HADDOCK Docking Software [33]. The docking simulation produced a docking score of −128.5 ± 7.1 (c2).
3.2. Aptamers Binding to Peanut Allergens
Ab aptamer specific to peanut allergens (AYA22A) was specifically developed using an in silico method for Ara h1/2 and screened using an enzyme-linked immunosorbent assay (ELISA). To achieve this, Ara h2 protein was immobilized on a MaxiSorp plate; then, as mentioned in Supplementary Table S1, biotinylated aptamers were introduced and incubated with the immobilized protein. Detection of the bound aptamers was accomplished using streptavidin-HRP and a TMB substrate, with the absorbance measured at 450 nm. Among the selected 23 AYA22A aptamers developed by our in silico method, we observed four aptamers (AYA22AR321, AYA22AR211, AYA22AR524, and AYA22AR55) that bound to peanut protein Ara h2 according to the ELISA screening (Supplementary Figure S1A). Therefore, we randomly selected three aptamers (AYA22AR321, AYA22AR211, and AYA22AR524) based on their binding to native and recombinant Ara h2 proteins. We further evaluated the binding of peanut allergen-specific aptamers (AYA22A) AYA22AR211, AYA22AR321, and AYA22AR524 to native Ara h2, which exhibited strong binding capabilities to Ara h2 protein. However, the non-specific control aptamers (1–4) did not show binding capacity for Ara h2, as depicted in Figure 2A. To determine the specificity of the aptamers with robust binding affinities, a competition ELISA-based binding assay was employed (Figure 2B). In this approach, non-biotinylated aptamers in one-hundred-fold excess relative to their biotinylated counterparts were introduced to the wells containing the respective biotinylated aptamers. If binding is specific, the non-biotinylated aptamers outcompete the biotinylated ones, leading to a reduction in absorbance at 450 nm. As demonstrated in Figure 2B, all three tested aptamers were effectively outcompeted by the corresponding non-biotinylated aptamers in one-hundred-fold excess, confirming their specific binding to the Ara h2 protein. Considering that the major allergens in peanuts are generally Ara h1 and Ara h3 (members of the cupin superfamily of proteins), as well as Ara h2 and Ara h6 (members of the prolamin superfamily), we investigated if Ara h2-targeted aptamers bound to other peanut allergens. We found that all three aptamers, namely AYA22AR211, AYA22AR321, and AYA22AR524, exhibited strong binding capabilities for Ara h1 and h2 but did not bind to Ara h6 (Supplementary Figure S1B(B1–B3)). Therefore, these selected aptamers (AYA22A) from both cupin and prolamin superfamily proteins show significant promise for applications in detecting and therapeutically intervening in peanut allergens.
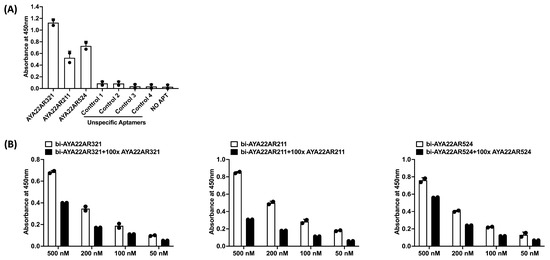
Figure 2.
Characterization of aptamers binding to Ara h2 peanut protein. (A) ELISA-based binding assay illustrating the binding between Ara h2 peanut protein and Ara h2-specific aptamers (the top three enriched aptamers are AYA22AR211, AYA22AR321, and AYA22AR524), along with non-specific aptamers (controls 1–4). Ara h2 peanut protein was immobilized on a 96-well MaxiSorp plate, and 100 nM of biotinylated aptamers was incubated with the immobilized protein in duplicate. Absorbance was measured after incubation with streptavidin horseradish peroxidase bound to the biotinylated aptamer in the presence of a TMB substrate. The reaction was terminated using 1 M H2SO4, and the absorbance was measured using a spectrophotometer at 450 nm. (B) Competition assay for aptamer binding to Ara h2 with an excess of non-biotin-labeled aptamers at different concentrations. The specific binding of aptamers (AYA22AR211, AYA22AR321, and AYA22AR524) that displayed binding to Ara h2 was tested in the presence of a 100-fold excess of the corresponding non-labeled aptamer. All data are representative of at least two independent experiments. Error bars represent mean ± SD of duplicate assays.
3.3. Aptamers Targeting Ara h1 and Ara h2 Effectively Inhibit IgE-Mediated Degranulation in an In Vitro RBL-2H3 Cell Allergy Model
To evaluate the efficacy of aptamers targeting the peanut allergens, we utilized an in vitro cell culture model employing RBL-2H3 cells to assess IgE-mediated degranulation. Given the scarcity of rat/mouse anti-Ara h2-specific IgE, we included rat serum, rat-anti-mouse IgE, and monoclonal mouse IgE dansyl (IgE dansyl clone 27–74, which is specific to hapten 5-dimethylaminonaphthalene-1-sulfonyl) in our experiments. RBL-2H3 cells were stimulated with Ara h2 protein. Our results demonstrate that IgE dansyl exhibited non-specific recognition of Ara h2 peanut allergen (Supplementary Figure S2A). This non-specific binding was likely due to cross-reactivity, where IgE against one allergen can bind to other allergens with similar molecular structures [46]. Notably, monoclonal mouse IgE dansyl showed comparable binding to Ara h2, unlike rat serum and rat-anti-mouse IgE. This suggests that IgE dansyl’s recognition extends beyond its hapten specificity, indicating a broader cross-reactive potential with peanut allergens. As peanut crude extract is known to contain major allergens, we further investigated its potential to induce anaphylactic degranulation mediated by IgE from RBL-2H3 cells. The crude extract was prepared using EMEM media (pH = 7.2), and serial dilutions of 1:8 (1.36 mg/mL), 1:16 (0.69 mg/mL), 1:32 (0.34 mg/mL), 1:64 (0.17 mg/mL), 1:128 (0.08 mg/mL), and 1:256 (0.04 mg/mL) were utilized, as shown in Supplementary Figure S3A. We observed that peanut crude extract induced IgE-mediated degranulation from RBL-2H3 cells at all tested dilutions. To confirm the specificity of this degranulation, we compared peanut crude extract-triggered degranulation with that induced by natural Ara h2 and recombinant Ara h2 (Supplementary Figure S3B). Evaluation of different dilutions revealed that peanut crude extract and natural Ara h2 and recombinant h2 induced significant degranulation at concentrations of 1:32 (0.34 mg/mL) and 1:64 (0.17 mg/mL), and 0.016 mg/mL (1000 pM) and and 0.08 mg/mL (5000 pM), respectively. Notably, peanut crude extract-triggered degranulation was more pronounced than that induced by natural or recombinant Ara h2. Based on these findings, peanut crude extract at a serial dilution of 1:32 was selected for assessment of the therapeutic intervention of AYA22A in the in vitro RBL-2H3 cell allergy model. The concentration of AYA22A used in the study was predetermined (Supplementary Figure S4).
All three AYA22A aptamers effectively inhibited cell degranulation at concentrations of 5 µM, 4 µM, and 3 µM, whereas AYA22AR211 and AYA22AR524 were not effective at 2 µM. Although 4 µM was also sufficient to block degranulation, 5 µM was selected due to its consistent and slightly higher efficacy across multiple experimental conditions. This approach ensures the robustness of the results and reduces the variability in downstream analyses. Therefore, a concentration of 5 µM was chosen for further investigation. Furthermore, Figure 3 illustrates that all three AYA22A aptamers significantly blocked peanut crude extract-triggered IgE-mediated RBL-2H3 cell degranulation. Similarly, AYA22AR321 and AYA22AR524 significantly inhibited Ara h2-triggered IgE-mediated RBL-2H3 cell degranulation as compared to non-specific aptamers (Supplementary Figure S5), further confirming the effectiveness of AYA22A aptamers in targeting peanut allergens. These results suggest that peanut allergen-specific aptamers (AYA22AR321, AYA22AR211, and AYA22AR524) have the potential to mitigate allergic responses by interfering with IgE-mediated degranulation, underscoring their therapeutic relevance in peanut allergy management.
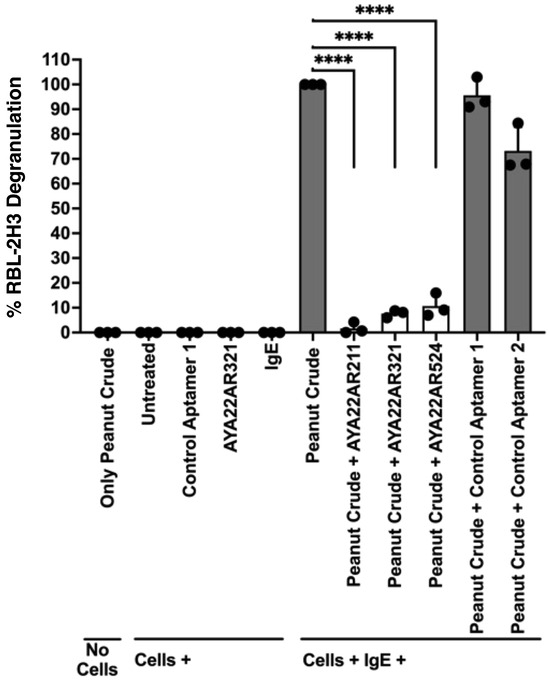
Figure 3.
Ara h2-specific aptamers mitigate the degranulation induced by peanut crude proteins from the RBL-2H3 cell line. RBL-2H3 cells (0.5 × 106) in 100 µL EMEM media without FBS were seeded into a 96-well plate with or without sensitization using 1 µg/mL of IgE dansyl (clone 27–74) and incubated at 37 °C for 1 h. Following the sensitization incubation, RBL-2H3 cells were washed twice with EMEM media without FBS and subsequently stimulated with aptamers under the indicated conditions at 37 °C for 1 h in 200 µL cell culture media. Crude peanut protein extract obtained from a crude peanut sample at a dilution of 1:64 with cell culture media was then added in the presence or absence of Ara h2-specific aptamers (5 µM) and non-specific aptamers (5 µM), along with appropriate control conditions, in 100 µL cell culture media. This mixture was further incubated at 37 °C for 30 min. Degranulation was assessed spectroscopically by measuring the activity of granule-stored enzyme -hexosaminidase secreted into the supernatant on the p-nitrophenyl-N-acetyl--d-glucosamine substrate. Percent (%) degranulation was calculated using the following formula: (Experimental -hexosaminidase release − vehicle control -hexosaminidase release)/(Triton-X-100 -hexosaminidase release − vehicle control -hexosaminidase release) × 100. All data are representative of at least two independent experiments. Error bars represent mean ± standard deviation (SD). Statistical significance was determined using an unpaired Student’s t test. **** p < 0.0001.
