IgE-Binding Epitopes of Pis v 1, Pis v 2 and Pis v 3, the Pistachio (Pistacia vera) Seed Allergens
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Human Sera
2.2. Analytical Methods
2.3. Immunoblotting
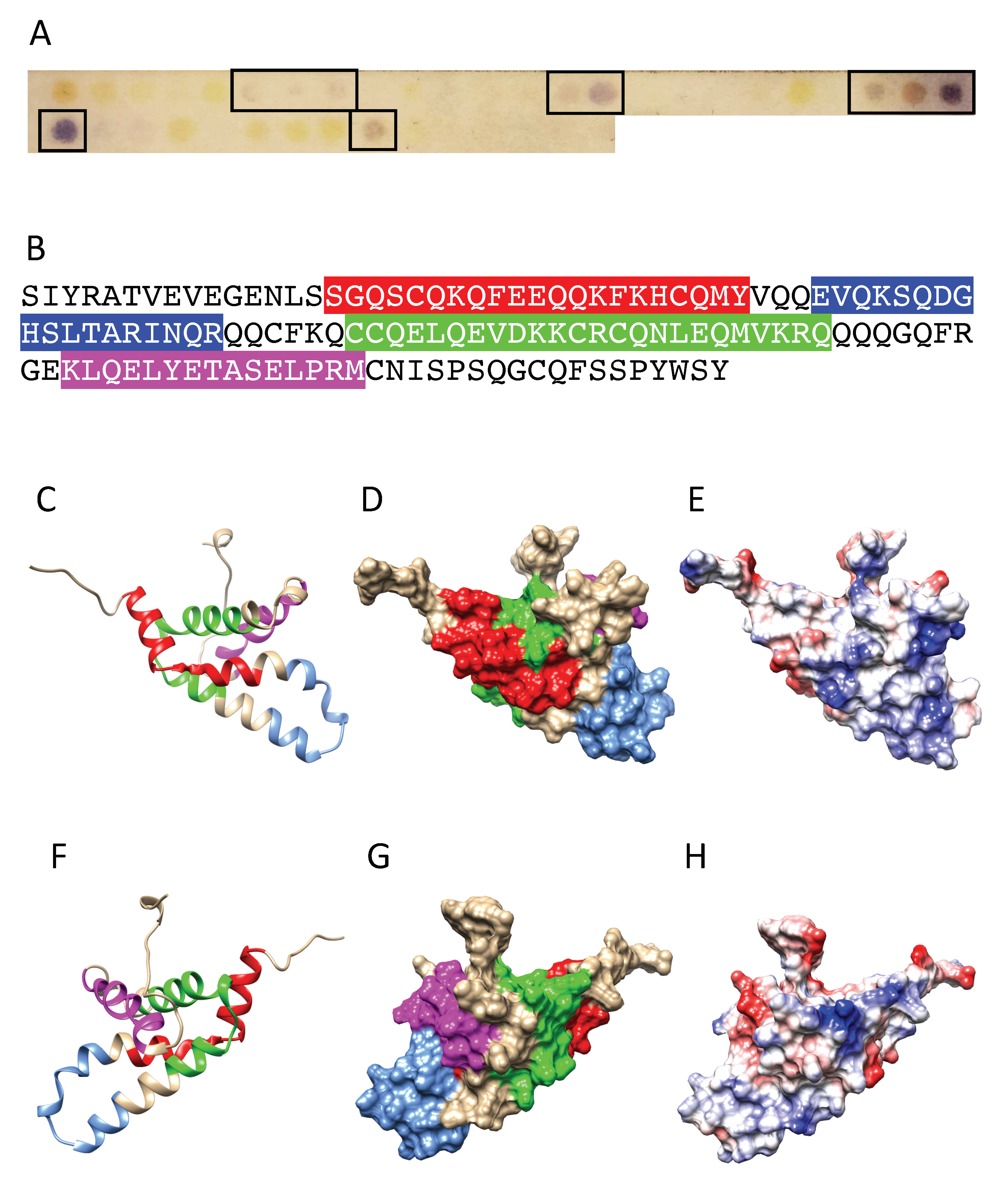
2.4. Peptide Synthesis and IgE-Binding Epitope Mapping
2.5. Bioinformatics
3. Results
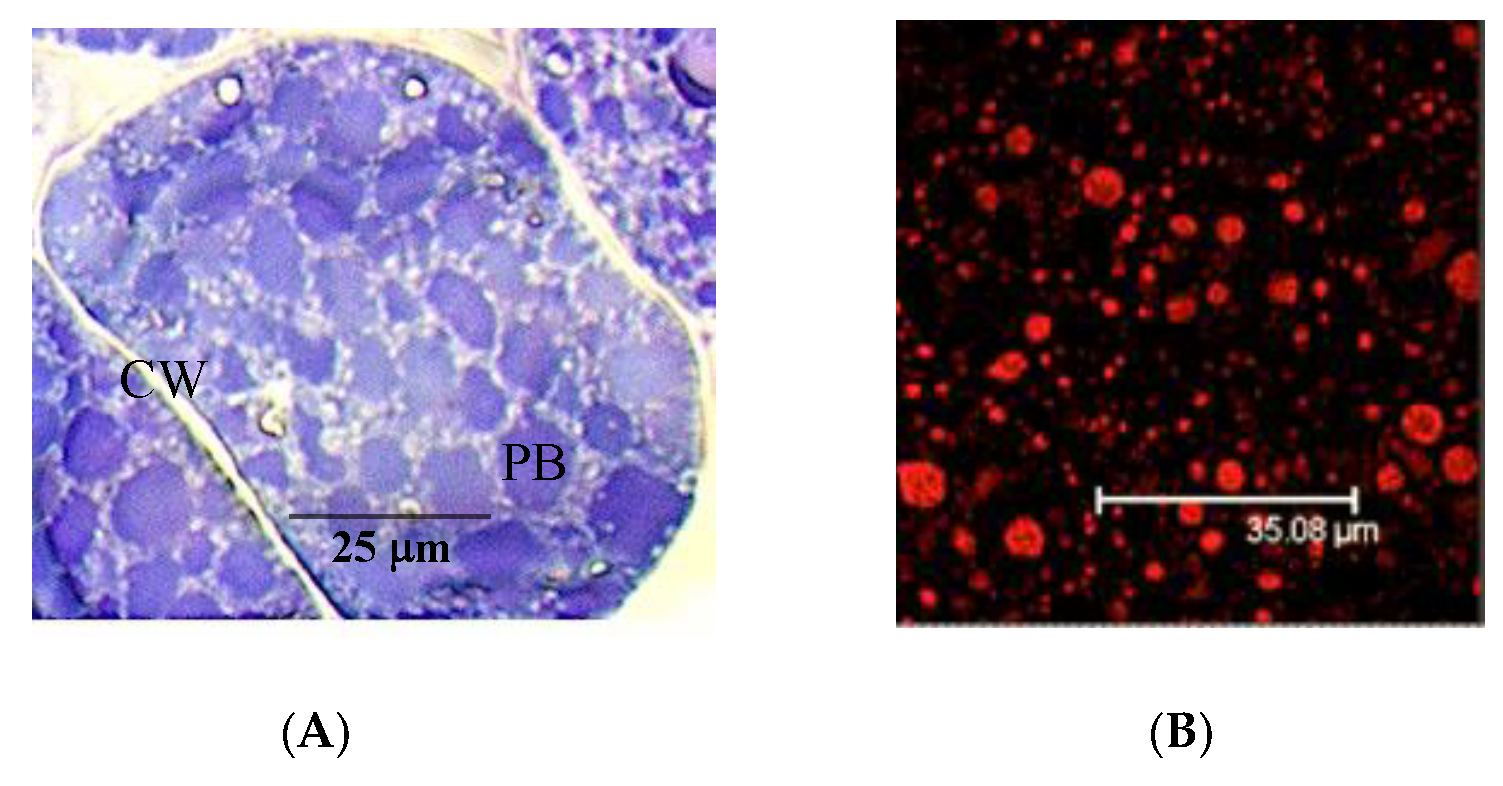
3.1. The Pistachio Allergens
3.2. Pis v 1 and Closely Related 2S Albumins
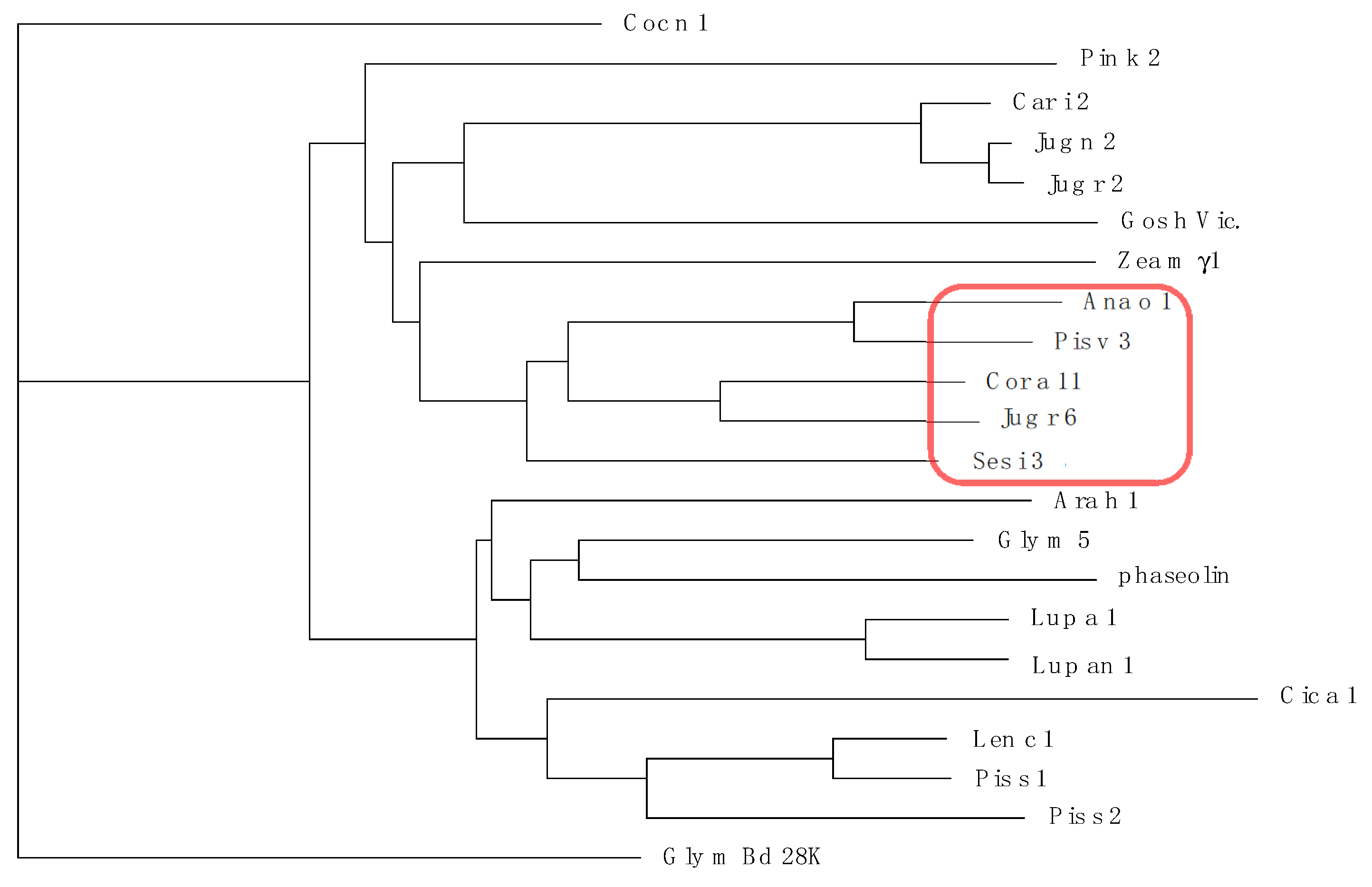
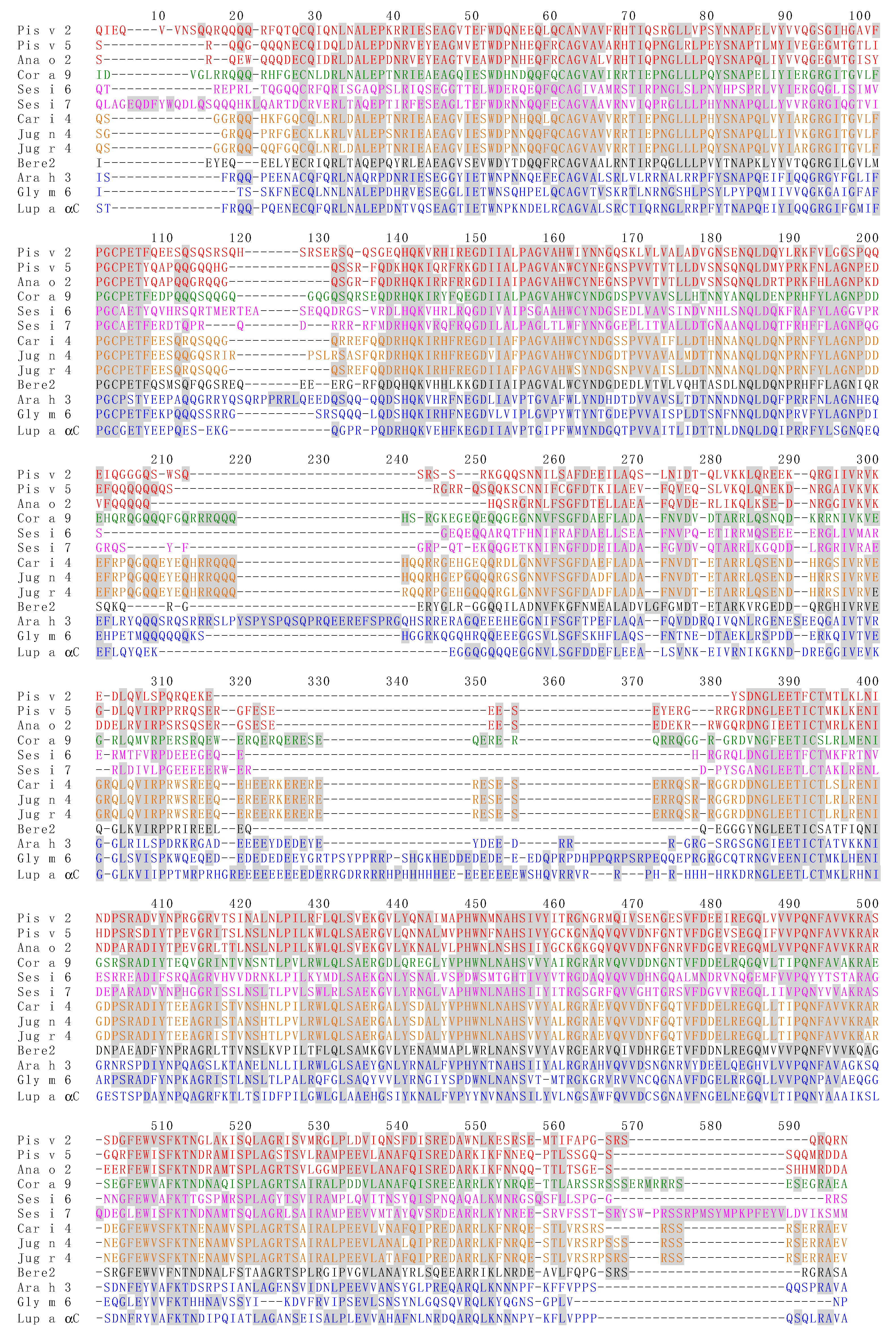
3.3. Pis v 3 and Closely Related 7S Globulins (Vicilins)
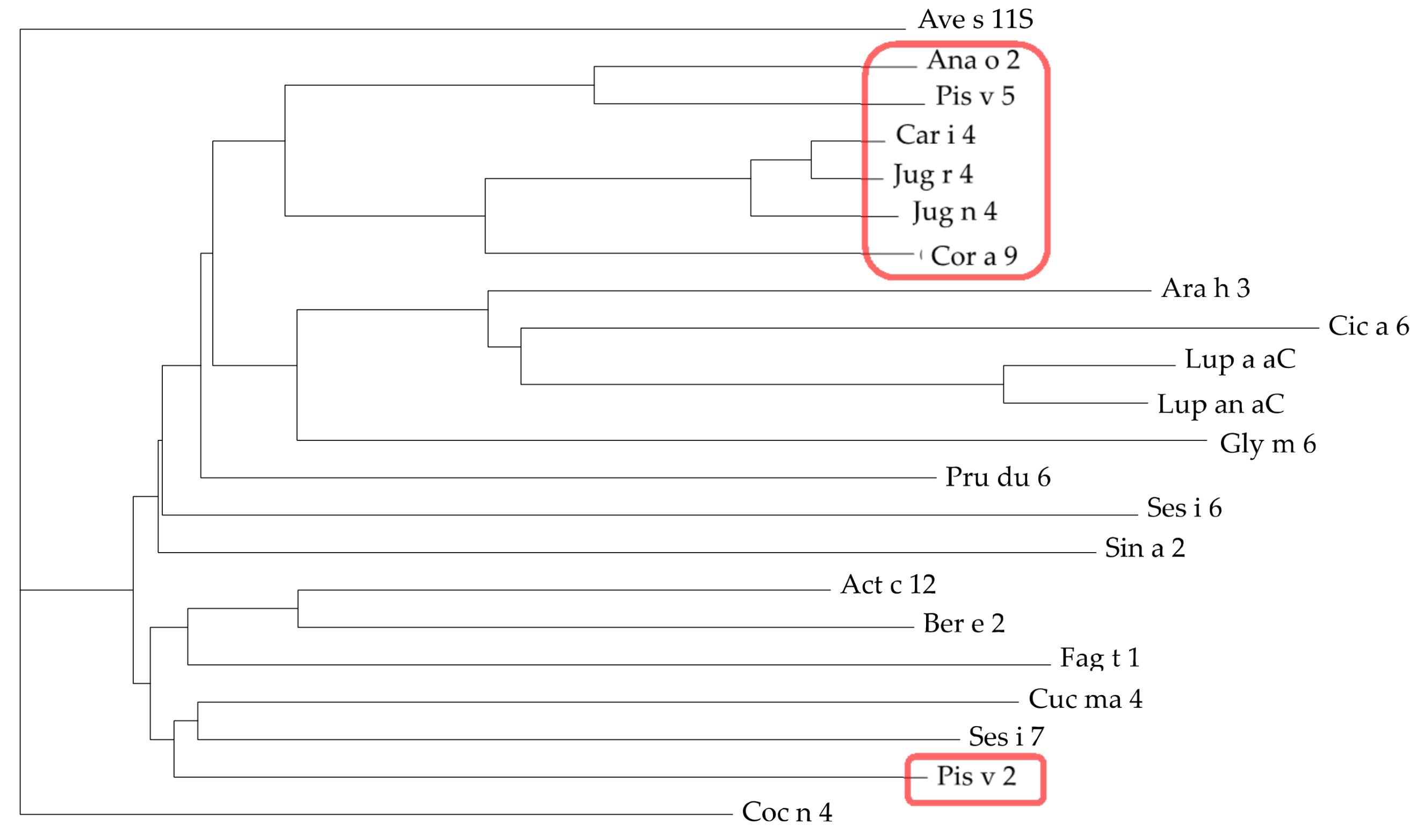
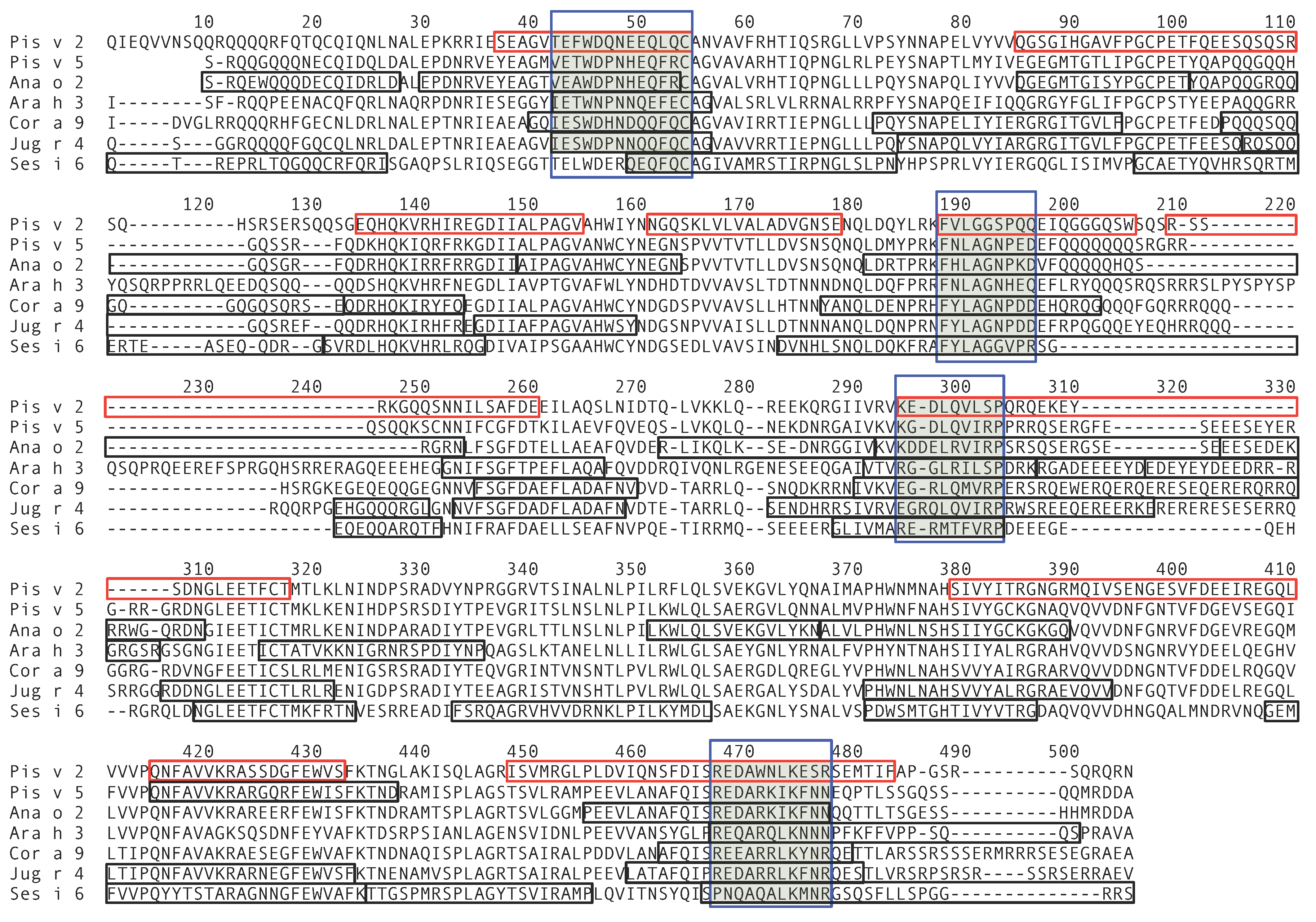
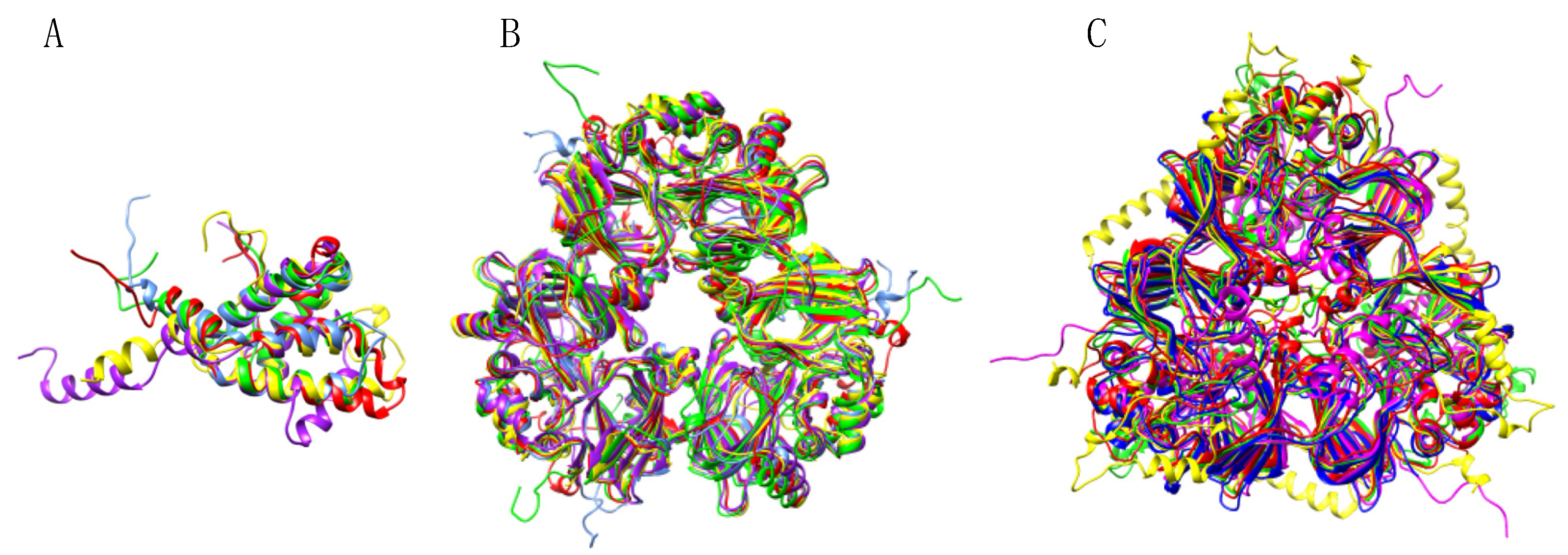
3.4. Pis v 2 and Closely Related 11S Globulins (Legumins)
3.5. Structural Similarities Observed among the Tree Nut Epitopic Regions
3.6. Predicted Resistance of IgE-Binding Epitopic Regions to Proteolysis
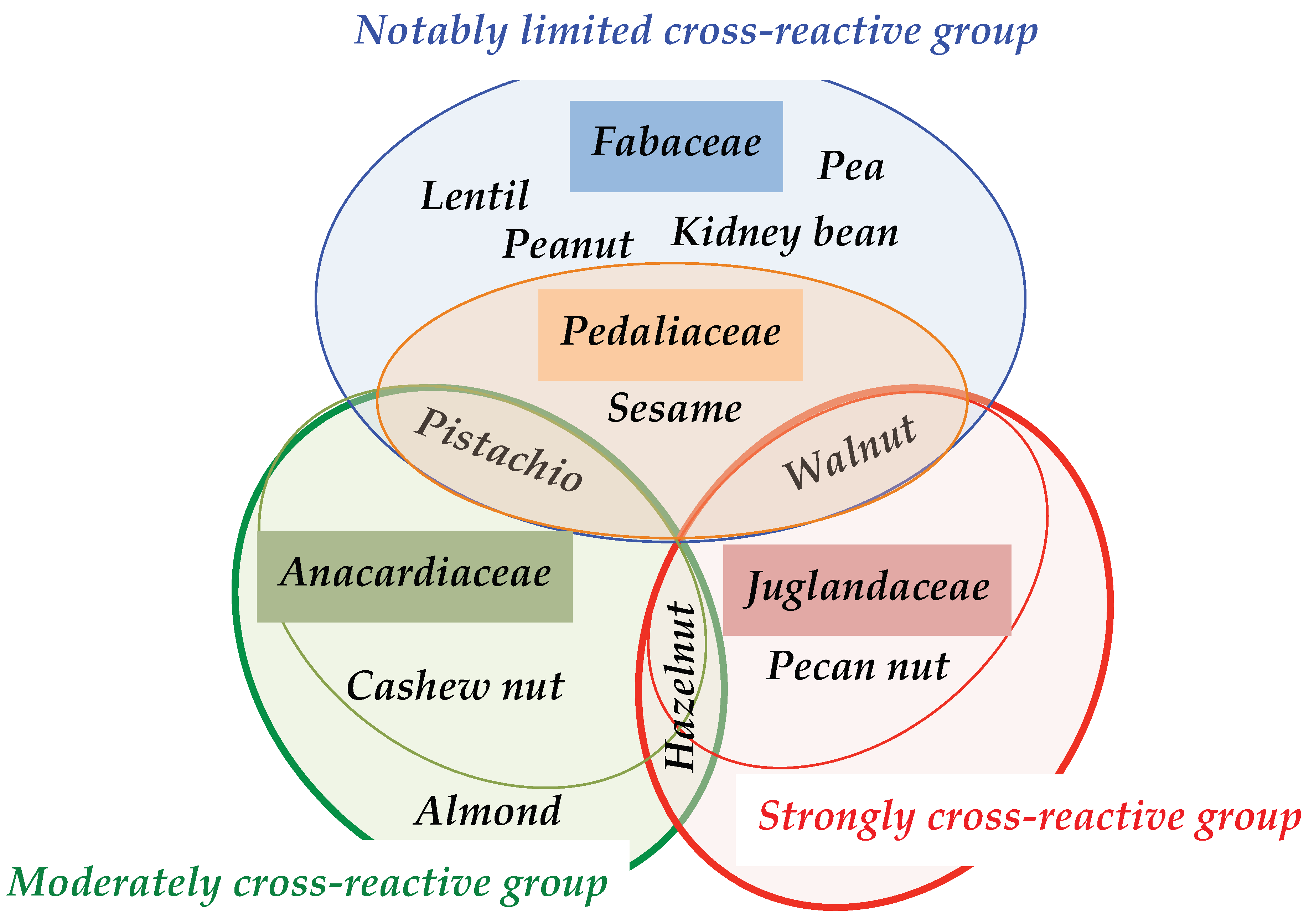
4. Discussion
Author Contributions
Funding
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Abbreviations of Allergens
| Act c 12 | Actinidia chinensis (kiwi) 11S globulin/legumin allergen |
| Ana o 1 | Anacardium occidentale (cashew) 7S globulin/vicilin allergen |
| Ana o 2 | Anacardium occidentale (cashew) 11S globulin/legumin allergen |
| Ana o 3 | Anacardium occidentale (cashew) 2S albumin allergen |
| Ara d 2/6 | Arachis duranensis (peanut) 2S albumin allergens |
| Ara h 1 | Arachis hypogaea (peanut) 7S globulin/vicilin allergen |
| Ara h 2/6/7 | Arachis hypogaea (peanut) 2S albumin allergens |
| Ara h 3 | Arachis hypogaea (peanut) 11S globulin/legumin allergen |
| Ara i 2/6 | Arachis ipaensis (peanut) allergens |
| Ave s 11S | Avena sativa (oat) 11S globulin/legumin allergen |
| Ber e 1 | Bertholletia excelsa (Brazil nut) 2S albumin allergen |
| Ber e 2 | Bertholletia excels (Brazil nut) 11S globulin/legumin allergen |
| Bra j 1 | Brassica juncea (brown mustard) 2S albumin allergen |
| Bra n 1 | Brassica napus (rapeseed) 2S albumin allergen |
| Bra r 1 | Brassica rapa (turnip) 2S albumin allergen |
| Car i 1 | Carya illinoinensis (Pecan nut) 2S albumin allergen |
| Car i 2 | Carya illinoinensis (Pecan nut) 7S globulin/vicilin allergen |
| Car i 4 | Carya illinoinensis (Pecan nut) 11S globulin/legumin allergen |
| Cic a 1 | Cicer arietinum (chickpea) 2S albumin allergen |
| Cic a 6 | Cicer arietinum (chickpea) 11S globulin/legumin allergen |
| Coc c 1 | Cocos nucifera (coconut) 7S globulin/vicilin allergen |
| Coc n 4 | Cocos nucifera (coconut) 11S globulin/legumin allergen |
| Cor a 14 | Corylys avellana (hazelnut) 2S albumin allergen |
| Cor a 11 | Corylus avellana (hazelnut) 7S globulin/vicilin allergen |
| Cor a 9 | Corylus avellana (hazelnut) 11S globulin/legumin allergen |
| Cuc ma 5 | Cucubita maxima (squach) 2S albumin allergen |
| Cuc ma 4 | Cucurbita maxima (squach) 11S globulin/legumin allergen |
| Fag e 2 | Fagopyrum esculentum (buckwheat) 2S albumin allergen |
| Fag t 1 | Fagopyrum tataricum (Tartary buckwheat) 11S globulin/legumin allergen |
| Fag t 2 | Fagopyrum tataricum (Tartary buckwheat) 2S albumin allergen |
| Gly m 8 | Glycine max (soybean) 2S albumin allergen |
| Gly m 5 | Glycine max (soybean) 7S globulin/vicilin allergen |
| Gly m 6 | Glycine max (soybean) 11S globulin/legumin allergen |
| Gly m Bd 28K | Glycine max (soybean) 7S globulin/vicilin allergen |
