Abstract
The family name Hay (plus associated spelling variants) is a prominent Anglo-Norman-in-origin surname that has been well-documented as a Scottish noble lineage since the 12th century CE. Their historical significance, linked to the rise in the Anglo-Norman era (1093–1286 CE) in Scotland, and the historical complexities of surname adoption post-Norman conquest of England, justifies the need for a comprehensive understanding of the genetic history of the Hay noble lineage. This study focuses on examining the patterns of paternal inheritance in lineages with the Hay surname. We conducted a comprehensive analysis of Y-chromosome data that is publicly available on the Family Tree DNA (FTDNA) platform, and specific FTDNA surname projects, as well as looking in more detail at three well-documented male-line descendants of William II de la HAYA, 1st of Erroll (d. 1201) that have been verified to a high degree of confidence. Our results reveal that all descendants of William II de la HAYA, 1st of Erroll (d. 1201) derive from the multigenerational Y-SNPs R1a-YP6500 (plus equivalent SNPs BY33394/FT2017) and R1a-FTT161. Furthermore, subclades of R1a-FTT161 have been identified that confirm direct male-line descent from two of William II de la HAYA’s sons. Subclade R1a-BY199342 (plus equivalents) confirms direct male-line descent from David de la HAYA, 2nd of Erroll (d. 1241), and subclade R1a-FTA7312 confirms direct male-line descent from Robert de la HAYA of Erroll. The result also confirms that the Hay noble lineage shares the Y-SNP R1a-YP4138 (estimated to have occurred in 832 CE) with several non-Hay test takers that have surnames of Norman origin, therefore providing further evidence to support the Norman origin hypothesis for these surnames. In addition to the identification of multigenerational Y-SNPs associated with documented Hay noblemen, this study has observed significant Y-DNA haplogroup diversity among males with the surname Hay (plus associated spelling variants: Hays, Haye, Hayes, Hey and Haya). Our results show that only 22% of the men sampled (n = 109) with the surname Hay (plus associated spelling variation) are descended from the 12th-century progenitor of the noble Hay lineage of Scotland. Therefore, this confirms that a significant proportion of males with the surname Hay do not descend from the noble progenitor of the Scottish Hay lineage of Erroll.
Keywords:
Y-DNA; genetic-genealogy; Hay; Scottish nobles; Y-SNPs; surname; genealogy; Anglo-Norman; Norman 1. Introduction
The lands of northern Britain that are now referred to as Scotland have not always been a unified realm. Between the early seventh century until the mid-ninth century, northern Britain was separated into four distinct kingdoms (Woolf 2007). The kingdom of Strathclyde was ruled by the Britons and incorporated the southwest region of modern-day Scotland; however, a large portion of this kingdom was once incorporated into the Anglo-Saxon kingdom of Northumbria by the early eighth century, which also included the Lothians and the southeast border regions (Woolf 2007). The lands that primarily encompassed the northeastern third of Scotland, stretching roughly from the Firth of Forth in the south to Caithness and Orkney in the north, were considered the kingdom of Pictavia, and was ruled by the Picts (Woolf 2007). The kingdom of Dál Riata was ruled by the Scots, and their kingdom included the western lands north of Loch Lomond and the Hebrides Islands (Woolf 2007).
The four kingdoms remained relatively stable until a significant event took place in 844 CE under the leadership of King Kenneth I (Cináed mac Alpin, died in 858 CE), who successfully unified the kingdom of Dál Riata and Pictavia into a single kingdom, often referred to as Alba (Snow 2001). Although the formation of the kingdom of Alba is generally considered the birth of Scotland, Broun (2015) argues that this is a significant oversight and oversimplification of the historical events that took place. Broun (1997) argues that it would not be until the thirteenth century, during the Anglo-Norman era, that the term Scotia (Scotland) was generally used when referring to the whole kingdom.
1.1. The Anglo-Norman Era in Scotland (1093–1286 CE)
The revolutionary change in the governance of Scotland, termed the ‘Anglo-Norman era’, was mainly facilitated by King David I of Scotland during his reign (c. 1124–1153 CE) (Barrow 1985). However, the start of Norman influence in Scotland started with the Treaty of Abernethy, where King Malcolm III of Scotland swore fealty to King William I of England in 1072 CE (Lynch 1992). King William I of England died on 9 September 1087 CE and was succeeded by his son William II (commonly referred to as William Rufus). Malcolm III of Scotland went on to break the terms of the Treaty of Abernethy by invading northern England several times in the early 1090s CE (Lynch 1992). On the 13th November 1093 CE, King Malcolm III of Scotland invaded Berwick in northern England with his son Edward (Lynch 1992). King Malcolm III of Scotland advanced to Alnwick and was ambushed by English forces led by Robert de Mowbray, Earl of Northumbria. Both King Malcolm III of Scotland and his son Edward were killed, leaving the Scottish forces without leadership, forcing their withdrawal back to the safety of Scotland, and facilitating the start of the Anglo-Norman era (c. 1093–1286 CE) (Lynch 1992).
King David I of Scotland (1084–1153 CE) had a new vision for Scotland: an aspiration to replicate the Norman feudal system imposed in England by King William I of England (c. 1066 CE) (Barrow 1985). To achieve his goal of adopting feudal tenure during his reign (c. 1124–1154 CE), King David I of Scotland attracted Anglo-Normans (people of higher social status of Anglo-Norman, French, and Flemish origin) to Scotland by offering Scottish lands, royal official appointments, noble titles, and knighthoods in return for feudal services (Barrow 1985). This invitation radically changed the governance of Scotland and impacted the demographics of the Scottish population to an unknown degree (Hammond 2006).