3.4. AYA22A Aptamers Demonstrate Stability and Efficacy in Reducing Degranulation Across Various Temperatures and Processing Conditions
Next, we evaluated the functional activity of peanut allergen-specific aptamers (AYA22A) in effectively diminishing peanut crude-induced degranulation across various temperatures. Stability tests were conducted by incubating a combination of peanut crude extract (serial dilution 1:32) and AYA22AR321 or a control aptamer at different temperatures (4 °C, 37 °C, or room temperature) for varying durations (7, 14, 21, 28, and 35 days) (Supplementary Figure S6 and Figure 4). The results demonstrate the high binding efficiency of the aptamer to the allergen, as evidenced by its ability to decrease the anaphylactic potential of the allergen in the extract and reduce degranulation induced by the peanut crude allergens. An efficiency test revealed that the aptamer remained stable at all tested temperatures, retaining its specific binding and bonding properties over an extended period. We aimed to evaluate the impact of high temperatures on the allergen–aptamer complex, assuming that aptamers themselves were not subjected to these extreme temperatures, as indicated in Supplementary Figure S7. However, our experiment primarily demonstrated the denaturation of peanut allergen proteins under these conditions, which prevented direct assessment of aptamer stability in relation to allergen binding and subsequent degranulation. Our aim was to assess the aptamers’ ability to reduce cell degranulation in response to bakery-processed peanuts triggering IgE-mediated anaphylactic reactions. Our observations revealed that light roasting of peanuts [36] did not diminish their anaphylactic potential, as it induced degranulation and the release of beta-hexosaminidase from IgE-mediated RBL-2H3 cells. However, the ability of aptamers AYA22AR321, AYA22AR211, and AYA22AR524, designed for peanut allergens, were re-confirmed, as they effectively neutralized the anaphylactic reaction induced by lightly roasted peanut crude extract (Supplementary Figure S8). These findings highlight the potential of AYA22A aptamers as therapeutic agents to mitigate allergic responses induced by peanut allergens, particularly under conditions involving thermal processing such as light roasting.
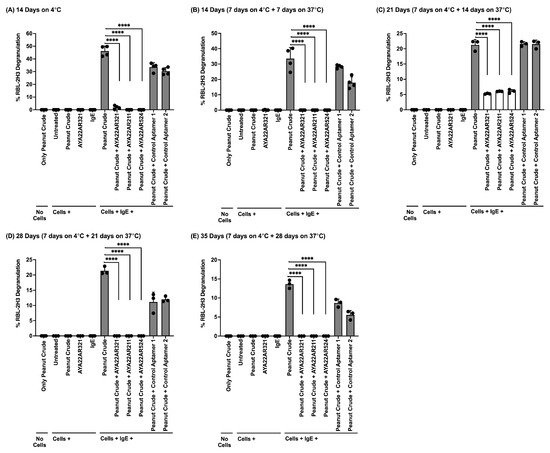
Figure 4.
AYA22A aptamers maintained efficacy and stability despite prolonged incubation periods. Crude peanut protein extract obtained from a crude peanut sample at a dilution of 1:64 with cell culture media was added in the presence or absence of Ara h2-specific aptamers (5 µM) and non-specific aptamers (5 µM), along with appropriate control conditions, in 5 mL cell culture media. This mixture was then incubated at various time points and temperatures (A–E) for 14, 21, 28, or 35 days. RBL-2H3 cells (0.5 × 106) in 100 µL EMEM media without FBS were seeded into a 96-well plate with or without sensitization using 1 µg/mL of IgE dansyl (clone 27–74) and incubated at 37 °C for 1 h. Following the sensitization incubation, RBL-2H3 cells were washed twice with EMEM media without FBS and subsequently stimulated with aptamers under the indicated conditions (A–E) at 37 °C for 1 h in 200 µL cell culture media. Degranulation was assessed spectroscopically by measuring the activity of granule-stored enzyme -hexosaminidase secreted into the supernatant on the p-nitrophenyl-N-acetyl--d-glucosamine substrate. Percent (%) degranulation was calculated using the following formula: (Experimental -hexosaminidase release − vehicle control -hexosaminidase release)/(Triton-X-100 -hexosaminidase release − vehicle control -hexosaminidase release)) × 100. All data are representative of at least two independent experiments. Individual dots represent data points from RBL-2H3 cells within the same assay. Error bars represent mean ± standard deviation (SD). Statistical significance was determined using an unpaired Student’s t test. **** p < 0.0001.
3.5. Peanut Seed Coated with AYA22A Mitigates Degranulation
To investigate the potential of AYA22A aptamers in reducing peanut allergenicity, we explored the efficacy of coating peanut seeds by soaking them with the AYA22AR321 aptamer to mitigate their anaphylactic potential. Dried peanut seeds were soaked with or without a 5 µM solution of AYA22AR321 for 30 min, then dried for 2 days. AYA22AR321-coated and uncoated seeds were processed to isolate peanut crude, as depicted in Supplementary Figure S9A, and further used to assess the impact of AYA22AR321 coating and soaking on degranulation. The treated seeds, both coated by soaking with 5 µM AYA22AR321, demonstrated a significant reduction in degranulation at peanut crude serial dilutions of 1:64, 1:128, and 1:256 compared to untreated seeds (no aptamer coating) (Figure 5 and Supplementary Figure S9B,C). We next evaluated aptamer coating by extending the soaking time to 2 days, as depicted in Supplementary Figure S10. We noticed that 2 days of coating by soaking with AYA22AR321 peanut seed crude extract resulted in a non-significant decrease in degranulation. Moreover, we evaluated the effect of 30 min of coating by soaking with variable drying times to determine the efficacy and effectiveness of AYA22AR321 functionality in degranulation. As shown in Supplementary Figure S11, we noticed that AYA22AR321 significantly diminished RBL-2H3 degranulation on days 2 and 4 compared to untreated seeds. During the extended two-day incubation period with aptamers, significant changes were observed in both the media composition and peanut seeds. These changes included notable degradation and alterations in the structural integrity of the peanut seeds. These observations suggest potential implications for experimental outcomes, possibly influencing observed degranulation levels. Prolonged exposure may have facilitated enzymatic activity, which could have differentially impacted biological responses compared to the shorter 30 min incubation period shown in Figure 5. Further investigations are necessary to elucidate these factors and achieve a comprehensive understanding of the unexpected outcomes observed in Supplementary Figure S10. These findings indicate that coating peanut seeds by soaking with AYA22A aptamer AYA22AR321 effectively attenuates their anaphylactic potential, highlighting a promising strategy for mitigating peanut-induced allergic responses. Further investigation into the mechanisms underlying this attenuation and the potential applications of aptamer-coated peanuts in allergy prevention and management is warranted.
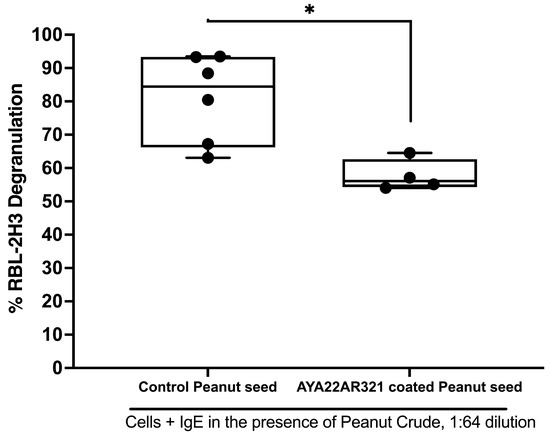
Figure 5.
Efficacy of AYA22AR321 aptamer coating of peanut seeds in reducing IgE-mediated degranulation in RBL-2H3 cells. Dried peanut seeds were soaked with or without 5 µM AYA22AR321 for 30 min, followed by 2 days of drying. AYA22AR321-coated and uncoated seeds were processed to isolate peanut crude, which was utilized in the degranulation assay. RBL-2H3 cells (0.5 × 106) were seeded in a 96-well plate, sensitized or not with 1 µg/mL IgE dansyl, and incubated at 37 °C for 1 h. Crude peanut protein extract (1:64 dilution) from coated and uncoated seeds was added to sensitized RBL-2H3 cells for 1 h at 37 °C. Degranulation was assessed spectroscopically by measuring the activity of granule-stored enzyme -hexosaminidase secreted into the supernatant on the p-nitrophenyl-N-acetyl--d-glucosamine substrate. Percent (%) degranulation was calculated using the following formula: (Experimental -hexosaminidase release − vehicle control -hexosaminidase release)/(Triton-X-100 -hexosaminidase release − vehicle control -hexosaminidase release) × 100. All data are representative of at least three independent experiments. Individual dots represent data points from RBL-2H3 cells within the same assay. Error bars represent mean ± standard deviation (SD). Statistical significance was determined using an unpaired Student’s t test. * p < 0.05.
3.6. AYA22A Aptamer Effectively Inhibits Peanut Butter-Induced Degranulation in RBL-2H3 Cells
Our investigation focused on assessing the efficacy of AYA22A aptamers in inhibiting peanut butter-induced degranulation, a common trigger for allergic reactions. We observed that extracts prepared from peanut crude and commercially available peanut butter could induce anaphylactic reactions, as evidenced by degranulation in RBL-2H3 cells. However, when peanut crude extract was spiked with 5 µM of AYA22AR321 aptamers, it demonstrated the ability to reduce degranulation in RBL-2H3 cells (Figure 6). Next, we evaluated the efficacy of AYA22AR321 aptamer spiked into Skippy and JIF brand peanut butter, followed by isolation of peanut crude to assess decreased RBL-2H3 degranulation, as depicted in Supplementary Figure S12. As shown in Supplementary Figure S13, AYA22AR321 was spiked at various concentrations, and these preparations were incubated at different temperatures. We noticed that AYA22AR321 at 5 µM significantly diminished degranulation in RBL-2H3 cells induced by peanut butter crude extract from both Skippy and JIF commercially available brands at 4 °C. However, at room temperature, AYA22AR321 at each concentration only significantly decreased degranulation in RBL-2H3 cells induced by peanut butter crude extract from JIF commercially available brand, while it could not diminish degranulation in RBL-2H3 induced by Skippy peanut butter under room-temperature conditions.
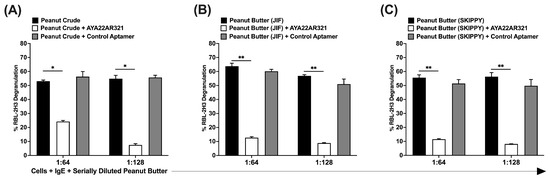
Figure 6.