| Gos h Vic | Gossypium hirsutum (cottonseed) 7S globulin/vicilin allergen |
| Hel a 2S | Helianthus annuus (sunflower) 2S albumin allergen |
| Jug n 1 | Juglans nigra (black walnut) 2S albumin allergen |
| Jug n 2 | Juglans nigra (black walnut) 7S globulin/vicilin allergen |
| Jug n 4 | Juglans nigra (black walnut) 11S globulin/legumin allergen |
| Jug r 1 | Juglans regia (walnut) 2S albumin allergen |
| Jug r 2/6 | Juglans regia (walnut) 7S globulin/vicilin allergens |
| Jug r 4 | Juglans regia (walnut) 11S globulin/legumin allergen |
| Jug r 4 | Juglans regia (walnut) 11S globulin/legumin allergen |
| Lup a δC | Lupinus albus (white lupine) delta-conglutin (2S albumin) allergen |
| Lup an δ1 | Lupinus angustifolius (narrowleaf lupine) conglutin delta-1 (2S albumin) allergen |
| Lup a 1 | Lupinus albus (white lupine) conglutin beta (7S globulin/vicilin) allergen |
| Lup an 1 | Lupinus angustifolius (narrowleaf lupine) conglutin beta (7S globulin/vicilin) allergen |
| Lup a αC | Lupinus albus (white lupine) conglutin alpha (11S globulin/legumin) allergen |
| Lup an αC | Lupinus angustifolius (narrowleaf lupine) conglutin alpha (11S globulin/legumin) allergen |
| phaseolin | Phaseolus vulgaris (kidney bean) 7S globulin/vicilin allergen |
| Pin k 2 | Pinus koraiensis (Korean pine) 2S albumin allergen |
| Pis s 1/2 | Pisum sativum (pea) 7S globulin/vicilin allergens |
| Pis v 1 | Pistacia vera (pistachio) 2S albumin allergen |
| Pis v 3 | Pistacia vera (pistachio) 7S globulin/vicilin allergen |
| Pis v 2/5 | Pistacia vera (pistachio) 11S globulin/legumin allergens |
| Pru du 6 | Prunus dulcis (almond) 11S globulin/legumin allergen |
| Ric c 1/3 | Ricinus communis (Castor bean) 2S albumin allergens |
| Ses i 1/2 | Sesamum indicum (sesame) 2S albumin allergens |
| Ses i 3 | Sesamum indicum (sesame) 7S globulin/vicilin allergen |
| Ses i 6/7 | Sesamum indicum (sesame) 11S globulin/legumin allergens |
| Sin a 1 | Sinapis alba (mustard) 2S albumin allergen |
| Sin a 2 | Sinapis alba (mustard) 11S globulin/legumin allergen |
| Zea m γ1 | Zea mays (maize) glutelin gamma (7S globulin/vicilin) allergen |
References
- Husain, Z.; Schwartz, R.A. Food allergy update: More than a peanut of a problem. Int. J. Dermatol. 2013, 52, 386–394. [Google Scholar] [CrossRef]
- Smeekens, J.M.; Bagley, K.; Kulis, M. Tree nut allergies: Allergen homology, cross-reactivity, and implications for therapy. Clin. Exp. Allergy 2018, 48, 762–772. [Google Scholar] [CrossRef]
- Costa, J.; Silva, I.; Vicente, A.A.; Oliveira, M.B.P.P.; Mafra, I. Pistachio nut allergy: An updated overview. Crit. Food. Sci. Nutr. 2019, 59, 546–562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geiselhart, S.; Hoffmann-Sommergruber, K.; Bublin, M. Tree nut allergens. Mol. Immunol. 2018, 100, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Teuber, S.S.; Beyer, K. Peanut, tree nut and seed allergies. Curr. Opin. Allergy Clin. Immunol. 2004, 4, 201–203. [Google Scholar] [CrossRef]
- Sicherer, S.H.; Muñoz-Furlong, A.; Godbold, J.H.; Sampson, H.A. US prevalence of self-reported peanut, tree nut, and sesame allergy: 11-year follow-up. J. Allergy Clin. Immunol. 2010, 125, 1322–1326. [Google Scholar] [CrossRef] [PubMed]
- Porcel, S.; Sánchez, A.B.; Rodríguez, E.; Fletes, C.; Alvarado, M.; Jiménez, S.; Hernández, J. Food-dependent exercise-induced anaphylaxis to pistachio. J. Investig. Allergol. Clin. Immunol. 2006, 16, 71–73. [Google Scholar]