1.2. The Documented Origins of the Hay Surname and the Earliest Hay Progenitor in Scotland
Among the foreign Anglo-Norman settlers of Scotland was the progenitor of the Scottish noble lineage Hay of Erroll, described as William de la HAYA II, 1st of Erroll (d. 1201 CE) (Paul 1904). William de la HAYA II, 1st of Erroll was a Norman knight who is documented as cupbearer (butler) of King William the Lion of Scotland, who succeeded King Malcolm IV on 9 December 1165 CE (Broun et al. 2007d). William de la HAYA II, 1st of Erroll (d. 1201 CE) is believed to be the son of William de la HAYA I (born in Cotentin, Normandy) and Julianna de Soulis, who was the sister of Ranulf de Soulis, Lord Liddesdale (Broun et al. 2007c) Butler of King Malcolm IV (c. 1153–1164 CE) (Broun et al. 2007b, 2007c, 2007d). Black (1946) argued that there are several village names starting with ‘La Haye [haie/haia]’ in Normandy, so one of these villages would be a good candidate for the origins of the surname Haya (e.g., La Haye-du-Puits or La Haye Bellefond, both in the Soules region). Ritchie (1954) also supported La Haye-du-Puits as a possible origin of the Hay noble lineage of Scotland; however, he offered La Haye-de-Herce and La Haye Malherbe as potential alternatives. Moncreiffe of that Ilk Iain and Armstrong (2010) argued that there is no doubt that the origins of the Scottish Hay noble lineage were from Haye-Hue (Haia-Hugonis, now La Haye-Bellefond), because the La Haye-Hue of Normandy bear the same three escutcheons coat of arms used by the Hays of Erroll, and they also marched with de Soules [Soulis] near St Lô in the Cotentin peninsula of Normandy. Barrow (1973) also supported Haye-Hue (La Haye-Bellefond) as the place of origin of the Hay noble lineage of Scotland. The documented links between La Haye-Hue of Normandy, de Soulis (Normandy and Liddesdale), and Hay of Erroll are not likely to be coincidental. The Hay noble lineage firmly established their presence in Scotland, taking control of significant lands located in the Scottish Lowlands (Broun et al. 2007a), and played a substantial role in Scottish history thereafter. For example, Sir Gilbert de la HAY (d. 1333) was the fifth feudal Baron of Erroll, Lord High Constable of Scotland, and companion of King Robert the Bruce of Scotland. Sir Gilbert de la HAY was one of the Scottish noblemen who signed the Declaration of Arbroath on 6 April 1320 CE (Barron 1997).
The evidence that links William de la HAYA I to Normandy is that ‘la Haya’ is likely a Norman toponymic name, deriving its origin from a place called ‘la Haye/Haie’ in Normandy. McClure (2015) and Ormrod et al. (2020) argued the case that surnames such as ‘de la Haye’ and ‘de la Mare’ often denote high-ranking people from estates in Normandy (e.g., a notable individual from la Haie ‘the enclosure’ or la Mare ‘the pool’). However, Ormrod et al. (2020) are also appropriately cautious in linking these locations to one specific origin or family. Alternatively, McClure (2015) also suggests that the surname could have its origins from French Walloon and Huguenot migration. There are several villages in Normandy that have such place names, and Norman placenames were also introduced to Britain via the linguistic legacy of the Norman Conquest of 1066 CE. When William the Conqueror and his followers introduced Norman French culture, governance, and language to Britain, placenames often reflected the new Norman elite’s language, ownership, heritage, or religious affiliations, and were either imposed on new settlements or modified from existing Anglo-Saxon placenames. A good example is the placename Barnard in Barnard Castle, County Durham, a place deriving its name from Bernard Baliol I (died before 1167 CE), a baron of Norman descent (The Institute for Name-Studies 2025b). Other examples include Norman French placenames derived from descriptive words, such as Belvoir (bel voir) in Leicestershire, meaning ‘beautiful view’ (The Institute for Name-Studies 2025a) and Richmond in North Yorkshire, which is derived from ‘riche mont’, meaning ‘strong hill’ (The Institute for Name-Studies 2025a; Smith 1928). However, it is possible that these names were transferred from places in Normandy. Therefore, there is a strong possibility that many unrelated people adopted these surnames independently. Durie (2022) also highlights the complexities involved in identifying surname origins in his essay on clans, families, and kinship structures in Scotland. He refers to the Hay noble lineage as being among the most influential families in Scotland; however, he does not discuss their potential origins prior to Scotland. Durie (2022) also highlights that having a surname that is specifically associated with nobility does not guarantee direct paternal descent from that noble lineage.
It is important to distinguish between the Hay noble lineage of Erroll and the surname Hay in its wider usage. The surname is demonstrably polygenetic, having arisen independently in several linguistic and geographic contexts. The Dictionary of American Family Names (2nd ed., OUP) lists alternative etymologies that include Norman toponymic (de la Haye = ‘the enclosure’), English topographic (Hay(e) = ‘enclosure’), nickname (heigh/hey = ‘high’), and Irish forms (Hayes, variant of O’hAodha) (Hanks 2003). The surname also appears among families of continental European, Middle Eastern, and East-Asian origin. Therefore, references in this paper to a “noble progenitor” pertain solely to the Scottish noble family of Erroll and should not be interpreted as implying that all modern bearers of the surname Hay share a common or noble ancestor.
1.3. Surname Changes
Surname changes did historically take place with no documentary evidence to explain why/how the change occurred. Instances of surname changes were often the result of adoptions, divorce, out-of-wedlock conceptions, and extra-marital conceptions, so these situations would often remain undocumented (Avni et al. 2023). The genealogical term for these occurrences is a ‘non-paternity event’ (NPE) or a ‘false paternal event’ (FPE) (Avni et al. 2023), while Larmuseau et al. (2017) uses the term ‘extra pair paternity’ (EPP). Avni et al. (2023) suggested that the chances of an EPP taking place were between 1 and 5% in each generation, with the chances of discovering an EPP increasing with each successive generation (e.g., there is a 10–50% chance of discovering an EPP over ten generations). Moreover, King et al. (2014) revealed multiple instances of non-paternity in King Richard III’s family tree through the DNA analysis of material collected from his skeletal remains.