AYA22AR321 diminishes the degranulation reactions induced by peanut butter extracts. Peanut crude extracts were prepared from peanut seeds (A), as well as commercially available peanut butter (B,C). RBL-2H3 cells (0.5 × 106) were seeded in a 96-well plate, sensitized or not with 1 µg/mL IgE dansyl, and incubated at 37 °C for 1 h. Crude peanut protein extract (at 1:64 and 1:128 dilutions) from commercially available peanut butter and seeds in the presence or absence of AYA22AR321 (5 µM) and control aptamer (5 µM) was added to sensitized RBL-2H3 cells for 1 h at 37 °C. Degranulation was assessed spectroscopically by measuring the activity of granule-stored enzyme -hexosaminidase secreted into the supernatant on the p-nitrophenyl-N-acetyl--d-glucosamine substrate. Percent (%) degranulation was calculated using the following formula: (Experimental -hexosaminidase release − vehicle control -hexosaminidase release)/(Triton-X-100 -hexosaminidase release − vehicle control -hexosaminidase release) × 100. All data are representative of at least three independent experiments. Error bars represent mean ± standard deviation (SD). Statistical significance was determined using an unpaired Student’s t test. * p < 0.05, ** p < 0.01.
Furthermore, we evaluated the efficacy and effectiveness of AYA22AR321 by further spiking it at a higher concentration of 5 µM into the peanut crude, as mentioned in Supplementary Figure S14 and Figure 7. Surprisingly, we found that spiked-in AYA22AR321 significantly mitigated degranulation from RBL-2H3 cells under each condition of peanut crude incubation (Figure 7). AYA22AR321 exhibited efficient and stable binding to the peanut allergens present in peanut butter crude, thereby inhibiting its degranulation potential. This binding property underscores the potential of AYA22A as a therapeutic agent in mitigating allergic responses to peanut allergens. Moreover, we sought to determine the effective concentration of AYA22A in peanut butter and assess the stability of AYA22A under various processing conditions in aptamer functionality. As shown in Supplementary Figure S15A, the stability of the aptamer–peanut butter crude combination was evaluated during vacuum dry centrifugation followed by incubation at 4 °C, room temperature, and 37 °C. Then, the combination was further processed to isolate Preps I, II, and III, followed by a degranulation assay, as indicated in Supplementary Figure S15B–G. The results indicate that AYA22AR321 spiked in at a higher concentration (50 µM) stably bound to peanut proteins present in peanut butter at room temperature and significantly inhibited degranulation in RBL-2H3 cells, mostly at 1:10 and 1:60 dilutions, compared to the unspiked aptamer condition, highlighting the robustness of this interaction. Collectively, these results demonstrate the promising potential of AYA22A aptamers in mitigating peanut-induced allergic responses by effectively reducing degranulation in RBL-2H3 cells. Future work could focus on optimizing the spiking concentration of aptamers in peanut butter to maximize their inhibitory effect and elucidating the underlying mechanisms of AYA22A’s interaction with peanut allergens. Additionally, clinical studies are warranted to evaluate the therapeutic efficacy and safety of AYA22A aptamers in managing peanut allergy.
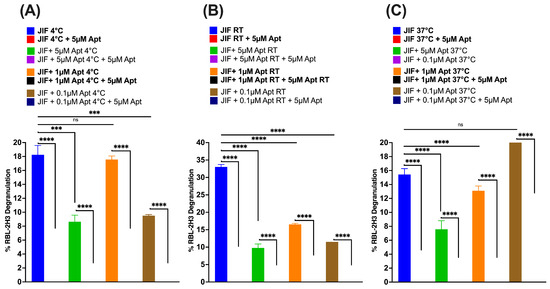
Figure 7.
Evaluation of AYA22AR321 efficacy in mitigating peanut butter crude extract-induced degranulation. The efficacy and effectiveness of AYA22AR321 were studied by spiking it at a higher concentration of 5 µM into the peanut butter crude extract collected from previously mixed AYA22AR321 at different concentrations and incubated under various conditions using a degranulation assay, as mentioned in Supplementary Figure S14. (A) 4 °C; (B) room temperature (RT); (C) 37 °C. RBL-2H3 cells (0.5 × 106) were seeded in a 96-well plate, sensitized or not with 1 µg/mL IgE dansyl, and incubated at 37 °C in a 5% CO2 incubator for 1 h. AYA22AR321-spiked crude protein from peanut butter, indicated at a 1:64 dilution, was added to sensitized RBL-2H3 cells for 1 h at 37 °C. Degranulation was assessed spectroscopically by measuring the activity of granule-stored enzyme -hexosaminidase secreted into the supernatant on the p-nitrophenyl-N-acetyl--d-glucosamine substrate. Percent (%) degranulation was calculated using the following formula: (Experimental -hexosaminidase release − vehicle control -hexosaminidase release)/(Triton-X-100 -hexosaminidase release − vehicle control -hexosaminidase release) × 100. All data are representative of at least two independent experiments. Error bars represent mean ± standard deviation (SD) in triplicate within the same assay. Statistical significance was determined using an unpaired Student’s t test. *** p < 0.001.**** p < 0.0001, and “ns” denotes non-significant.
3.7. Peanut Crude Extract from the Seed of AYA22A-Watered Laboratory Peanut Plants Decreases Degranulation
We investigated the potential of peanut crude extract obtained from laboratory peanut plants watered with AYA22A aptamers (AYA22AR321, AYA22AR211, and AYA22AR524) to reduce degranulation, a key process in allergic reactions. Initially, we assessed the uptake of aptamers by plants using an aptamer (AYA2012004_L (control aptamer 1) and AYA22AR321), along with a control group (untreated). Laboratory peanut plants were watered daily with aptamers for 4 days, and real-time PCR analysis revealed the presence of AYA2012004_L in the root, stem, and some parts of the leaf but not in the seed (Supplementary Figure S16A,B). Our findings that the aptamer is taken up by plants and detected in different parts of plants are in agreement with previous investigations [47,48]. The lack of AYA2012004_L aptamer detection in peanut seeds aligns with the results of the degranulation assays conducting using AYA22A aptamers AYA22AR321, AYA22AR211, and AYA22AR524 (Supplementary Figure S16C(c1–c4)), indicating that the 4-day watering regimen was insufficient for the aptamers to be effectively taken up into the mature peanut seeds. Continuing our investigations (Supplementary Figure S17), we assessed the uptake and accumulation of aptamers by plants from the seedling stage until maturity (0 to 140 days) with 100 days of daily watering with 5 µM aptamers, including AYA22AR321, AYA22AR211, AYA22AR524, control aptamer 4, and control aptamer 5 (Supplementary Table S1), as well as untreated water. Figure 8A illustrates a comparative analysis demonstrating a significant reduction in RBL-2H3 cell degranulation induced by peanut crude extracts from AYA22A aptamer-watered peanut plants’ seeds as compared to seeds from control plants (water only) and plants watered with control aptamers. The degranulation assays confirmed the accumulation of AYA22A aptamers in peanut seeds, where they efficiently bound to peanut proteins, thereby reducing the potential for degranulation. Moreover, we observed that the decrease in RBL-2H3 cell degranulation induced by laboratory-grown peanut seeds’ crude (Figure 8A) was comparable to the effect observed when spiking AYA22A aptamers and control aptamers into commercial peanut seeds’ extracted crude, as compared to unspiked peanut crude-induced degranulation in RBL-2H3 (Figure 8C). Furthermore, we investigated the additive effect of AYA22A aptamers on laboratory-grown peanut seeds. Consequently, we spiked 5 µM of the respective AYA22A aptamers into the peanut crude extract obtained from the laboratory-grown peanut seeds (Figure 8B). Remarkably, the spiked-in aptamers significantly further mitigated degranulation from RBL-2H3 cells. We also evaluated the effect of laboratory-developed aptamer-watered peanut seed with or without aptamer coating on degranulation in RBL-2H3, as illustrated in Supplementary Figure S18. We noticed that seeds from aptamer-watered plants, regardless of aptamer coating, exhibited decreased degranulation compared to control plants. This suggests that both aptamer-coated and uncoated seeds from aptamer-watered plants effectively reduced cell degranulation, further supporting the role of AYA22AR321 aptamer in mitigating allergic responses.
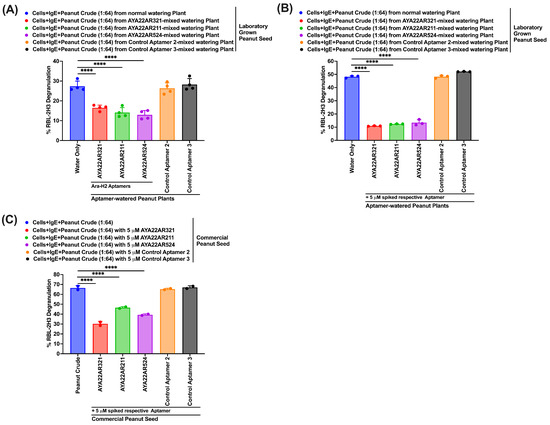
Figure 8.
Laboratory peanut plants treated with AYA22A aptamers exhibit decreased degranulation in RBL-2H3 cells. Peanut crude extract was isolated from laboratory-grown peanut seeds taken from plants that were watered in the absence or presence of AYA22A, as well as commercial peanut seeds as controls. Degranulation assays were conducted with or without spiking aptamers (A–C). RBL-2H3 cells (0.5 × 106) were seeded, sensitized with 1 µg/mL IgE dansyl, and incubated at 37 °C for 1 h. AYA22A aptamer-spiked and unspiked peanut crude proteins were added to sensitized RBL-2H3 cells in a 1:64 dilution for 1 h at 37 °C. Spectroscopic assessment of -hexosaminidase activity measured degranulation. Percent (%) degranulation was calculated using the following formula: (Experimental -hexosaminidase release − vehicle control -hexosaminidase release)/(Triton-X-100 -hexosaminidase release − vehicle control -hexosaminidase release) × 100. All data are representative of at least two independent experiments. Individual dots represent data points from RBL-2H3 cells within the same assay. Error bars represent mean ± SD. Statistical significance was determined using an unpaired Student’s t test **** p < 0.0001.
Collectively, these findings demonstrate the ability of laboratory peanut plants to take up and accumulate AYA22A aptamers, particularly in the seeds. The presence of aptamers in the seed correlated with a reduction in degranulation potential, highlighting the potential for utilizing aptamer-watered plants as a novel approach to reducing allergenicity in peanuts. Our study provides valuable insights into the use of aptamer-watered plants to mitigate peanut allergenicity. Future work should focus on optimizing the concentration and duration of aptamer watering to enhance their uptake and efficacy in reducing allergenicity. Additionally, further investigations are needed to elucidate the mechanisms underlying aptamer uptake and accumulation in peanut plants and to evaluate the safety and efficacy of this approach in larger-scale agricultural settings.