- Rancé, F.; Bidat, E.; Bourrier, T.; Sabouraud, D. Cashew allergy: Observations of 42 children without associated peanut allergy. Allergy 2003, 58, 1311–1314. [Google Scholar] [CrossRef]
- Fernandez, C.; Fiandor, A.; Martinez-Garate, A.; Martinez-Quesada, J. Allergy to pistachio: Cross-reactivity between pistachio nut and other Anacardiaceae. Clin. Exp. Allergy 1995, 25, 1254–1259. [Google Scholar] [CrossRef]
- Ahn, K.; Bardina, L.; Grishina, G.; Beyer, K.; Sampson, H.A. Identification of two pistachio allergens, Pis v 1 and Pis v 2, belonging to the 2S albumin and 11S globulin family. Clin. Exp. Allergy 2009, 39, 926–934. [Google Scholar] [CrossRef]
- Willison, L.N.; Tawde, P.; Robotham, J.M.; Penney, R.M.; Teuber, S.S.; Sathe, S.K.; Roux, K.H. Pistachio vicilin, Pis v 3, is immunoglobulin E-reactive and cross-reacts with the homologous cashew allergen, Ana o 1. Clin. Exp. Allergy 2008, 38, 1229–1238. [Google Scholar] [CrossRef]
- Ayuso, R.; Grishina, G.; Ahn, K.; Bardina, L.; Beyer, K.; Sampson, H.A. Identification of a MnSOD-like protein as a new major pistachio allergen. J. Allergy Clin. Immunol. 2007, 119, 115. [Google Scholar] [CrossRef]
- Burks, A.W.; Cockrell, G.; Stanley, J.S.; Helm, R.M.; Bannon, G.A. Recombinant peanut allergen Ara h I expression and IgE binding in patients with peanut hypersensitivity. J. Clin. Investig. 1995, 96, 1715–1721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viquez, O.M.; Summer, C.G.; Dodo, H.W. Isolation and molecular characterization of the first genomic clone of the major peanut allergen, Ara h 2. J. Allergy Clin. Immunol. 2001, 107, 713–717. [Google Scholar] [CrossRef] [PubMed]
- Rabjohn, P.; Helm, E.M.; Stanley, J.S.; West, C.M.; Sampson, H.A.; Burks, A.W.; Bannon, G.A. Molecular cloning and epitope analysis of the peanut allergen Ara h 3. J. Clin. Investig. 1999, 103, 535–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bastiaan-Net, S.; Reitsma, M.; Cordewener, J.H.G.; van der Valk, J.P.M.; America, T.A.H.P.; Dubois, A.E.J.; Gerth van Wijk, R.; Savelkoul, H.F.J.; de Jong, N.W.; Wichers, H.J. IgE cross-reactivity of cashew nut allergens. Int. Arch. Allergy Immunol. 2019, 178, 19–32. [Google Scholar] [CrossRef] [PubMed]
- Dreskin, S.C.; Koppelman, S.J.; Andorf, S.; Nadeau, K.C.; Kalra, A.; Braun, W.; Negi, S.S.; Chen, X.; Schein, C.H. The importance of the 2S albumins for allergenicity and cross-reactivity of peanuts, tree nuts, and sesame seeds. J. Allergy Clin. Immunol. 2020. [Google Scholar] [CrossRef]
- Sirvent, S.; Akotenou, M.; Cuesta-Herranz, J.; Vereda, A.; Rodríguez, R.; Villalba, M.; Palomares, O. The 11S globulin Sin a 2 drom yellow mustard seeds shows IgE cross-reactivity with homologous counterparts from tree nuts and peanut. Clin. Transl. Allergy 2012, 2, 23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polk, B.; Dinakarpandian, D.; Nanda, M.; Barnes, C.; Dinakar, C. Association of tree nut and coconut sensitizations. Ann. Allergy Asthma Immunol. 2016, 117, 412–416. [Google Scholar] [CrossRef]
- Dubiela, P.; Kabasser, S.; Smargiasso, N.; Geiselhart, S.; Bublin, M.; Hafner, C.; Mazzucchelli, G.; Hoffmann-Sommergruber, K. Jug r 6 is the allergenic vicilin present in walnut responsible for IgE cross-reactivity to other three nuts and seeds. Sci. Rep. 2018, 8, 11366. [Google Scholar] [CrossRef] [Green Version]