Traditional genealogical research heavily relies on documentary evidence to prove genealogies (Glynn 2022; Greytak et al. 2019). The documentary evidence strongly suggests that William de la HAYA II has direct paternal descendants surviving to the present day (Lundy 2019a; Paul 1904). However, traditional genealogical evidence alone is limited, because it cannot identify an EPP occurring in any of the intervening generations of a pedigree (Durie 2022) unless they were documented. Based on the evidence provided by Avni et al. (2023), there is a good chance that someone with the surname Hay does not descend from William de la HAYA II. Furthermore, to the best of the authors’ knowledge, William de la HAYA I’s proposed link to Normandy is only supported by limited toponymic evidence, so the pursuit of further evidence to explore his origins is well-justified.
1.4. Genetic Genealogy
The advancements in DNA sequencing technology and the development of haplotype-based methods to detect population substructure have proven to provide insight into the degree of shared ancestry between populations (Fournier et al. 2023; Guan 2014; Nait Saada et al. 2020; Shaw et al. 2024). A recent study by Morez et al. (2023) implemented FineSTRUCTURE clustering analysis and identity-by-descent (IBD) on an imputed diploid dataset of ancient Pictish and present-day Scottish genomes, revealing partial population replacement that took place in eastern Scotland during the Anglo-Norman era. Morez et al. (2023) demonstrated that there was less genetic affinity between present-day eastern Scottish samples and the ancient Pictish genomes they sequenced, while present-day western Scottish samples shared significantly more genetic affinity with the ancient Pictish genomes. Furthermore, Morez et al. (2023) observed a clear genetic signature that differentiated the eastern Scottish population (showing a greater genetic affinity with populations with Anglo-Norman ancestry) from the western Scottish population (showing a greater genetic affinity with ancient samples of Iron-Age-British and Pictish ancestry). Moreover, Gretzinger et al. (2022) observed a noticeable increase in Iron-Age-French ancestry among the present-day English population when compared to the ancient Iron-Age-British samples that have been sequenced. Gretzinger et al. (2022) concluded that Iron-Age-French-like ancestry was likely introduced to England during the early Medieval Age by people with origins likely from France and this continued through to the late Medieval Age with the Normans.
King and Jobling (2009a, 2009b) argued that because of the correlation between surnames being traditionally inherited paternally, and Y-DNA being biologically passed on from father to son virtually unchanged, the study of Y-DNA has been of great value to genealogical studies investigating direct paternal ancestry. The consensus among researchers is that the comparison of mutations found within the Y-chromosome’s short tandem repeat markers (Y-STRs) and single-nucleotide polymorphisms (Y-SNPs) between two or more males can accurately predict the degree of their patrilineal relationship, show how lineages evolved over time, and predict when lineages arose and diverged (Chen et al. 2021; Erzurumluoglu et al. 2018; King and Jobling 2009a, 2009b; Sole-Morata et al. 2015; Denise Syndercombe Court 2021). Y-SNP mutations on the Y-chromosome define Y-DNA haplogroups, since the variants are assumed to be synapomorphies that define specific branches in the phylogeny (Kivisild 2017). For example, the Y-SNP M417 is a mutation that defines the haplogroup R1a1a1, a major subclade (branch) of the haplogroup R1a (Lall et al. 2021; Underhill et al. 2015). The R1a-M417 haplogroup is especially significant because it represents the progenitor of a major expansion within the haplogroup R1a, and it is one of the main Y-DNA haplogroups associated with the spread of Indo-European languages (Klyosov and Rozhanskii 2012). Genetic genealogical studies by Holton and Macdonald (2020), Stead (2023), Holton (2023), and DePew et al. (2024) have identified Y-SNPs that occurred in a specific individual or a narrow lineal group of patrilineal-related individuals that lived within a known genealogical timeframe. A Y-SNP that can be specifically designated to an individual is termed a SNP-Progenitor (SNP-Progen), while Y-SNPs that can be identified as occurring in a lineal group of patrilineal-related individuals are known as Multigenerational-SNPs (Multigen-SNPs) (Holton and Macdonald 2020; Holton 2023; Stead 2023). The identification of these Y-SNPs is useful to genetic genealogists because they can confirm or reject hypotheses based on ambiguous documented evidence. Furthermore, once these Y-SNPs have been positively confirmed, they can be used to identify descendants who are not fortunate enough to be able to prove their descent with documentary evidence.
This study aimed to analyse Y-SNP data from FTDNA’s BigY700 tests to investigate whether the Y-DNA testing of well-documented living patrilineal descendants of William de la HAYA II supports the genealogical consensus that his direct paternal lineage has survived to the present. The Y-SNP data from testing the BigY700 allow for genetic genealogists to estimate a time to the most recent common ancestor (TMRCA) when two or more individuals share a specific Y-SNP, but their common ancestor is unknown. The mutation rates provide a date range for the occurrence of all Y-SNPs that are shared between two or more test takers, and if the common ancestor between two or more matching test takers is known, their shared Y-SNPs can be assigned to specific individuals. Another aim is to identify SNP-Progens or Multigen-SNPs associated with the Hay noble lineage, so any EPP events can be identified using Y-DNA testing. Furthermore, the study aims to use the data for SNP-Progens or Multigen-SNPs to help identify Hay males of direct paternal descent from the Scottish noble lineage who cannot prove their lineage due to gaps in the documentary records. The data from this study also has the potential to provide evidence to help reach a reliable conclusion on the proposed Norman origins of William de la HAYA I. Furthermore, the analysis of Y-DNA haplogroups found among Family Tree DNA test takers of reported British ancestry bearing the surname Hay (plus associated spelling variants: Hays, Haye, Hayes, Hey and Haya) will be assessed to reach conclusions on the level of haplogroup diversity. An analysis of Y-DNA haplogroup diversity will provide data to evaluate the hypothesis made by Durie (2022), who suggested that surnames in Scotland were largely adopted for many reasons, and that a surname of noble origin does not mean an individual is descended from the noble progenitors of that surname.