3.8. AYA22AR321 Was Detected in Peanut Crude Extracted from the Seed of AYA22AR321-Watered Laboratory Peanut Plants
We evaluated the uptake of AYA22AR321 in peanut crude extracted from laboratory-grown peanut plants watered with AYA22AR321, employing an enzyme-linked immunosorbent assay (ELISA). Initially, we established the aptamer detection ELISA by utilizing a biotinylated 3′ and/or 5′ complementary sequence of the AYA22AR321 aptamer, followed by HRP-conjugated streptavidin. Supplementary Figure S19 illustrates that the 3′ complementary sequence exhibited specific detection of AYA22AR321 spiked into peanut crude. Consequently, we employed the 3′ complementary AYA22AR321 aptamer sequence for further detection of the aptamer in various parts of laboratory-grown AYA22AR321-watered peanut plants, including roots, stem, leaves, and seeds. Figure 9 demonstrates the detection of AYA22AR321 exclusively in peanut seeds, with no detection observed in leaves, roots, or stem of AYA22AR321-watered peanut plants. Thus, our findings indicate the selective uptake of AYA22AR321 by peanut seeds, suggesting a targeted accumulation mechanism. Future studies could focus on elucidating the molecular pathways involved in the transport and accumulation of aptamers within plant tissues, as well as exploring the potential implications of aptamer uptake in agricultural practice to enhance plant traits or mitigate allergenicity.
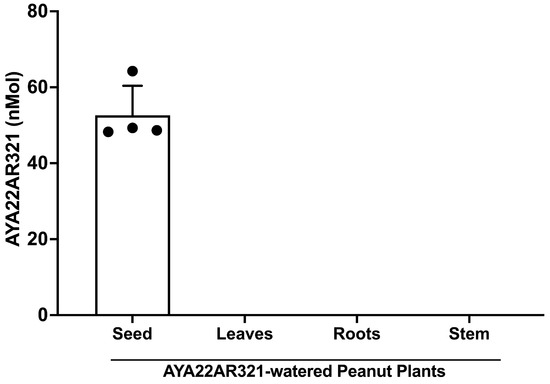
Figure 9.
Detection of AYA22AR321 in aptamer-watered peanut plants by ELISA. A Overnight, a MaxiSorp plate was coated with preparations of AYA22AR321-watered peanut plant crude extract from seeds, leaves, roots, and stems at a concentration of 0.25 mg/mL. AYA22AR321 was detected using biotinylated 3′ complementary DNA at a concentration of 2 µM. Colorimetric detection was achieved using HRP-conjugated streptavidin in conjunction with TMB. The reaction was terminated using 1 M H2SO4, and the absorbance was was measured using a spectrophotometer at 450 nm. All data are representative of at least two independent experiments. Individual dots represent data points within the same assay. Error bars represent mean ± SD.
3.9. AYA22A Aptamers Downregulate the Production of Th2 Cytokines and Upregulate Th1 Cytokines in an In Vitro Allergy Model
In our in vitro allergy model, we investigated the effect of AYA22A aptamers on the production of T helper 2 (Th2) and T helper 1 (Th1) cytokines. Previous studies have shown that major allergens from birch pollen (Bet v 1), peach (Pru p 3), peanut (Ara h 1 or Ara h 2), house dust mite (Der p 1), and grass pollen (Phl p 5) are presented by dendritic cells (DCs) through actin-dependent pathways and upregulate surface markers (i.e., CD80, CD83, CD86, and HLA-DR) after stimulation [49,50,51,52]. These studies revealed that allergen-specific stimulation of monocyte-derived dendritic cells (Mo-DCs) induces T-cell proliferation and upregulation of Th2-type cytokines. To investigate the impact of AYA22A aptamers on the ability of rAra h2 protein or peanut crude extract to induce Th1 or Th2 responses, we generated Mo-DCs from two healthy donors’ monocytes. The dendritic cells were pulsed with or without a cell stimulation cocktail (IL-1b, IL-6, TNF, and PGE2) as described in the Section 2. Surface markers CD11c, CD86, PD-L1, CD14, CD80, and HLA-DR were studied by flow cytometry. We observed that stimulated dendritic cells (DCs) expressed significantly higher levels of CD80 and HLA-DR, while the levels of CD86 and PD-L1 were elevated but did not reach statistical significance (Supplementary Figure S20).
Next, to study the effect of AYA22A-aptamers on the interaction of peanut allergens (rAra h2 or peanut crude extracts) with T cells, we pulsed Mo-DCs in the presence or absence of AYA22A aptamers (AYA22AR211 or AYA22AR321) with a cell stimulation cocktail. We observed that this stimulation in the presence or absence of AYA22AR211 does not affect the total percentage of CD4 T cells or promote CD4 proliferation. However, we noticed that AYA22AR211 significantly diminished the frequency of IL-5 and IL-13 in total CD4 T cells and also showed similar decreased trends of IL-5 and IL-13 in proliferated CD4 T cells. We could only observe the diminished frequency of IL-4 in non-proliferated cells in the presence of AYA22AR211 but not the frequency of IL-13 (Supplementary Figure S21). Next, we sought to determine if the AYA22A aptamers can diminish Th2 cytokines induced by peanut crude. Therefore, we compared the neutralizing effect of AYA22AR321 on Th2 cytokines induced by peanut allergens (rAra h2) versus peanut crude. We noticed that peanut crude increased the percentage frequency (Figure 10A), as well as the gMFI (Figure 10B), of IL-5, IL-13, and IL-4, which were effectively diminished by AYA22AR321 as compared to aptamer control. We also observed a decreased pattern of cytokines IL-4, IL-5, and IL-13 in the %frequency, as well as in the gMFI, caused by AYA22AR321 induced by rAra h2 (Figure 10A,B). Moreover, we observed that AYA22A aptamers bind to peanut allergens and attenuate allergen presentation on DCs, which is an important step in the antigen presentation of peanut allergenicity. We pulsed the Mo-DCs with a cell stimulation cocktail including peanut crude and rAra h2 in the presence or absence of AYA22AR321 and a control aptamer. We noticed that AYA22AR321 effectively diminished the expression of Ara h2 under both conditions (when incubated with peanut crude and with rAra h2) compared to peanut crude and rAra h2 alone or in the presence of a control aptamer (Supplementary Figure S22A,B). In addition, we determined the in vitro effect of AYA22A aptamers (AYA22AR321, AYA22AR211, and AYA22AR524) on autologous PBMCs, as illustrated in Supplementary Figure S23A. Results demonstrated that AYA22A aptamers, mostly AYA22AR321, induced the production Th1-type cytokines such as IFN (1.8 fold change), along with IL-1b (2 fold change), TNF (1.5 fold change), IL-10 (1.7 fold change), IL12p70 (1.5 fold change), and IL-17 (1.2 fold change) (Supplementary Figure S23B). A recent study showed that TNF-, IFN-, IL-1, and IL-17 induce Mesenchymal Stem/Stromal Cells (MSCs) and increase the expression of immunoregulatory molecules in MSCs [53]. These events in MSCs can downregulate local inflammatory responses, reduce inflammatory damage, and reverse airway remodeling by restoring the Th1/Th2 balance, reversing the Th17/Tregs imbalance, inhibiting DC maturation and antigen presentation, suppressing the degranulation response of Mast cells, restraining innate lymphoid cell group 2 (ILC2)-mediated pathology, promoting the shift of M1 macrophages to M2 macrophages, inducing the differentiation of Treg cells, and repairing epithelial injury through paracrine secretion or direct cell–cell contact [54]. Notably, we also observed that AYA22AR321, AYA22AR211, and AYA22AR524 attenuate the frequency of IL-4+CD4+ and IL-5+CD4+ T cells and increase the frequency of IFN+CD4+ T cells in autologous PBMCs co-cultured in an an in vitro allergy model (Supplementary Figure S23C). Collectively, our findings highlight the potential of AYA22A aptamers to modulate allergic responses by targeting key immune cells and cytokine pathways. Future research could focus on elucidating the mechanisms underlying the immunomodulatory effects of AYA22A aptamers and exploring their therapeutic potential in allergic diseases.
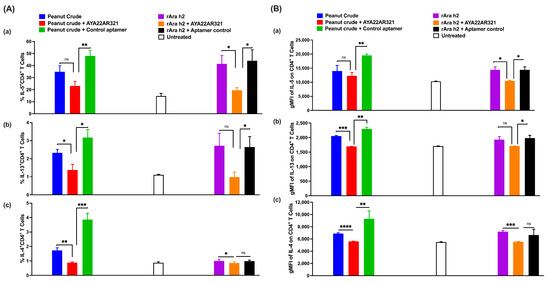
Figure 10.
AYA22AR321 decreases the expression of Th2 cytokines. Mo-DCs from healthy donors were pulsed with or without a cell stimulation cocktail (IL-1b, IL-6, TNFa, and PGE2) in the presence and absence of peanut crude protein and rAra h2 for 48 h, then co-cultured with autologous enriched T cells for 48 h, as described in the Materials and Methods. (A) % frequencies of Th2 cytokines on CD4+ T cells studied by flow cytometry—(a) IL-5; (b) IL-13; (c) IL-4. (B) gMFI of (a) IL-5, (b) IL-13, and (c) IL-4. All data are representative of at least two independent experiments. Error bars represent mean ± SD of triplicate assays within the same experiment. Statistical significance was determined using an unpaired Student’s t test. * p < 0.05, ** p < 0.01, *** p < 0.001, and **** p < 0.0001, and “ns” denotes non-significant.
3.10. AYA22A Aptamers Neutralize the Effect of CD23 Expression on B Cells and Diminish CD63 Expression
We aimed to investigate whether AYA22A aptamers can neutralize the effect of CD23 expression on B cells and decrease CD63 expression. We utilized an autologous in vitro allergic model of co-cultured PBMCs, as depicted in Supplementary Figure S24. Our findings reveal that AYA22AR321 reduced the expression of CD23 on B cells in both donors across different peanut crude concentrations compared to untreated conditions (Figure 11A). Although the decrease in CD23 expression on B cells was not statistically significant in either donor, a consistent pattern of reduced CD23 expression in the presence of AYA22AR321 was observed. CD23 surface density on B cells is closely associated with IgE levels and plays a crucial role in IgE-facilitated allergen uptake [43]. The observed decrease in CD23 expression could potentially be attributed to aptamer binding to Ara h2, potentially interfering with IgE-facilitated allergen uptake. Furthermore, it is noteworthy that CD23 provides a non-inflammatory pathway for IgE–allergen complexes, emphasizing the importance of further investigation in this area. Additionally, our results demonstrate that only AYA22AR321 significantly decreased CD63 expression on autologous B cells compared to untreated conditions. However, AYA22AR321, AYA22AR211, and AYA22AR524 all non-significantly reduced CD63 expression compared to both untreated and control aptamer-treated conditions (Figure 11). This finding may offer new insights into the regulation of high-affinity memory B cells and long-lived plasma cells in allergy, suggesting the need for further exploration. Thus, our study highlights the potential of AYA22A aptamers in modulating key surface markers on B cells implicated in allergic responses. Future investigations should focus on elucidating the underlying mechanisms of aptamer action and exploring their therapeutic potential in allergy management.