- Too, J.J.Y.; Shek, L.P.C.; Rajakulendran, M. Cross-reactivity of pink peppercorn in cashew and pistachio allergic individuals. Asia Pac. Allergy 2019, 9, e25. [Google Scholar] [CrossRef] [PubMed]
- Azcona, O.M.; Romero, L.; Bartolome, B.; Balboa, V.; Vila, L. Clinical cross-reactivity among mango, pistachio nut, and cashew nut in allergic children. J. Allergy Clin. Immunol. Pract. 2020, 8, 3234–3236.e2. [Google Scholar] [CrossRef] [PubMed]
- Agne, P.S.; Bidat, E.; Agne, P.S.; Rancé, F.; Paty, E. Sesame seed allergy in children. Eur. Ann. Allergy Clin. Immunol. 2004, 36, 300–305. [Google Scholar] [PubMed]
- Smith, P.K.; Krohn, R.I.; Hermanson, G.T.; Mallia, A.K.; Gartner, F.H.; Provenzano, M.D.; Fujimoto, E.K.; Goeke, N.M.; Olson, B.J.; Klenk, D.C. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1987, 150, 76–85. [Google Scholar] [CrossRef]
- Tastet, C.; Lescuyer, P.; Diemer, H.; Luche, S.; van Dorsselaer, A.; Rabilloud, T. A versatile electrophoresis system for the analysis of high- and low-molecular-weight proteins. Electrophoresis 2003, 24, 1787–1794. [Google Scholar] [CrossRef] [PubMed]
- Shevchenko, A.; Wilm, M.; Vorm, O.; Jensen, O.N.; Podtelejnikov, A.V.; Neubauer, G.; Shevchenko, A.; Mortensen, P.; Mann, M. A strategy for identifying gel-separated proteins in sequence databases by MS alone. Biochem. Soc. Trans. 1996, 24, 893–896. [Google Scholar] [CrossRef] [Green Version]
- Frank, R. Spot-synthesis: An easy technique for the positionally addressable parallel chemical synthesis on a membrane support. Tetrahedron 1992, 48, 9217–9232. [Google Scholar] [CrossRef]
- Laune, D.; Molina, F.; Ferrieres, G.; Villard, S.; Bes, C.; Rieunier, F.; Chardes, T.; Granier, C. Application of the Spot method to the identification of peptides and amino acids from the antibody paratope that contribute to antigen binding. J. Immunol. Methods 2002, 267, 53–70. [Google Scholar] [CrossRef]
- Notredame, C.; Higgins, D.G.; Heringa, J. T-coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000, 302, 1331–1338. [Google Scholar] [CrossRef] [Green Version]
- Krieger, E.; Koraimann, G.; Vriend, G. Increasing the precision of comparative models with YASARA NOVA—A self-parameterizing force field. Proteins 2002, 47, 393–402. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The protein data bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ullah, A.; Barros Mariutti, R.; Massod, R.; Putinhon Caruso, I.; Gravatim Costa, G.H.; de Freita, C.M.; Santos, C.R.; Zanphorlin, L.M.; Rossini Mutton, M.J.; Murakami, M.T.; et al. Crystal structure of mature 2S albumin from Moringa oleifera seeds. Biochem. Biophys. Res. Commun. 2015, 468, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Mueller, G.A.; Gosavi, R.A.; Pomés, A.; Wünschmann, S.; Moon, A.F.; London, R.E.; Pedersen, L.C. Ara h 2: Crystal structure and IgE binding distinguish two subpopulations of peanut allergic patients by epitope diversity. Allergy 2011, 66, 878–885. [Google Scholar] [CrossRef] [Green Version]
- Rundqvist, L.; Tengel, T.; Zdunek, J.; Björn, E.; Schleucher, J.; Alcocer, M.J.C.; Larsson, G. Solution structure, copper binding and backbone dynamics of recombinant Ber e 1—The major allergen from Brazil nut. PLoS ONE 2012, 7, e46435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.-F.; Jiang, P.; Zhu, D.-Y.; Hu, Y.; Max, M.; Wang, D.C. Crystal structure of mabinlin II: A novel structural type of sweet proteins and the main structural basis for its sweetness. J. Struct. Biol. 2008, 162, 50–62. [Google Scholar] [CrossRef]