2. Materials and Methods
Holton and Macdonald (2020) highlight the significant challenges faced when researching well-documented living descendants of medieval noble lineages. However, they confirmed that there are many examples of individuals of various patrilineal ancestries who have the good fortune of being well-documented, having extant lines of descent to the present day (Holton and Macdonald 2020). Research by Larmuseau et al. (2014), Stead (2023), and DePew et al. (2024) also demonstrates the existence of well-documented lineages back to the high-to-late Middle Ages (c. 1000–1500 CE).
2.1. Traditional Genealogical Documentary Research
The methodology implemented in this study utilised a mixed disciplinary approach that combined traditional genealogical research, genetic genealogical research, and archaeogenetic research methods. Holton and Macdonald (2020) demonstrated that authoritative documentary sources can be used to identify living descendants of noble lineages. Therefore, a range of publicly available authoritative documentation and primary and secondary sources (Table 1) were used to identify three potential test takers with proven patrilineal descent from William de la HAYA II, 1st of Erroll.

Table 1.
Examples of the sources consulted to prove the patrilineal ancestries of the three test takers used for this study.
Based on the outcome of the traditional genealogy research, it was hypothesised that all three potential test takers’ should share a Y-SNP, dated to have occurred between 1000 and 1150 CE, because their genealogies suggest that they descend from William de la HAYA II, 1st of Erroll. However, the genealogies of two of the potential test takers suggested that they shared a more recent ancestor (John HAY, 1st of Tweeddale, 1595–1653 CE) when compared to the third test taker; therefore, they likely share at least one more Y-SNP in common that occurred between 1150 and 1595 CE. Furthermore, it is important to highlight that the two well-documented test takers that descended from John HAY, 1st of Tweeddale could be confidently documented back to Sir John HAY (1200–1262 CE), and he is hypothesised to be the grandson of William de la HAYA II via his son Robert (Douglas and Wood 1813; Paul 1904).
To confirm that the well-documented individuals were the direct paternal descendants of the noble Hay lineage of Scotland, Y-DNA testing was required to investigate whether the individuals shared Y-SNPs that could be dated to around the time period of their proposed common noble Hay ancestor. The identified living descendants were invited to participate in this study after the receipt of signed consent, with the recruitment period starting on the 6th of February 2024 and ending on the 7th of October 2024. Ethical approval for this research was obtained from the University of Strathclyde’s Ethics Committee, reference number: UEC23/94. After signed consent was received, a non-invasive BigY700 test kit was posted out to each participant for cheek cell sample collection, using the cotton mouth swabs provided in the test kit. Once received, each participant completed the DNA sample collection and then posted the sample back to the FTDNA laboratory for sequencing. The BigY700 test by Family Tree DNA (FTDNA) was the test of choice for this project for several reasons (Davis et al. 2019). Firstly, FTDNA was chosen due to their large Y-haplotree, currently consisting of 81,000 branches, 716,000 variants, and 566,000 SNP-tested users in its Y-DNA database (FTDNA 2025c). Secondly, FTDNA has strict policies that ensure test taker privacy and the security of data (FTDNA 2024); they also provide a ‘closed project’ option for researchers that ensures a secure platform to carry out data analysis. The sequenced data provided by FTDNA include several private variants for each test taker; therefore, these data have not been made available because private variants could be used to disclose a participant’s identity.
2.2. Y-DNA Testing Strategy
The BigY700 targets all the Y-SNPs present in the SNP rich regions that are most relevant for genetic genealogical research, with up to 70 reads per position covered; however, any Y-SNP that returns a minimum of 10 derived (positive) reads is reported (Davis et al. 2019). FTDNA’s age estimates for SNPs that feature within their Y-DNA phylogenetic tree are calculated using a method similar to the methodology developed by (McDonald 2021). The approach calculates coalescence ages (times to most recent common ancestor, or TMRCAs) using a new, probabilistic statistical model that includes Y-SNP, Y-STR, and ancillary historical data (McDonald 2021). McDonald (2021) demonstrated that this methodology provided highly accurate estimates by using the Y-DNA data of well-documented direct patrilineal Royal Stewart test takers.
In this study, the Y-SNP data of the three documented test takers were compared against their specific genealogies to identify SNP-Progens or Multigen-SNPs. FTDNA’s age estimates for specific Y-SNPs were accepted as the most accurate estimates, since their calculations were based on McDonald’s methodology (Davis et al. 2019; McDonald 2021). The example in Figure 1 demonstrates how a SNP-Progen and Multigen-SNP(s) are identified by combining the documented genealogical information with the Y-SNP data of three fictitious test takers. When two or more Y-DNA test takers have well-documented ancestry that triangulates back to a direct paternal ancestor living during the medieval era, Multigen-SNPs and SNP-Progens can also be identified with a high degree of confidence.
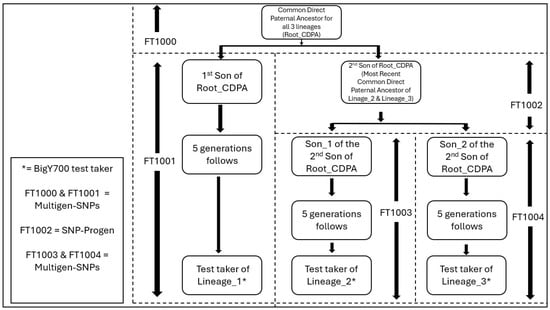
Figure 1.
Diagram demonstrating how SNP-Progen and Multigen-SNPs are identified.