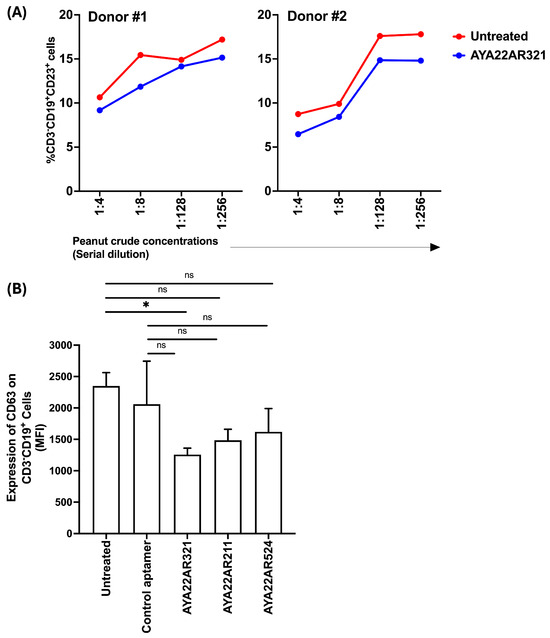
Figure 11.
AYA22A aptamers attenuate the expression of CD23 and CD63 on B cells. PBMCs from healthy donors were pulsed with or without peanut crude protein for 24 h incubation, then further co-cultured with autologous PBMCs for 3 days of incubation, as depicted in Supplementary Figure S24 (a schematic representation of experimental design for the expression of CD23 and CD63 on human B cells). The expression of (A) CD23 (low-affinity binding IgE receptor) and (B) CD63 on B cells was assessed by flow cytometry. All data are representative of at least two independent experiments. Error bars represent the mean ± SD of triplicate assays. Statistical significance was determined using an unpaired Student’s t test. * p < 0.05, and “ns” denotes non-significant.
3.11. AYA22A Aptamer Therapeutically Decreases CD63 Expression in Human Basophil Cells
In this study, we aimed to evaluate the therapeutic efficacy of the AYA22A aptamer in mitigating Ara h2-mediated degranulation markers on peripheral basophils. To achieve this, we employed an in vitro cell model utilizing PBMCs isolated from healthy donors’ blood in the presence or absence of AYA22AR321, as illustrated in Figure 12A–C. Our findings revealed a significant reduction in CD63 expression on CD123+HLA−DR−CD3−CD19− basophils when exposed to Ara h2 or peanut crude, following co-culture with AYA22AR321 (Figure 12A–C). These results demonstrate the preventative and therapeutic potential of AYA22AR321 in attenuating the degranulation response induced by Ara h2 or peanut crude extract, highlighting its ability to modulate basophil activation. Importantly, our findings suggest a promising avenue for further exploration of the therapeutic application of the AYA22A aptamer in allergic responses mediated by Ara h2. Additionally, investigation into its efficacy in in vivo and clinical settings would further contribute to our understanding of its therapeutic utility.
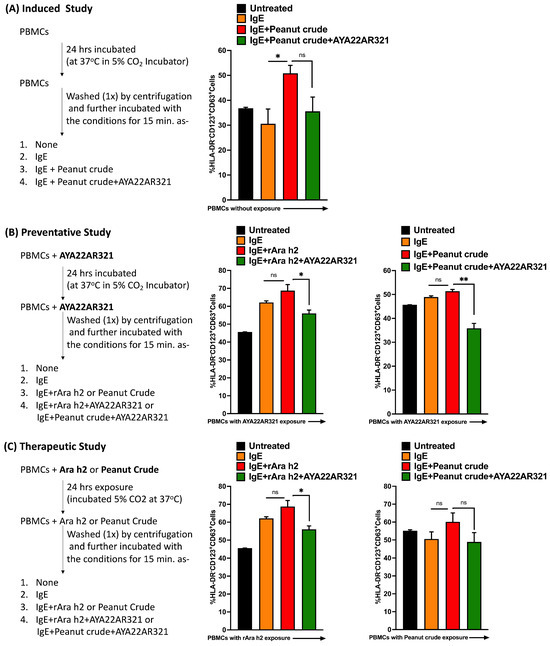
Figure 12.
AYA22AR321 prevents and diminishes the expression of CD63 on human basophils. PBMCs from healthy donors were incubated as illustrated in Figures (A) for the induced in vitro study, (B) for the preventative in vitro study, and (C) for the therapeutic study. The expression of CD63 (activation marker of human basophils) on human basophils was assessed by flow cytometry. All data are representative of at least two independent experiments. Error bars represent the mean ± SD of triplicate assays. Statistical significance was determined using an unpaired Student’s t test. * p < 0.05, ** p< 0.01, and “ns” denotes non-significant.
4. Discussion
Peanut allergy represents a growing health concern among both children and adults in the United States. Clinically, it can manifest with a spectrum of symptoms, ranging from skin reactions to potentially life-threatening systemic responses. However, the precise factors driving the increased prevalence of peanut allergy in recent decades remain incompletely understood. The United States Food and Drug Administration (US FDA) has approved the first peanut oral immunotherapy, Palforzia, which aims to desensitize individuals and reduce the risk of peanut allergic reactions [55,56]. Additionally, Omalizumab, a humanized lgG1 monoclonal antibody, has emerged as another therapeutic option. Omalizumab functions by binding to lgE, thereby hindering its interaction with high-affinity lgE receptors on the surfaces of mast cells, basophils, and eosinophils [57,58,59]. While Omalizumab, along with other therapeutic modalities like epicutaneous immunotherapy (EPIT) and sublingual immunotherapy (SLIT), is undergoing clinical investigation, its efficacy remains to be firmly established, especially when compared to peanut oral immunotherapy (pOIT) such as Palforzia [60,61]. Therefore, the need for treatment of peanut allergy is unmet.
In this study, utilizing computational biology tools, we designed and developed a series of peanut allergen-specific aptamers (AYA22A) that not only bind to the Ara h2 and Ara h1 proteins but also demonstrate an ability to impede the degranulation process triggered by peanut crude protein. It is noteworthy that peanut crude protein encompasses 16 allergens, including various isoforms identified by the Allergen Nomenclature Sub-Committee of the International Union of Immunological Societies [8,62]. These allergens are classified into four predominant food allergen families, namely the Cupin superfamily (Ara h1,3), the Prolamin superfamily (Ara h2,6,7,9), the Profilin family (Ara h5), and Bet v-1-related proteins (Ara h8), in addition to two other families, namely Oleosin (Ara h10,11) and Defensin (Ara h12,13). Among these, six major peanut allergens (Ara h1, 2, 3, 5, 8, and 9) are recognized as primary triggers of allergic responses across diverse geographical regions. Notably, Ara h1, Ara h2, and Ara h3 are implicated in allergic reactions within the USA and are frequently associated with severe symptoms. Our findings demonstrate that the AYA22A aptamers effectively mitigate degranulation induced by whole-peanut crude proteins, suggesting their potential as promising preventive and therapeutic agents for peanut allergies [63]. Maleki et al. [64,65,66] showed that thermal processing/roasting plays an important role in enhancing the allergenic properties of peanuts. In our investigation, we examined the efficacy of AYA22A aptamers under different thermal conditions and evaluated their performance during peanut food processing. Our systematic assessment encompassed the ability of AYA22A aptamers to counteract degranulation across a spectrum of conditions in the presence of peanut products. Additionally, employing stability analyses, we established that AYA22A aptamers consistently suppressed peanut crude-induced degranulation for a period of 35 days, regardless of storage temperature (4 °C, room temperature, and 37 °C). Interestingly, as observed in Figure 4, longer incubation periods led to increased inhibition of RBL-2H3 degranulation, even in the absence of AYA22A aptamers. This suggests that extended incubation may result in the degradation or denaturation of allergenic proteins within the crude extract, thereby reducing their capacity to trigger degranulation. Our data demonstrate that the AYA22A aptamers retained their inhibitory effect on RBL-2H3 degranulation over the incubation periods of 14, 21, 28, and 35 days. This sustained efficacy implies that the aptamers are not only stable but also robust in their inhibitory function over extended durations. The stability analyses confirmed that AYA22A aptamers consistently suppressed peanut crude-induced degranulation across a range of storage temperatures (4 °C, room temperature, and 37 °C) for up to 35 days. The observed increase in inhibition with longer incubation times could be attributed to the progressive breakdown of allergenic proteins in the peanut crude extract, which diminishes their ability to induce degranulation. However, the aptamers’ efficacy in inhibiting degranulation was not compromised over time, indicating their potential as a reliable interventions in preventing allergic reactions. Thus, our findings underscore the potential of AYA22A aptamers in providing long-term stability and effectiveness in mitigating allergic responses, even under varying storage conditions and extended incubation periods.
Paungfoo-Lonhienne et al. [47] elucidated the phenomenon of exogenous DNA uptake by plants, demonstrating the ingress of DNA into root hairs pollen tubes, providing further evidence that high-molecular-weight exogenous biomolecules can penetrate plant cells. We sought to investigate this hypothesis. Therefore, by using a real-time PCR method, we detected the uptake of DNA aptamers by the root, leaf, and stem tissues of peanut plants; then, we proceeded to ascertain if AYA22A aptamers, known for their specificity to peanut allergens Ara h1 and Ara h2, could co-localize with peanut seed storage proteins. Leveraging our in-house cultivated peanut plant seeds, we successfully detected AYA22A aptamers using ELISA. We also noticed that peanut seed from our laboratory peanut plants successfully mitigated degranulation. To the best of our knowledge, this pioneering endeavor represents the inaugural evidence of peanut plants assimilating allergen-specific aptamers, seamlessly incorporating them into seed proteins, and efficaciously attenuating allergen-induced degranulation. Notably, we observed that AYA22A aptamers were taken up by peanut plants and co-localized in peanut seeds when the aptamers were administered by watering for approximately 100 days, starting from the sprouting stage. Our short-term treatment study, in which fully developed peanut plants with fruits were watered with aptamers, aligns with the findings of Paungfoo-Lonhienne et al. [47]. However, the uptake of aptamers by peanut plants and their subsequent co-localization to seed cells may depend on the plant’s physiological and developmental biology.
In a study by Li et al. [67], various strains of mice were administered a peanut Ara h2 plasmid DNA vaccine to modulate a prolonged immune response. This sensitization led to the production of antigen-specific lgG1 and lgG2a but not lgE. However, when this DNA vaccine was injected into the skin, it triggered an allergic reaction instead of providing protection against allergy [67,68]. As a result, it failed in a phase II clinical trial. In contrast, our DNA aptamer is employed during the peanut growing or peanut product processing stage, without direct contact with humans. This approach reduces the likelihood of inducing allergies on its own, as it does not involve direct exposure to human immunity. Our findings suggested that AYA22A aptamers hold promise as a potential prevention and desensitization measure that can be used before, after, or before and after the peanut manufacturing process. This provides manufacturers with valuable options for selecting the most effective approach to prevent peanut allergies in their products.