- Jain, A.; Kumar, A.; Salunke, D.M. Crystal structure of the vicilin from Solanum melongena reveals existence of different anionic ligands in structurally similar pockets. Sci. Rep. 2016, 6, 23600. [Google Scholar] [CrossRef] [Green Version]
- Wlodawer, A.; Dauter, Z.; Porebski, P.; Minor, W.; Stanfield, R.; Jaskolski, M.; Pozharski, E.; Weichenberger, C.X.; Rupp, B. Detect, correct, retract: How to manage incorrect structural models. FEBS J. 2018, 285, 444–466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shikhi, M.; Nair, D.T.; Salunke, D.M. Structure-guided identification of function: Role of Capsicum annuum vicilin during oxidative stress. Biochem. J. 2018, 475, 3057–3071. [Google Scholar] [CrossRef]
- Zhang, Y.; Lee, B.; Du, W.-X.; Zhang, Y.; Wang, S.; Fan, Y.; Yi, J.; McHugh, T.H. Identification and characterization of a new Pecan [Carya illinoinensis (Wangenh.) K. Koch] allergen, Car i 2. J. Agric. Food Chem. 2016, 64, 4146–4151. [Google Scholar] [CrossRef]
- Jin, T.; Wang, Y.; Chen, Y.-W.; Fu, T.-J.; Kothary, M.H.; McHugh, T.H.; Zhang, Y. Crystal structure of Korean pine (Pinus koraiensis) 7S seed storage protein with copper ligands. J. Agric. Food Chem. 2014, 62, 222–228. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Kesari, P.; Dhindwal, S.; Choudhary, A.K.; Katiki, M.; Verma, A.; Ambatipudi, K.; Tomar, S.; Sharma, A.K.; Mishra, G.; et al. A novel function for globulin in sequestering plant hormone: Crystal structure of Wirightia tinctoria 11S globulin in coimplex with auxin. Sci. Rep. 2017, 7, 4705. [Google Scholar] [CrossRef] [Green Version]
- Jin, T.; Wang, C.; Zhang, C.; Wang, Y.; Chen, Y.-W.; Guo, F.; Howard, A.; Cao, M.-J.; Fu, T.-J.; McHugh, T.H.; et al. Crystal structure of cocosin, a potential food allergen from coconut (Cocos nucifera). J. Agric. Food Chem. 2017, 65, 7560–7568. [Google Scholar] [CrossRef]
- Tandang-Silvas, M.R.G.; Fukuda, T.; Fukuda, C.; Prak, K.; Cabanos, C.; Kimura, A.; Itoh, T.; Mikami, B.; Utsumi, S.; Maruyama, N. Conservation and divergence of plant seed 11S globulins based on crystal structures. Biochim. Biophys. Acta 2010, 1804, 1432–1442. [Google Scholar] [CrossRef]
- Jin, T.; Albillos, S.M.; Guo, F.; Howard, A.; Fu, T.-J.; Kothary, M.H.; Zhang, Y.-Z. Crystal structure of prunin-1, a major component of the almond (Prunus dulcis) allergen amandin. J. Agric. Food Chem. 2009, 57, 8643–8651. [Google Scholar] [CrossRef]
- Oda, Y.; Matsunaga, T.; Fukuyama, K.; Miyazaki, T.; Morimoto, T. Tertiary and quaternary structures of 0.19 α-amylase inhibitor from wheat kernel determined by X-ray analysis at 2.06 Å resolution. Biochemistry 1997, 36, 13503–13511. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, K.; Schweimer, K.; Reese, G.; Randow, S.; Suhr, M.; Becker, W.-M.; Vieths, S.; Rösch, P. Structure and stability of 2S albumin-type peanut allergens: Implications for the severity of peanut allergic reactions. Biochem. J. 2006, 395, 463–472. [Google Scholar] [CrossRef]