Figure 1 shows how three BigY700 test takers are related, thus demonstrating how SNP-Progens and Multigen-SNPs are identified. Test taker of Lineage_1* descends from the 1st son of the common direct paternal ancestor for all 3 lineages (Root_CDPA). Test taker of Lineage_2* and Test taker of Lineage_3* both descend from the 2nd son of Root_CDPA. The situation provides Y-SNP data that can allow us to identify Multigen_SNPs that cannot be assigned to a specific progenitor in the pedigree (FT1000, FT10011, FT1003, and FT1004), and one SNP-Progen (FT1002) in the example provided. Based on the information shown in Figure 1, everyone in the phylogenetic pathway will have their Y-SNP ancestral path written, as shown in Table 2.

Table 2.
Y-SNP ancestral path for the individuals shown in Figure 1 (* = BigY700 test taker).
2.3. Analysis of Y-DNA Haplogroup Diversity Among FTDNA Test Takers with the Surname Hay
To investigate the degree of Y-DNA haplogroup diversity among individuals with the surname Hay, this study utilised the Y-DNA data of 109 test takers that are publicly available on the Hay surname FTDNA project (FTDNA 2025b). In situations where a test taker has not carried out any specific Y-SNP testing and they have only tested Y-STR markers, FTDNA only provides a very general Y-DNA haplogroup prediction. The STR data of the test takers that had tested between 67 and 111 STR markers (n = 29) were inputted into the NEVGEN tool that was developed by Gentula and Nevski (2015) to predict a more refined Y-DNA haplogroup, as implemented by Zhabagin et al. (2024). Once all the data were collated, test takers were grouped based on the closest major Y-DNA haplogroup they were associated with, so that Y-DNA haplogroup diversity among Hay test takers at FTDNA could be evaluated.
3. Results
3.1. Y-DNA Haplogroup Diversity Among Test Takers with the Surname Hay (Plus Spelling Variants)
The first set of results demonstrates the Y-DNA haplogroup diversity observed among the test takers with the surname Hay (plus spelling variants) that are participating in the Hay FTDNA project. Figure 2 shows that Y-DNA haplogroups R1a, R1b, I1, I2, J1, and Q were all represented.
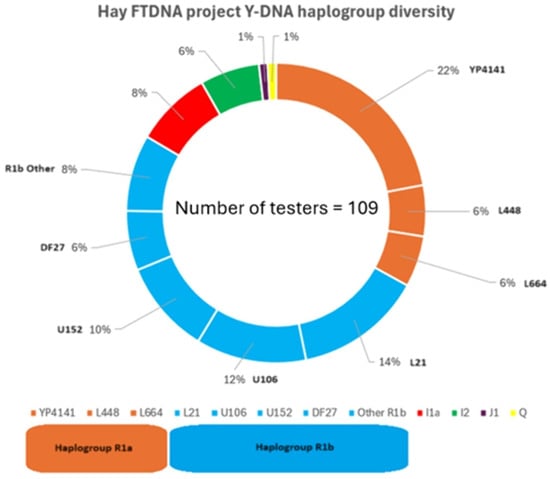
Figure 2.
Y-DNA haplogroup diversity among test takers with the surname Hay (plus associated spelling variants) within the Hay FTDNA project.
The evidence also demonstrates significant diversity within specific haplogroups (e.g., R1a > YP4141; R1a > L448 and R1a > L664) with haplogroup R1b proving to be the most diverse haplogroup represented. The Y-DNA diversity observed in this study is similar to previous research completed on Clan Forbes (Stead 2023).
However, several of the included spelling variants (Haye, Hayes, Hey, Haya) have independent origins; their presence within the dataset almost certainly contributes to the observed haplogroup diversity. Consequently, these data should be interpreted as representing the diversity among all test takers using the Hay-type surnames at FTDNA, rather than as evidence of genetic heterogeneity within the noble lineage itself.
3.2. The Y-DNA Results of the Three Well-Documented Noble Hay Descendants
The BigY700 test results of the three well-documented noble Hay descendants showed that they all shared the following Y-SNP ancestral path (see Table 3): R1a-M420 > YP4141 > YP4132 > YP4169 > YP4208 > YP4131 > YP4138 > YP6500 > FTT161.

Table 3.
Basic Y-SNP ancestral path and specific Y-SNP details.
Bonito et al. (2021) suggested that the palindromic regions of the Y-chromosome are highly prone to gene conversion events; therefore, the identification of SNPs may be complex when performing short-read targeted NGS. Hallast et al. (2023) identifies the four heterochromatic subregions in the human Y-chromosome—the (peri-)centromeric region, DYZ18, DYZ19, and Yq12—as being highly repetitive; therefore, these regions are less suitable for the identification of reliable Y-SNPs using NGS. Figure 3 is a map of the Y-chromosome that identifies the positions of the Y-SNPs shown in Table 3. These Y-SNPs are located within the non-recombining portion of the Y-chromosome (NRY), where Y-SNPs occur at a relatively slow rate and without the instability caused by gene conversion, deletions, and duplications. Furthermore, these Y-SNPs are not located in any of the regions that are deemed unreliable for Y-SNP calling; therefore, these Y-SNPs are regarded to be highly stable over generations, making them reliable for phylogenetic studies.