Furthermore, we investigated the immunological response associated with peanut allergy [69,70,71,72,73]. Our results indicate that AYA22A aptamers can confer immunity by neutralizing CD23 [44] and CD63 [45] expression on B cells and modulating dendritic cells in the presence of peanut crude proteins. Moreover, AYA22A aptamers were found to stimulate the production of Th1 cytokines in autologous Mo-DC T-cell co-culture, as well as autologous PBMC co-cultures, resulting in decreased levels of Th2 cytokines (IL-4, IL-5, and IL-13). These findings suggest that peanut allergen-mediated acquired immunity can be effectively targeted by AYA22A aptamers. Its high affinity and specificity for the Ara h1 and Ara h2 proteins make it a promising candidate for clinical intervention in peanut allergy treatment. Our findings closely align with the immunological responses observed following the administration of subcutaneous immunotherapy (SLIT) or oral immunotherapy (OIT) [14,74,75,76]. In light of these findings, it is suggested that AYA22A aptamers may serve as a potential immunotherapy approach for the treatment of peanut allergies.
Peanut allergy research has spanned several decades, with significant milestones and advancements occurring over the years. The field has evolved from the initial recognition of peanut allergies to a deeper understanding of epidemiology and the development of diagnosis tools. Early strategies focused on strict avoidance, but over time, immunotherapies emerged as potential treatments. As our understanding of the mechanism underlying the peanut allergies improved, landmark studies and new guidelines, such as LEAP (Learning Early About Peanut Allergy) and ITN (Immune Tolerance Network) studies, demonstrated that early introduction of peanuts to high-risk infants significantly reduced the prevalence of peanut allergies and induced lasting tolerance into adolescence [77,78].
However, for individuals who did not benefit from early exposure or developed allergies later in life, there remains a critical need for the therapeutic management of established peanut allergies. Our study focused on the molecular interaction between aptamers and targeted peanut allergens, providing additional mechanistic insight. By demonstrating how aptamers can inhibit degranulation of immune cells, this study contributes to a deeper understanding of potential therapeutic targets and interventions. This study complements the preventive approach established by the LEAP and ITN studies, contributing to a comprehensive approach to address this significant health issue.
Conventionally, aptamer selection relies on the SELEX (Systematic Evolution of Ligands by Exponential Enrichment) method. However, in this investigation, AYA22A aptamers were synthesized by employing an exhaustive computational strategy, predicting secondary and tertiary structures, and conducting docking simulations to probe interactions with Ara h1 and Ara h2 proteins. This computational approach minimizes reliance on chemical reagents, thereby mitigating costs while ensuring manageable computational overhead. Moreover, computational methodologies afford the opportunity to delve into and refine the subtleties of aptamer–ligand interactions beyond experimental limitations.
We elucidated the design and fabrication of aptamers tailored to target Ara h1 and Ara h2 proteins, which exhibit promising prospects in the prevention and treatment of peanut allergy. Furthermore, we validated the inhibitory efficacy of these aptamers against allergen-induced degranulation. Our observations advocate for the therapeutic potential of AYA22A aptamers (AYA22AR321, AYA22AR211, and AYA22AR524) as effective, safe, and cost-efficient remedies for comprehensive peanut allergy management. Subsequent investigations will delve into the assessment of degranulation inhibition by AYA22A aptamers using in vivo rodent models.
5. Conclusions
In conclusion, peanut allergy poses a significant health challenge, with increasing prevalence and potentially severe consequences. Current treatments, such as Palforzia and Omalizumab, aim to mitigate allergic reactions, yet their efficacy remains under scrutiny. Our study introduces AYA22A aptamers as a novel avenue for peanut allergy intervention. Through computational design and experimental validation, we demonstrated their ability to effectively counteract allergen-induced immune responses, offering promise as preventive and therapeutic agents. Notably, our findings align with emerging immunotherapy approaches, suggesting AYA22A aptamers as a potential adjunct or alternative treatment for peanut allergies. Further research into their clinical application and efficacy in animal models is warranted.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/allergies4030008/s1, Figure S1: Selection and Binding Affinity of AYA22A Aptamers; Figure S2: IgE-Mediated Recognition and Degranulation Responses to Ara h2 and Peanut Crude Extract in RBL-2H3 Cells; Figure S3: Assessment of peanut allergens-induced degranulation from the RBL-2H3 cell line; Figure S4: Assessment of AYA22A aptamers’ effectiveness on mitigation of the degranulation induced by peanut crude proteins from the RBL-2H3 cell line; Figure S5: AYA22A aptamers inhibit degranulation induced by natural Ara h2 (Peanut allergenic protein) from RBL-2H3 cell-line; Figure S6: Effectiveness of Ara h2 aptamers maintained despite prolonged incubation periods; Figure S7: Assessment of roasting conditions of peanut crude extract on degranulation from RBL-2H3 cell-line; Figure S8: Roasted peanut crude extract induces IgE-mediated degranulation from RBL-2H3 cell-line; Figure S9: Efficacy of AYA22AR321 Aptamer Coating on Peanut Seeds in Reducing Degranulation; Figure S10: Efficacy of AYA22AR321 Aptamer Coating on Peanut Seeds in Reducing Degranulation; Figure S11: Efficacy of AYA22AR321 Aptamer Coating on Peanut Seeds in Reducing Degranulation; Figure S12: Schematic illustration of the preparation of peanut crude extract from commercially available peanut butter with or without spiking of AYA22AR321; Figure S13: Effectiveness of AYA22AR321 in decreasing degranulation induced by peanut butter crude proteins; Figure S14: Schematic illustration of the preparation of peanut crude extract from commercially available peanut butter with or without spiking of AYA22AR321; Figure S15: Schematic illustration of the preparation of peanut crude extract from commercially available peanut butter with or without spiking of AYA22AR321; Figure S16: Assessment of Aptamer Uptake in Laboratory-Grown Peanut Plants; Figure S17: Schematic illustration of laboratory-grown peanut plants; Figure S18: Assessment of the efficacy of AYA22AR321 –watered peanut plant seed in the presence of Aptamer Coating in Reducing Degranulation; Figure S19: Determine the ELISA protocol for the detection of Ara h2 specific aptamer spiking in peanut crude protein; Figure S20: Estimation of Dendritic cell activation markers; Figure S21: AYA22AR321 decreases the expression of the Th2 cytokines; Figure S22: AYA22AR321 decreases the expression of the Th2 cytokines; Figure S23: AYA22A aptamers downregulate the production of Th2 Cytokines and upregulate Th1 Cytokines; Figure S24: Schematic representation of experimental design. Table S1: The sequences of the selected AYA22A aptamers; Table S2: Materials and key resources used in the methods.
Author Contributions
M.A.A. participated in the conceptualization and design of the research project, reviewed the data, and supervised the research and reporting of the results. T.T. and L.A.-M. designed the experiments. T.T., R.R.N., T.O., V.P. and N.G. conducted the experiments and data analyses. J.Z. and T.T. conducted the literature review and wrote the manuscript. L.A.-M., T.T., K.Z., J.Z. and K.M. reviewed the data. K.M. conducted bioinformatics analysis. T.T. and N.G. oversaw the research operation. L.A.-M. supervised the research. All authors contributed to manuscript editing and discussion of the results reported in the final manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Ayass Bioscience, LLC, a privately held Texas limited liability company.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
This article has accompanying Supplementary Files. All data generated or analyzed during this study are included in the published article (and its Supplementary Material Files).
Acknowledgments
We thank Larisa Marder for all illustrative work.
Conflicts of Interest
Ayass Bioscience, LLC is a privately-held Texas limited liability company (“Ayass Bioscience”). The research project was funded by Ayass Bioscience. The principal investigator, Dr. Ayass is the founder and president of Ayass Bioscience. All staff who worked on this research project were employees of Ayass Bioscience. The value of compensation paid to such employees did not influence the results. Moreover, the participation of employees of Ayass Bioscience did not affect the design, conduct, reporting, or outcome of the research. The employees of Ayass Bioscience who performed the research related to the study do not have a proprietary interest in any product or services, such as patients rights, trademark, copyright, or licensing agreement related to the research or any outcome. Any and all work product derived from the research project and proprietary rights related such work product belong exclusively to Ayass Bioscience.