- Jin, T.; Guo, F.; Chen, Y.-W.; Howard, A.; Zhang, Y.-Z. Crystal structure of Ara h 3, a major allergen in peanut. Mol. Immunol. 2009, 46, 1796–1804. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemistry of protein structures. J. Appl. Cryst. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Melo, F.; Feytmans, E. Assessing protein structures with a non-local atomic interaction energy. J. Mol. Biol. 1998, 277, 1141–1152. [Google Scholar] [CrossRef]
- Benkert, P.; Biasini, M.; Schwede, T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 2011, 27, 343–350. [Google Scholar] [CrossRef] [PubMed]
- Arnold, K.; Bordoli, L.; Kopp, J.; Schwede, T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics 2006, 22, 195–201. [Google Scholar] [CrossRef] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pomés, A.; Davies, J.M.; Gadermaier, G.; Hilger, C.; Holzhauser, T.; Lidholm, J.; Lopata, A.L.; Mueller, G.A.; Nandy, A.; Radauer, C.; et al. WHO/IUIS Allergen Nomenclature Sub-Committee. Mol. Immunol. 2018, 100, 3–13. [Google Scholar] [CrossRef]
- Böhm, M.; Bohne-Lang, A.; Frank, M.; Loss, A.; Rojas-Macias, M.A.; Lütteke, T. Glycosciences.DB: An annotated data collection linking glycomics and proteomics data (2018 update). Nucleic Acids Res. 2019, 47, D1195–D1201. [Google Scholar] [CrossRef] [Green Version]
- Sordet, C.; Culerrier, R.; Granier, C.; Didier, A.; Rougé, P. Expression of Jug r 1, the 2S albumin allergen from walnut (Juglans regia), as a correctly folded and functional recombinant protein. Peptides 2009, 30, 1213–1221. [Google Scholar] [CrossRef]
- Pastorello, E.A.; Varin, E.; Farioli, L.; Pravettoni, V.; Ortolani, C.; Trambaioli, C.; Fortunato, D.; Giuffrida, M.G.; Rivolta, F.; Robino, A.; et al. The major allergen of sesame seeds (Sesamum indicum) is a 2S albumin. J. Chromatogr. B Biomed. Sci. Appl. 2001, 756, 85–93. [Google Scholar] [CrossRef]
- Rougé, P.; Brunet, E.; Borges, J.P.; Jauneau, A.; Saggio, B.; Bourrier, T.; Rancé, F.; Didier, A.; Barre, A. Proteins with cupin motif as major seed allergens. Rev. Fr. Allergol. 2011, 51, 36–40. [Google Scholar] [CrossRef]
- Aalberse, R.C.; Mueller, G.A.; Derksen, N.I.L.; Aalberse, J.A.; Edwards, L.L.; Pomés, A.; Lidholm, J.; Rispens, T.; Briza, P. Identification of the amino acid fragment of Ara h 1 as a major target of the IgE-binding activity in the basic peanut protein fraction. Clin. Exp. Allergy 2020, 50, 401–405. [Google Scholar] [CrossRef]
- Burks, A.W.; Shin, D.; Cickrell, G.; Stanley, J.S.; Helm, R.M.; Bannon, G.A. Mapping and putational analysis of the IgE-binding epitopes on Ara h 1, a legume vicilin protein and a major allergen in peanut hypersensitivity. Eur. J. Biochem. 1997, 245, 334–339. [Google Scholar] [CrossRef]
- Shin, D.S.; Compadre, C.M.; Maleki, S.J.; Kopper, R.A.; Sampson, H.; Huang, S.K.; Burks, A.W.; Bannon, G.A. Biochemical and structural analysis of the IgE binding sites on Ara h 1, an abundant and highly allergic peanut protein. J. Biol. Chem. 1998, 273, 13753–13759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nesbit, J.B.; Dchein, C.H.; Braun, B.A.; Gipson, S.A.Y.; Cheng, H.; Hurlburt, B.K.; Maleki, S.J. Epitopes with similar physicochemical properties contribute to cross reactivity between peanut and tree nuts. Mol. Immunol. 2020, 122, 223–231. [Google Scholar] [CrossRef] [PubMed]