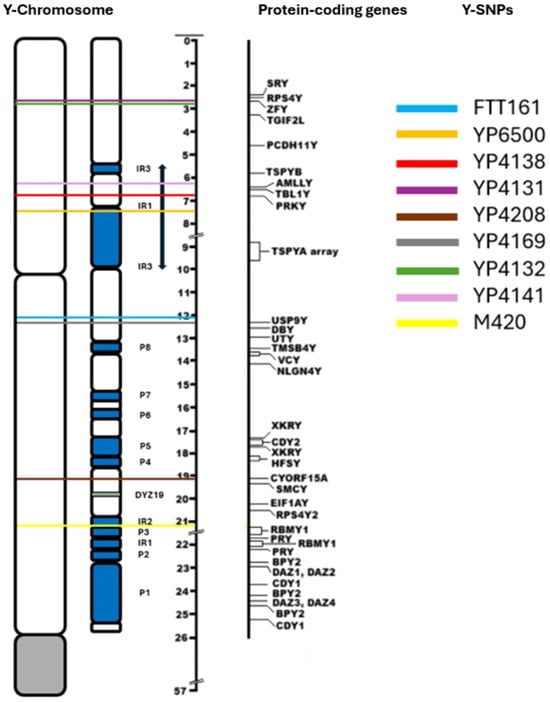
Figure 3.
Y-chromosome map adapted from Jobling and Tyler-Smith (2003), showing Y-SNPs associated with the noble Hay lineage.
The sequences in Table 4 show the bases surrounding the Y-SNPs associated with the ancestral path for the noble Hay lineage. It can be observed that each derived Y-SNP is surrounded by non-repetitive sequences; therefore, this increases the likelihood that these Y-SNPs are stable and phylogenetically relevant.

Table 4.
Sequencing data of the bases surrounding the Y-SNPs identified in the Y-SNP pathway for the Hay noble lineage.
The first line in each row of Table 4 shows the 17 bases of sequence surrounding either side of each Y-SNP found in the Y-SNP pathway for the Hay noble lineage. The sequences are in the Genome Reference Consortium Human Build 38 (GRCh38) format. The 18th base in each sequence is boxed using square brackets to indicate the location of the specific SNP (e.g., FTT161 = [A] ancestral allele; [G] derived allele).
The Y-SNP FTT162 is phylogenetically positioned downstream of FTT161, and it defines a noble Hay branch that has a significant number of descendants: n = 47 test takers at the YFull database (YFull 2025). This Y-SNP is in the highly repetitive heterochromatic region of the Y-chromosome (see Figure 4). This region is regarded as having large portions of noncoding DNA that are typically prone to mutations caused by replication slippage (Lemos et al. 2010; Lovett et al. 1993; Viguera et al. 2001); therefore, Y-SNPs occurring within this region of the Y-chromosome need to be treated with caution, as they may not be phylogenetically relevant (Hallast et al. 2015).
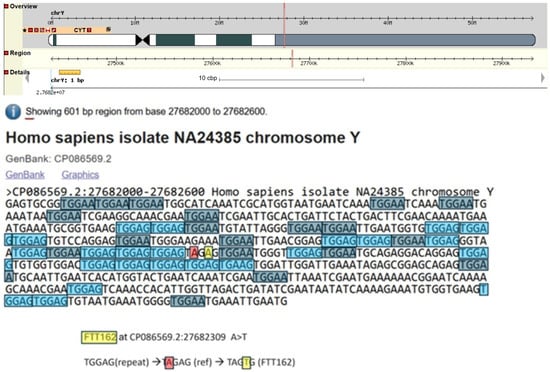
Figure 4.
Y-SNP FTT162: location: CP086569.2:27682309: A > T and surrounding sequence (ISOGG 2024; NBCI 2025).
Since FTT162 is in a less stable region of the Y-chromosome, we analysed the surrounding sequence on either side of this SNP to assess its stability and the reliability of being phylogenetically relevant. Figure 4 highlights the repetitive sequences across a 600 bp region, with FTT162 in the middle of the sequence and annotated in yellow. The way that FTT162 is likely to have formed is by a change from the TGGAG repeat to TAGAG (TGGAG → TAGAG; G > A; annotated in red), and this terminated the TGGAG repeated sequence that resulted in stabilising this section of the sequence. After the TAGAG section had changed, the resulting stability provided a unique background for the FTT162 SNP (A > T; annotated in yellow) to occur. When FTT162 occurred, it changed the dynamics of the surrounding repeats so that two out of five bases were different to the left and to the right of the repeat units, either side of FTT162. Therefore, these changes would have increased the stability at FTT162, making it an unlikely place for the DNA polymerase to slip, thus restricting any conversion to either of the neighbouring repeats: TGGAG or TGGAA, respectively. For the reasons discussed above, we have no concerns about the stability and reliability of this Y-SNP.
The three well-documented Hay test takers also match a significant number of test takers (n = 24/109 at the Hay FTDNA project) with the surname Hay (plus associated spelling variants), who also tested positive for at least FTT161 (FTDNA 2025b). At this point, it is unclear whether YP6500 (plus equivalent SNPs BY33394/FT2017) are Y-SNPs specific to the Hay noble lineage or whether they occurred in an ancestor that predates William de la HAYA, I of Normandy. Furthermore, the Y-SNPs BY33394 and FT2017 are assumed to be equivalent to YP6500, as it is unclear whether they appeared simultaneously in the evolutionary timeline or whether their occurrences were totally independent of each other. New data often change the status of assumed equivalent Y-SNPs by proving that they occurred independently of other Y-SNPs. Therefore, it is never a guarantee that any Y-SNP is equivalent to other Y-SNPs. Once new data provide enough evidence to show that assumed equivalent Y-SNPs did occur independently, their position within the phylogenetic tree changes accordingly to represent their correct chronological order. The results show that there are some non-Hay surnames among the test takers (n = 12) who are positive for FTT161 and downstream SNPs; these non-Hay surnames are likely the result of an EPP taking place. The results strongly suggest that FTT161 is a Y-SNP that defines the noble Hay lineage, with the downstream Y-SNPs of FTT161 identifying descent from William II de la HAYA, 1st of Erroll (d. 1201). Most test takers with the surname Hay on the Hay FTDNA project (n = 85/109) are not direct paternal descendants of the Hay noble progenitor (FTDNA 2025b). Figure 5 shows the phylogenetic block-tree of R1a-YP4138, which splits to form the subclades YP6500 (ancestor of the Hay noble lineage) and three parallel subclades, CTS11317, BY190800, and BY234055, respectively. The subclades CTS11317, BY190800, and BY234055 have several test takers with surnames of Norman origin (Manwarren/Manering and Travers) (King 1874), and several test takers with surnames of English origin. The non-Hay surnames associated with the parallel subclades of YP6500 are expected, since the age estimate for R1a > YP4138 is 582 < 800 > 1027 CE (95% probability) (FTDNA 2025a); this is a timeframe generally accepted as being before the earliest known adoption of surnames in Europe (Petersen et al. 2022).