Abbreviations
The following abbreviations are used in this manuscript:
| PA | peanut allergy |
| pOIT | peanut oral immunotherapy |
| Ara h1 | Arachis hypoga 1 |
| Ara h2 | Arachis hypoga 2 |
| Ara h3 | Arachis hypoga 3 |
| SELEX | systematic evoluation of ligands by exponential enrichment |
| FBS | fetal bovin serum |
| GM-CSF | granulocyte–macrophage colony-stimulating factor |
| EPIT | epicutaneous immunotherapy |
| SLIT | sublingual immunotherapy |
References
- Jones, S.; Burks, A. Food allergy. N. Engl. J. Med. 2017, 377, 1168–1176. [Google Scholar] [CrossRef]
- Mahr, T.; Lieberman, J.; Haselkorn, T.; DamLe, V.; Ali, Y.; Chidambaram, A.; Griffin, N.; Sublett, J. Characteristics of peanut allergy diagnosis in a US health care claims database (2011–2017). J. Allergy Clin. Immunol. Pract. 2021, 9, 1683–1694. [Google Scholar] [CrossRef]
- Warren, C.; Lei, D.; Sicherer, S.; Schleimer, R.; Gupta, R. Prevalence and characteristics of peanut allergy in US adults. J. Allergy Clin. Immunol. 2021, 147, 2263–2270. [Google Scholar] [CrossRef]
- McCann, W.; Hass, S.; Norrett, K.; Cameron, A.; Etschmaier, M.; Duhig, A.; Yu, S. The Peanut Allergy Burden Study: Real-world impact of peanut allergy on resource utilization and productivity. World Allergy Organ. J. 2021, 14, 100525. [Google Scholar] [CrossRef]
- Al-Muhsen, S.; Clarke, A.; Kagan, R. Peanut allergy: An overview. CMAJ 2003, 168, 1279–1285. [Google Scholar]
- Allen, K.J.; Turner, P.J.; Pawankar, R.; Taylor, S.; Sicherer, S.; Lack, G.; Rosario, N.; Ebisawa, M.; Wong, G.; Mills, E.N.C.; et al. Precautionary labelling of foods for allergen content: Are we ready for a global framework? World Allergy Organ. J. 2014, 7, 10. [Google Scholar] [CrossRef]
- Sampson, H. Food allergy. JAMA 1997, 278, 1888–1894. [Google Scholar] [CrossRef]
- Hemmings, O.; Du Toit, G.; Radulovic, S.; Lack, G.; Santos, A. Ara h 2 is the dominant peanut allergen despite similarities with Ara h 6. J. Allergy Clin. Immunol. 2020, 146, 621–630. [Google Scholar] [CrossRef]
- Flinterman, A.; Knol, E.; Lencer, D.; Bardina, L.; Hartog Jager, C.; Lin, J.; Pasmans, S.; Bruijnzeel-Koomen, C.; Sampson, H.; Hoffen, E.; et al. Peanut epitopes for IgE and IgG4 in peanut-sensitized children in relation to severity of peanut allergy. J. Allergy Clin. Immunol. 2008, 121, 737–743. [Google Scholar] [CrossRef]
- Rambasek, T.; Kavuru, M. Omalizumab dosing via the recommended card versus use of the published formula. J. Allergy Clin. Immunol. 2006, 117, 708–709. [Google Scholar] [CrossRef]
- Fu, T.; Abbott, U.; Hatzos, C. Digestibility of food allergens and nonallergenic proteins in simulated gastric fluid and simulated intestinal fluid a comparative study. J. Agric. Food Chem. 2002, 50, 7154–7160. [Google Scholar] [CrossRef]
- Klemans, R.; Broekman, H.; Knol, E.; Bruijnzeel-Koomen, C.; Otten, H.; Pasmans, S.; Knulst, A. Ara h 2 is the best predictor for peanut allergy in adults. J. Allergy Clin. Immunol. Pract. 2013, 1, 632–638. [Google Scholar] [CrossRef]
- Sen, M.; Kopper, R.; Pons, L.; Abraham, E.; Burks, A.; Bannon, G. Protein structure plays a critical role in peanut allergen stability and may determine immunodominant IgE-binding epitopes. J. Immunol. 2002, 169, 882–887. [Google Scholar] [CrossRef]
- Schworer, S.; Kim, E. Sublingual immunotherapy for food allergy and its future directions. Immunotherapy 2020, 12, 921–931. [Google Scholar] [CrossRef]
- Kim, E.; Yang, L.; Ye, P.; Guo, R.; Li, Q.; Kulis, M.; Burks, A. Long-term sublingual immunotherapy for peanut allergy in children: Clinical and immunologic evidence of desensitization. J. Allergy Clin. Immunol. 2019, 144, 1320–1326. [Google Scholar] [CrossRef]
- Wang, J.; Li, X. Chinese herbal therapy for the treatment of food allergy. Curr. Allergy Asthma Rep. 2012, 12, 332338. [Google Scholar] [CrossRef]
- Hu, J.; Chen, J.; Ye, L.; Cai, Z.; Sun, J.; Ji, K. Anti-IgE therapy for IgE-mediated allergic diseases: From neutralizing IgE antibodies to eliminating IgE+ B cells. Clin. Transl. Allergy 2018, 8, 27. [Google Scholar] [CrossRef]
- Fowler, J.; Lieberman, J. Update on clinical research for food allergy treatment. Front. Allergy 2023, 4, 1154541. [Google Scholar] [CrossRef]
- Keefe, A.; Pai, S.; Ellington, A. Aptamers as therapeutics. Nat. Rev. Drug Discov. 2010, 9, 537–550. [Google Scholar] [CrossRef]
- Darmostuk, M.; Rimpelova, S.; Gbelcova, H.; Ruml, T. Current approaches in SELEX: An update to aptamer selection technology. Biotechnol. Adv. 2015, 33, 1141–1161. [Google Scholar] [CrossRef]
- Thiel, K.; Giangrande, P. Therapeutic applications of DNA and RNA aptamers. Oligonucleotides 2009, 19, 209–222. [Google Scholar] [CrossRef]
- Jayasena, S. Aptamers: An emerging class of molecules that rival antibodies in diagnostics. Clin. Chem. 1999, 45, 1628–1650. [Google Scholar] [CrossRef]
- Wieg, T.; Williams, P.; Dreskin, S.; Jouvin, M.; Kinet, J.; Tasset, D. High-affinity oligonucleotide ligands to human IgE inhibit binding to Fc epsilon receptor I. J. Immunol. 1996, 157, 221–230. [Google Scholar]
- Poongavanam, M.; Kisley, L.; Kourentzi, K.; Landes, C.; Willson, R. Ensemble and single-molecule biophysical characterization of D17. 4 DNA aptamer–IgE interactions. Biochim. Biophys. Acta-(Bba)-Proteins Proteom. 2016, 1864, 154–164. [Google Scholar] [CrossRef]
- Ayass, M.; Tripathi, T.; Griko, N.; Okyay, T.; Ramankutty Nair, R.; Zhang, J.; Zhu, K.; Melendez, K.; Pashkov, V.; Abi-Mosleh, L. Dual Checkpoint Aptamer Immunotherapy: Unveiling Tailored Cancer Treatment Targeting CTLA-4 and NKG2A. Cancers 2024, 16, 1041. [Google Scholar] [CrossRef]
- Passante, E.; Ehrhardt, C.; Sheridan, H.; Frankish, N. RBL-2H3 cells are an imprecise model for mast cell mediator release. Inflamm. Res. 2009, 58, 611–618. [Google Scholar] [CrossRef]
- Kulczycki, A., Jr.; Isersky, C.; Metzger, H. The interaction of IgE with rat basophilic leukemia cells: I. Evidence for specific binding of IgE. J. Exp. Med. 1974, 139, 600–616. [Google Scholar] [CrossRef]
- Barsumian, E.; Isersky, C.; Petrino, M.; Siraganian, R. IgE-induced histamine release from rat basophilic leukemia cell lines: Isolation of releasing and nonreleasing clones. Eur. J. Immunol. 1981, 11, 317–323. [Google Scholar] [CrossRef]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef]
- Antczak, M.; Popenda, M.; Zok, T.; Sarzynska, J.; Ratajczak, T.; Tomczyk, K.; Adamiak, R.; Szachniuk, M. New functionality of RNAComposer: An application to shape the axis of miR160 precursor structure. Acta Biochim. Pol. 2016, 63, 737–744. [Google Scholar] [CrossRef]
- Popenda, M.; Szachniuk, M.; Antczak, M.; Purzycka, K.; Lukasiak, P.; Bartol, N.; Blazewicz, J.; Adamiak, R. Automated 3D structure composition for large RNAs. Nucleic Acids Res. 2012, 40, e112. [Google Scholar] [CrossRef]
- Honorato, R.; Koukos, P.; Jiménez-Garcia, B.; Tsaregorodtsev, A.; Verlato, M.; Giachetti, A.; Rosato, A.; Bonvin, A. Structural biology in the clouds: The WeNMR-EOSC ecosystem. Front. Mol. Biosci. 2021, 8, 729513. [Google Scholar] [CrossRef]
- Van Zundert, G.; Rodrigues, J.; Trellet, M.; Schmitz, C.; Kastritis, P.; Karaca, E.; Melquiond, A.; Dijk, M.; De Vries, S.; Bonvin, A. The HADDOCK2. 2 web server: User-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 2016, 428, 720–725. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Židek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Morita, Y.; Siraganian, R. Inhibition of IgE-mediated histamine release from rat basophilic leukemia cells and rat mast cells by inhibitors of transmethylation. J. Immunol. 1981, 127, 1339–1344. [Google Scholar] [CrossRef]
- Koppelman, S.; Smits, M.; Tomassen, M.; De Jong, G.; Baumert, J.; Taylor, S.; Witkamp, R.; Veldman, R.; Pieters, R.; Wichers, H. Release of major peanut allergens from their matrix under various pH and simulated saliva conditions—Ara h2 and ara h6 are readily bio-accessible. Nutrients 2018, 10, 1281. [Google Scholar] [CrossRef]
- Maleki, S.; Viquez, O.; Jacks, T.; Dodo, H.; Champagne, E.; Chung, S.; Landry, S. The major peanut allergen, Ara h 2, functions as a trypsin inhibitor, and roasting enhances this function. J. Allergy Clin. Immunol. 2003, 112, 190–195. [Google Scholar] [CrossRef]
- Yoo, J.; Kim, N.; Seo, J.; Kim, S.; Lee, S.; Kim, S.; Kim, H.; Lee, S.; Kim, M. Inhibitory effects of mulberry fruit extract in combination with naringinase on the allergic response in IgE-activated RBL-2H3 cells. Int. J. Mol. Med. 2014, 33, 469–477. [Google Scholar] [CrossRef]
- Staats, H.; Kirwan, S.; Choi, H.; Shelburne, C.; Abraham, S.; Leung, G.; Chen, D. A mast cell degranulation screening assay for the identification of novel mast cell activating agents. Medchemcomm 2013, 4, 88–94. [Google Scholar] [CrossRef]
- Ayass, M.; Tripathi, T.; Griko, N.; Pashkov, V.; Dai, J.; Zhang, J.; Herbert, F.; Ramankutty Nair, R.; Okyay, T.; Zhu, K.; et al. Highly efficacious and safe neutralizing DNA aptamer of SARS-CoV-2 as an emerging therapy for COVID-19 disease. Virol. J. 2022, 19, 227. [Google Scholar] [CrossRef]
- Hiasa, M.; Abe, M.; Nakano, A.; Oda, A.; Amou, H.; Kido, S.; Takeuchi, K.; Kagawa, K.; Yata, K.; Hashimoto, T.; et al. GM-CSF and IL-4 induce dendritic cell differentiation and disrupt osteoclastogenesis through M-CSF receptor shedding by up-regulation of TNF-ɑconverting enzyme (TACE). Blood J. Am. Soc. Hematol. 2009, 114, 4517–4526. [Google Scholar]