- Archila, L.D.; Chow, I.-T.; McGinty, J.W.; Renand, A.; Jeong, D.; Robinson, D.; Farrington, M.L.; Kwok, W.W. Ana o 1 and Ana o 2 cashew allergens share cross-reactive CD4(+) T cell epitopes with other tree nuts. Clin. Exp. Allergy 2016, 46, 871–883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goetz, D.W.; Whisman, B.A.; Goetz, A.D. Cross-reactivity among edible nuts: Double immunodiffusion, crossed immunoelectrophoresis, and human specific IgE serologic surveys. Ann. Allergy Asthma Immunol. 2005, 95, 45–52. [Google Scholar] [CrossRef]
- De Leon, M.P.; Glaspole, I.N.; Drew, A.C.; Rolland, J.M.; O’Hehir, R.E.; Suphioglu, C. Immunological analysis of allergenic cross-reactivity between peanut and tree nuts. Clin. Exp. Allergy 2003, 33, 1273–1280. [Google Scholar] [CrossRef]
- De Leon, M.P.; Drew, A.C.; Glaspole, I.N.; Suphioglu, C.; Rolland, J.M.; O’Hehir, R.E. Functional analysis of cross-reactive immunoglobulin E antibodies: Peanut-specific immunoglobulin E sensitizes basophils to tree nut allergens. Clin. Exp. Allergy 2005, 35, 1056–1064. [Google Scholar] [CrossRef] [PubMed]
- Maleki, S.J.; Kopper, R.A.; Shin, D.S.; Park, C.-W.; Compadre, C.M.; Sampson, H.A.; Burks, A.W.; Bannon, G.A. Structure of the major peanut allergen Ara h 1 may protect IgE-binding epitopes from degradation. J. Immunol. 2000, 164, 5844–5849. [Google Scholar] [CrossRef] [Green Version]
- Robotham, J.M.; Wang, F.; Seamon, V.; Teuber, S.S.; Sathe, S.K.; Sampson, H.A.; Beyer, K.; Seavy, M.; Roux, K.H. Ana o 3, an important cashew nut (Anacardium occidentale L.) allergen of the 2S albumin family. J. Allergy Clin. Immunol. 2005, 115, 1284–1290. [Google Scholar] [CrossRef] [PubMed]
- Xia, L.; Willison, L.N.; Porter, L.; Robotham, J.M.; Teuber, S.S.; Sathe, S.K.; Roux, K.H. Mapping of a conformational epitope on the cashew allergen Ana o 2: A discontinuous large sub-unit epitope dependent upon homologous or heterologous small subunit association. Mol. Immunol. 2010, 47, 1808–1816. [Google Scholar] [CrossRef]
- Robotham, J.M.; Xia, L.; Willison, L.N.; Teuber, S.S.; Sathe, S.K.; Roux, K.H. Characterization of a cashew allergen, 11S globulin (Ana o 2), conformational epitope. Mol. Immunol. 2010, 47, 1830–1838. [Google Scholar] [CrossRef]
- Van Ree, R.; Cabanes-Macheteau, M.; Akkerdaast, J.; Milazzo, J.-P.; Loutelier-Bourhis, C.; Rayon, C.; Villalba, M.; Koppelman, S.; Aalberse, R.; Rodriguez, R.; et al. β(1,2)-xylose and α(1,3)-fucose residues have a strong contribution in IgE binding to plant glycoallergens. J. Biol. Chem. 2000, 275, 11451–11458. [Google Scholar] [CrossRef] [Green Version]
- Andorf, S.; Borres, M.P.; Block, W.; Tupa, D.; Bollyky, J.B.; Sampath, V.; Elizur, A.; Lidholm, J.; Jones, J.E.; Galli, S.J.; et al. Association of clinical reactivity with sensitization to allergen components in multifood-allergic children. J. Allergy. Clin. Immunol. Pract. 2017, 5, 1325–1334. [Google Scholar] [CrossRef] [PubMed]
- Van derValk, J.P.M.; El Bouche, R.; Gerth van Wijk, R.; de Groot, H.; Wichers, H.J.; Dubois, A.E.J.; de Jong, N.W. Low percentage of clinically relevant pistachio nut and mango co-sensitisation in cashew nut sensitised children. Clin. Transl. Allergy 2017, 7, 8. [Google Scholar] [CrossRef] [Green Version]
- Couch, C.; Franxman, T.; Greenhawt, M. Characteristics of tree nut challenges in tree nut allergic and tree nut sensitized individuals. Ann. Allergy Asthma Immunol. 2017, 118, 591–596.e3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fong, A.T.; Du Toit, G.; Versteeg, S.A.; van Ree, R. Pink peppercorn: A cross-reactive risk for cashew- and pistachio-allergic patients. J. Allergy Clin. Immuinol. Pract. 2019, 7, 724–725.e1. [Google Scholar] [CrossRef]