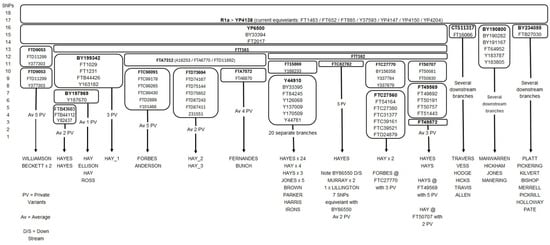
Figure 5.
R1a > YP4138 phylogenetic block-tree showing the Y-SNPs associated with the descendants of the Hay noble progenitor YP6500.
3.3. Combining Hay Y-SNPs with Well-Documented Pedigrees to Create the Most Likely Noble Phylogenetic Tree
Figure 6 was produced by combining the Y-SNP data with the ancestral pedigrees of the three well-documented Hay test takers. This process allowed us to assign specific Multigen-SNPs to a refined number of Hay ancestors.
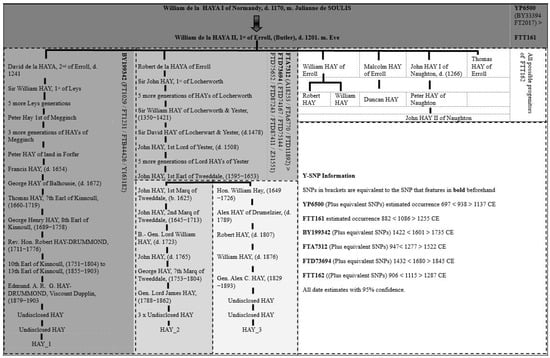
Figure 6.
Original Hay noble phylogenetic tree, using the pedigrees of the three well-documented noble Hay descendants.
The phylogenetic tree identifies several Multigen-SNPs associated with the noble Hay lineage. Test takers HAY_1, HAY_2, and HAY_3 are the three well-documented individuals who were identified during the traditional genealogical research phase of the study. The other test takers that match the three well-documented Hay individuals can also be placed in approximate positions within the Hay noble phylogenetic tree, based on their Y-SNPs; however, further documentary evidence or the testing of more well-documented descendants will be required for a more accurate positioning. Three clear subclades that are phylogenetically downstream of the Y-SNP FTT161 were identified in this study (BY199342, FTA7312, and FTT162). The combined Y-SNP and ancestral documentation data suggest lines of descent from three of the six sons of William II de la HAYA, 1st of Erroll. The Y-SNP BY199342 (plus equivalents) that were found in test taker HAY_1 can confidently be assigned to the descendants of David de la HAYA, 2nd of Erroll (d. 1241 CE), first son of William II de la HAYA, 1st of Erroll (d. 1201 CE). However, it is uncertain whether BY199342 occurred in David de la HAYA, 2nd of Erroll, or in one of his immediate direct descendants. Furthermore, HAY_2 and HAY_3 are negative for BY199342 (plus, equivalents) and positive for FTA7312 (plus equivalents), while likely descending from Robert de la HAYA of Erroll (the younger brother of David de la HAYA, 2nd of Erroll). The Y-SNP data and the pedigrees of the well-documented test takers show a clear genetic divergence that was previously hypothesised. Moreover, the results support the documentary evidence that suggests that HAY_1, HAY_2 and HAY_3 are descendants of William II de la HAYA, 1st of Erroll with a high degree of certainty.
3.4. An Alternative Hay Noble Phylogenetic Tree Based on the Addition of HAY_4
The observation of variation in mutation rates during specific time periods could explain a proposed documented pedigree for an existing FTDNA test taker with the surname HAY, who is positive for the subclade FTT162 (referred to as HAY_4 in this report). The documentary evidence for HAY_4 suggests possible descent from Robert de la HAYA of Erroll, younger brother of David de la HAYA, 2nd of Erroll. This proposed ancestry would need to span approximately two hundred years, totalling eight generations between William de la HAYA II, 1st of Erroll and Sir William HAY of Locherworth and Yester, Sheriff of Peebles (1350–1421 CE), which can be seen in Figure 7.
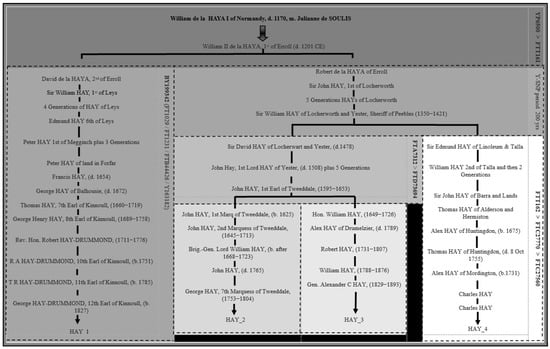
Figure 7.
Alternative Hay noble phylogenetic tree when HAY-4′s proposed pedigree is considered.
Although the alternative Hay phylogenetic tree proposed in Figure 7 is possible on a genetic level, it is important to highlight that there are alternative ancestral pedigrees for HAY_4, but these cannot be documented back to William II de la HAYA, 1st of Erroll (d. 1201) due to a lack of records. To validate HAY_4’s true line of descent back to William II de la HAYA, 1st of Erroll, the Y-DNA testing of a well-documented direct paternal descendant of the four remaining sons of William II de la HAYA, 1st of Erroll (William, Malcolm, Thomas and John HAY I of Naughton) would be required.