- Tripathi, T.; Yin, W.; Xue, Y.; Zurawski, S.; Fujita, H.; Hanabuchi, S.; Liu, Y.; Oh, S.; Joo, H. Central Roles of OX40L–OX40 Interaction in the Induction and Progression of Human T Cell–Driven Acute Graft-versus-Host Disease. Immunohorizons 2019, 3, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Selb, R.; Eckl-Dorna, J.; Neunkirchner, A.; Schmetterer, K.; Marth, K.; Gamper, J.; Jahn-Schmid, B.; Pickl, W.; Valenta, R.; Niederberger, V. CD23 surface density on B cells is associated with IgE levels and determines IgE-facilitated allergen uptake, as well as activation of allergen-specific T cells. J. Allergy Clin. Immunol. 2017, 139, 290–299. [Google Scholar] [CrossRef] [PubMed]
- Engeroff, P.; Caviezel, F.; Mueller, D.; Thoms, F.; Bachmann, M.; Vogel, M. CD23 provides a noninflammatory pathway for IgE-allergen complexes. J. Allergy Clin. Immunol. 2020, 145, 301–311. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Liu, J.; Truong, T.; Zukin, E.; Chen, W.; Saxon, A. Blocking allergic reaction through targeting surface-bound IgE with low-affinity anti-IgE antibodies. J. Immunol. 2017, 198, 3823–3834. [Google Scholar] [CrossRef] [PubMed]
- Kamath, S.; Bublin, M.; Kitamura, K.; Matsui, T.; Ito, K.; Lopata, A. Cross-reactive epitopes and their role in food allergy. J. Allergy Clin. Immunol. 2023, 151, 1178–1190. [Google Scholar] [CrossRef] [PubMed]
- Paungfoo-Lonhienne, C.; Lonhienne, T.; Mudge, S.; Schenk, P.; Christie, M.; Carroll, B.; Schmidt, S. DNA is taken up by root hairs and pollen, and stimulates root and pollen tube growth. Plant Physiol. 2010, 153, 799–805. [Google Scholar] [CrossRef] [PubMed]
- Purnamasari, R.; Sudadi, U.; Santosa, D. A Review: Is Cinderella’s story of self-DNA extracellular effect towards plant growth real? IOP Conf. Ser. Earth Environ. Sci. 2021, 824, 012026. [Google Scholar] [CrossRef]
- Humeniuk, P.; Dubiela, P.; Hoffmann-Sommergruber, K. Dendritic cells and their role in allergy: Uptake, proteolytic processing and presentation of allergens. Int. J. Mol. Sci. 2017, 18, 1491. [Google Scholar] [CrossRef]
- Castenmiller, C.; Nagy, N.; Kroon, P.; Auger, L.; Desgagnés, R.; Martel, C.; Mire, L.; Morel, B.; Roberge, J.; Stordeur, V.; et al. A novel peanut allergy immunotherapy: Plant-based enveloped Ara h 2 Bioparticles activate dendritic cells and polarize T cell responses to Th1. World Allergy Organ. J. 2023, 16, 100839. [Google Scholar] [CrossRef]
- Peyron, I.; Hartholt, R.; Pedró-Cos, L.; Alphen, F.; Ten Brinke, A.; Lardy, N.; Meijer, A.; Voorberg, J. Comparative profiling of HLA-DR and HLA-DQ associated factor VIII peptides presented by monocyte-derived dendritic cells. Haematologica 2018, 103, 172. [Google Scholar] [CrossRef] [PubMed]
- Ashjaei, K.; Bublin, M.; Smole, U.; Lengger, N.; Hafner, C.; Breiteneder, H.; Wagner, S.; Hoffmann-Sommergruber, K. Differential T-helper cell polarization after allergen-specific stimulation of autologous dendritic cells in polysensitized allergic patients. Int. Arch. Allergy Immunol. 2015, 166, 97–106. [Google Scholar] [CrossRef] [PubMed]
- López-Garcia, L.; Castro-Manrreza, M. TNF-ɑ and IFN-ɣ participate in improving the immunoregulatory capacity of mesenchymal stem/stromal cells: Importance of cell-cell contact and extracellular vesicles. Int. J. Mol. Sci. 2021, 22, 9531. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Li, Y.; Zeng, J.; Chang, N.; Cheng, Y.; Zhen, X.; Zhong, D.; Chen, R.; Ma, G.; Wang, Y. Mesenchymal Stem/Stromal Cells in Asthma Therapy: Mechanisms and Strategies for Enhancement. Cell Transplant. 2023, 32, 09636897231180128. [Google Scholar] [CrossRef] [PubMed]
- Borne, G.; Daniel, C.; Wagner, M.; Plaisance, C.; Nolen, A.; Kelkar, R.; Ahmadzadeh, S.; Myrcik, D.; Shekoohi, S.; Kaye, A.; et al. Palforzia for Peanut Allergy: A Narrative Review and Update on a Novel Immunotherapy. Cureus 2023, 15, e50485. [Google Scholar] [PubMed]
- From the Medical Letter on Drugs and Therapeutics. Peanut Allergen Powder (Palforzia). Available online: https://jamanetwork.com/journals/jama/article-abstract/2768131 (accessed on 15 March 2023).
- Busse, W.; Corren, J.; Lanier, B.; McAlary, M.; Fowler-Taylor, A.; Della Cioppa, G.; As, A.; Gupta, N. Omalizumab, anti-IgE recombinant humanized monoclonal antibody, for the treatment of severe allergic asthma. J. Allergy Clin. Immunol. 2001, 108, 184–190. [Google Scholar] [CrossRef]
- Belliveau, P. Omalizumab: A monoclonal anti-IgE antibody. Medscape Gen. Med. 2005, 7, 27. [Google Scholar]
- Massanari, M.; Holgate, S.; Busse, W.; Jimenez, P.; Kianifard, F.; Zeldin, R. Effect of omalizumab on peripheral blood eosinophilia in allergic asthma. Respir. Med. 2010, 104, 188–196. [Google Scholar] [CrossRef] [PubMed]
- Azzano, P.; Paquin, M.; Langlois, A.; Morin, C.; Parizeault, G.; Lacombe-Barrios, J.; Samaan, K.; Graham, F.; Paradis, L.; Des Roches, A.; et al. Determinants of omalizumab dose–related efficacy in oral immunotherapy: Evidence from a cohort of 181 patients. J. Allergy Clin. Immunol. 2021, 147, 233–243. [Google Scholar] [CrossRef]
- Tirumalasetty, J.; Barshow, S.; Kost, L.; Morales, L.; Sharma, R.; Lazarte, C.; Nadeau, K. Peanut allergy: Risk factors, immune mechanisms, and best practices for oral immunotherapy success. Expert Rev. Clin. Immunol. 2023, 19, 785–795. [Google Scholar]
- Zhou, Y.; Wang, J.; Yang, X.; Lin, D.; Gao, Y.; Su, Y.; Yang, S.; Zhang, Y.; Zheng, J. Peanut allergy, allergen composition, and methods of reducing allergenicity: A review. Int. J. Food Sci. 2013, 2013, 909140. [Google Scholar] [CrossRef] [PubMed]
- Palladino, C.; Breiteneder, H. Peanut allergens. Mol. Immunol. 2018, 100, 58–70. [Google Scholar] [CrossRef] [PubMed]
- Maleki, S.; Chung, S.; Champagne, E.; Raufman, J. The effects of roasting on the allergenic properties of peanut proteins. J. Allergy Clin. Immunol. 2000, 106, 763–768. [Google Scholar] [CrossRef] [PubMed]
- Moghaddam, A.; Hillson, W.; Noti, M.; Gartlan, K.; Johnson, S.; Thomas, B.; Artis, D.; Sattentau, Q. Dry roasting enhances peanut-induced allergic sensitization across mucosal and cutaneous routes in mice. J. Allergy Clin. Immunol. 2014, 134, 1453–1456. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Wang, S.; Sun, X.; Deng, Z.; Niu, B.; Chen, Q. Study on mechanism of increased allergenicity induced by Ara h 3 from roasted peanut using bone marrow-derived dendritic cells. Food Sci. Hum. Wellness 2023, 12, 755–764. [Google Scholar] [CrossRef]
- Li, X.; Huang, C.; Schofield, B.; Burks, A.; Bannon, G.; Kim, K.; Huang, S.; Sampson, H. Strain-dependent induction of allergic sensitization caused by peanut allergen DNA immunization in mice. J. Immunol. 1999, 162, 3045–3052. [Google Scholar] [CrossRef] [PubMed]
- National Library of Medicine. Clinical Trials: A Study to Evaluate Safety, Tolerability and Immune Response in Adults Allergic to Peanut after Receiving Intradermal or Intramuscular Administration of ASP0892 (ARA-LAMP-vax), a Single Multivalent Peanut (Ara h1, h2, h3) Lysosomal Associated Membrane Protein DNA Plasmid Vaccine. Available online: https://clinicaltrials.gov/study/NCT02851277 (accessed on 15 March 2023).
- Mayorga, C.; Palomares, F.; Cañas, J.; Pérez-Sánchez, N.; Núñez, R.; Torres, M.; Gómez, F. New insights in therapy for food allergy. Foods 2021, 10, 1037. [Google Scholar] [CrossRef] [PubMed]
- Ellenbogen, Y.; Jiménez-Saiz, R.; Spill, P.; Chu, D.; Waserman, S.; Jordana, M. The initiation of Th2 immunity towards food allergens. Int. J. Mol. Sci. 2018, 19, 1447. [Google Scholar] [CrossRef]
- Wisniewski, J.; Commins, S.; Agrawal, R.; Hulse, K.; Yu, M.; Cronin, J.; Heymann, P.; Pomes, A.; Platts-Mills, T.; Workman, L.; et al. Analysis of cytokine production by peanut-reactive T cells identifies residual Th2 effectors in highly allergic children who received peanut oral immunotherapy. Clin. Exp. Allergy 2015, 45, 1201–1213. [Google Scholar] [CrossRef]
- Czolk, R.; Klueber, J.; Sørensen, M.; Wilmes, P.; Codreanu-Morel, F.; Skov, P.; Hilger, C.; Bindslev-Jensen, C.; Ollert, M.; Kuehn, A. IgE-mediated peanut allergy: Current and novel predictive biomarkers for clinical phenotypes using multi-omics approaches. Front. Immunol. 2021, 11, 594350. [Google Scholar] [CrossRef]
- Aguilera-Insunza, R.; Venegas, L.; Iruretagoyena, M.; Rojas, L.; Borzutzky, A. Role of dendritic cells in peanut allergy. Expert Rev. Clin. Immunol. 2018, 14, 367–378. [Google Scholar] [CrossRef] [PubMed]
- Plewako, H.; Wosińska, K.; Arvidsson, M.; Björker, J.; Skov, P.; Hakansson, L.; Rak, S. Basophil interleukin 4 and interleukin 13 production is suppressed during the early phase of rush immunotherapy. Int. Arch. Allergy Immunol. 2006, 141, 346–353. [Google Scholar] [CrossRef] [PubMed]
- Sampath, V.; Nadeau, K.C. Newly identified T cell subsets in mechanistic studies of food immunotherapy. J. Clin. Investig. 2019, 129, 1431–1440. [Google Scholar] [CrossRef] [PubMed]
- Berin, M.; Agashe, C.; Burks, A.; Chiang, D.; Davidson, W.; Dawson, P.; Grishin, A.; Henning, A.; Jones, S.; Kim, E.; et al. Allergen-specific T cells and clinical features of food allergy: Lessons from CoFAR immunotherapy cohorts. J. Allergy Clin. Immunol. 2022, 149, 1373–1382. [Google Scholar] [CrossRef] [PubMed]
- Du Toit, G.; Roberts, G.; Sayre, P.; Bahnson, H.; Radulovic, S.; Santos, A.; Brough, H.; Phippard, D.; Basting, M.; Feeney, M.; et al. Randomized trial of peanut consumption in infants at risk for peanut allergy. N. Engl. J. Med. 2015, 372, 803–813. [Google Scholar] [CrossRef]
- Du Toit, G.; Huffaker, M.; Radulovic, S.; Feeney, M.; Fisher, H.; Byron, M.; Dunaway, L.; Calatroni, A.; Johnson, M.; Foong, R.; et al. Follow-up to adolescence after early peanut introduction for allergy prevention. NEJM Evid. 2024, 3, EVIDoa2300311. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).