- Cetinkaya, P.G.; Karaguzei, D.; Esenboga, S.; Sahiner, U.M.; Soyer, O.; Buyuktiryaki, B.; Birben, E.; Karaaslan, C.; Sekerel, B.E. Pistachio and cashew nut allergy in childhood: Predictive factors towards development of a decision tree. Asian Pac. J. Allergy Immunol. 2019. [Google Scholar] [CrossRef]
- Amat, F.; Benharoun-Stern, R.; Benissa, M.-R.; Just, J. Usefulness of rAna o 3 assessment before oral food challenge to pistachio. Pediatr. Allergy Immunol. 2020. [Online ahead of print]. [Google Scholar] [CrossRef]
- Cetinkaya, P.G.; Buyuktiryaki, B.; Soyer, O.; Sahiner, U.M.; Sackesen, C.; Sekerel, B.E. Phenotypical characterization of tree nuts and peanut allergies in esat Mediterranean children. Allergol. Immunopathol. 2020, 48, 316–322. [Google Scholar] [CrossRef]
- Saba, L.; Clerc-Urmès, I.; Delahaye, C.; Chevillot, E.; Jarlot-Chevaux, S.; Dumond, P.; Schweitzer, C.; Divaret-Chauveau, A. Predictive factors of allergy to pistachio in children allergic to cashew nut. Pediatr. Allergy Immunol. 2020, 31, 506–514. [Google Scholar] [CrossRef]













| Subjects | Sex/Age (Years) | ** Pistachio Specific IgE (KU/L) | *** Cashew Specific IgE (KU/L) |
|---|---|---|---|
| 1 | * M/14 | >100 | 83.8 |
| 2 | M/5 | 2.20 | 1.35 |
| 3 | M/8 | 0.36 | 0.25 |
| 4 | M/4 | 0.17 | 0.45 |
| 5 | M/9 | 1.65 | 1.51 |
| 6 | M/16 | 0.47 | 0.10 |
| 7 | *F/7 | 5.69 | 3.92 |
| 8 | F/8 | 2.54 | 1.44 |
| 9 | M/4 | 2.46 | 1.20 |
| Subjects | Sex/Age (Years) | * Sesame Specific Ige (KU/L) |
|---|---|---|
| 1 | M/6 | 0.10 |
| 2 | M/16 | 0.26 |
| 3 | F/10 | 0.74 |
| 4 | M/10 | 0.26 |
| 5 | M/5 | 1.53 |
| 6 | M/10 | 1.85 |
| 7 | M/10 | 14.2 |
| 8 | F/6 | 46.3 |
| 9 | F/9 | 84.4 |
| Models | 3D Structures from the PDb Used as Templates | Number of Models Built by Homology Modelling | Residues out of the Allowed Areas in the Ramachandran Plot | * Residues with Values over the Threshold in ANOLEA Plot | QMEAN Value |
|---|---|---|---|---|---|
| Pis v 1 | 2D2S, 2LVF, 3OB4, 5DOM, 5U87 | 5 | 0 | 20 over 122 residues | −1.19 |
| Pis v 2 | 2E9Q, 3QAC, 3FZ3, 5WPW, 5WXU | 19 | 3:S201(A),D249(A),D249(B) | 27 over 440 residues | −1.78 |
| Pis v 3 | 4LEJ, 5CAD, 5E1R, 5VF5, 5YJS | 9 | 2:K334(B),F502(C) | 23 over 405 residues | −0.21 |
| Ana o 3 | 1HSS, 1W2Q, 2DS2, 3OB4 | 5 | 0 | 20 over 114 residues | −1.55 |
| Ses i 3 | 4LEJ, 5CAD, 5E1R, 5VF5, 5YJS | 10 | 0 | 21 over 406 residues | −0.39 |
| Ses i 6 | 2E9Q, 3ZF3, 5WPW, 5WXU | 25 | 3:Q121(A),R234(C),S248(C) | 29 over 438 residues | −1.08 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barre, A.; Nguyen, C.; Granier, C.; Benoist, H.; Rougé, P. IgE-Binding Epitopes of Pis v 1, Pis v 2 and Pis v 3, the Pistachio (Pistacia vera) Seed Allergens. Allergies 2021, 1, 63-91. https://doi.org/10.3390/allergies1010006
Barre A, Nguyen C, Granier C, Benoist H, Rougé P. IgE-Binding Epitopes of Pis v 1, Pis v 2 and Pis v 3, the Pistachio (Pistacia vera) Seed Allergens. Allergies. 2021; 1(1):63-91. https://doi.org/10.3390/allergies1010006
Chicago/Turabian StyleBarre, Annick, Christophe Nguyen, Claude Granier, Hervé Benoist, and Pierre Rougé. 2021. "IgE-Binding Epitopes of Pis v 1, Pis v 2 and Pis v 3, the Pistachio (Pistacia vera) Seed Allergens" Allergies 1, no. 1: 63-91. https://doi.org/10.3390/allergies1010006