4. Discussion
The Y-DNA haplogroup diversity observed among the Hay FTDNA project test takers demonstrates a lack of homogeneity among the FTDNA test takers with the surname Hay (plus associated spelling variants). Only 22% of the FTDNA test takers with the surname Hay descend from a Hay nobleman, defined by the Y-SNPs YP6500 and FTT161 (plus subclades). A similar percentage of Y-DNA haplogroup diversity was also observed in a Clan Forbes genetic study, where Stead (2023) reported that 27% of FTDNA test takers within the Clan Forbes project were shown to be direct paternal descendants of the noble lineage. Furthermore, the results also support the hypothesis made by Durie (2022), who suggested that surnames in Scotland were largely adopted for many reasons, and that a surname of noble origin does not mean an individual is descended from the noble progenitor of that surname. More research needs to be conducted on other surnames associated with Scottish nobility for comparison to this study and the Clan Forbes genetic study (Stead 2023). With further data, more specific questions can be asked; for example, are there specific historical events that facilitated surname changes, and can this be evidenced in Y-DNA data?
A key clarification arising from this study is that the surname Hay is polygenetic. Multiple etymological pathways to Norman, English, Irish, and continental European origins are possible for producing the same or similar surname independently. Only one of these branches, the Scottish lineage of Hay of Erroll, can be associated with documented nobility. Our findings demonstrate that only 22% of modern Hay test takers share descent from this lineage; therefore, this is consistent with the expectation for a polygenetic surname. The results of this study reinforce that a ‘surname associated with a noble origin’ does not imply noble descent for all bearers. This clarification strengthens, rather than weakens, the interpretation of the Y-DNA results.
The Y-SNP YP4138, plus equivalents, are directly upstream of YP6500 in the phylogenetic tree. Subclades of YP4138 are found among several test takers with the Norman-in-origin surnames Manwarren and Travers, therefore, strongly supporting the Norman origins of YP4138 and its subclades. The age estimate for YP4138 is 582 < 800 > 1027 CE (95% probability) (FTDNA 2025a), which also aligns with the Norman origin hypothesis. Some test takers with the surname Travers have well-documented ancestry from the Travers noble lineage of Horton, Cheshire (Travers 1864) and Cork, Ireland (Lundy 2019b). Although the Travers lineage of Horton and Cork can only be confidently documented back to the mid-15th century, they could be descended from the Norman ancestor of Ralph Travers of Berney, c. 1197 (Lundy 2019b; Travers 1864). Further testing of well-documented individuals with surnames of Norman origin that match the noble Hay and Travers test takers could provide important Y-SNP data to help shed further light on their ancestral connections.
The research conducted in this study has provided strong evidence that the Multigen-SNPs YP6500 (plus equivalents) and FTT161 are key Y-SNPs that identify descent from the Hay noble lineage. Furthermore, two subclades of FTT161 (BY199342 and FTA7312) are Multigen-SNPs that indicate direct paternal descent from David de la HAYA, 2nd of Erroll and his younger brother Robert de la HAYA of Erroll. Any individual that matches these key Y-SNPs can be confirmed as a direct paternal descendant of the Hay noble lineage with a high degree of confidence, even if documentary evidence is absent. Furthermore, the Y-SNP data can also provide more specific ancestral designations (e.g., descent from David de la HAYA, 2nd of Erroll or his younger brother Robert). This dataset will become more refined when additional well-documented Hay test takers complete BigY700 testing, leading to more Multigen_SNPs being assigned to specific Hay noblemen, and potentially identifying specific SNP-Progens.
The technological and methodological advances made by the telomere-to-telomere (T2T) consortium have already had a huge impact on identifying new Y-SNPs that were not reported previously (Rhie et al. 2023). Although the BigY700 has identified several T2T Y-SNPs (e.g., FTT161 and FTT162) within the noble Hay phylogenetic tree (ISOGG 2024), these Y-SNPs did not become recognised until T2T research was made available to researchers. Therefore, it is recommended that several noble Hay test takers are eventually sequenced using the long-read sequencing methodology implemented by the T2T consortium, as the current research suggests that a higher resolution of the Y-chromosome can be achieved (Jain et al. 2022; Jain et al. 2018; Rhie et al. 2023). Furthermore, long-read sequencing will likely develop the noble Hay phylogenetic tree by identifying new Y-SNPs, resulting in more accurate Y-SNP dating.
To the best of our knowledge, this study is the first to identify Y-chromosomal Multigen_SNPs that are associated with a well-documented noble lineage from Scotland that diverges from a common direct paternal ancestor in the 12th century. This study is also the first to provide genetic evidence that supports the Anglo-Norman migration into Scotland during the reign of David I of Scotland. This study demonstrates that integrating Y-SNP data from test takers with well-documented noble lineages and traditional genealogical research can provide valuable evidence for addressing uncertainties surrounding theories of ancient origins. The results also offer new opportunities for men bearing the surname Hay (and its variants) who lack documentary evidence to evaluate potential descent from the noble Hay lineage. However, the observed Y-DNA haplogroup diversity among individuals with the Hay surname with ancestral links to Scotland suggests that such descent is less probable than is often assumed.
Author Contributions
Conceptualization, P.S.; methodology, P.S.; software, not applicable.; validation, P.S.; formal analysis, P.S.; investigation, P.S.; resources, P.S.; data curation, P.S.; writing—original draft preparation, P.S.; writing—review and editing, P.R.H. and A.F.M.; visualization, P.S.; supervision, P.R.H. and A.F.M.; project administration, P.S., P.R.H. and A.F.M.; funding acquisition, not applicable. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee of the University of Strathclyde, Glasgow, Scotland (protocol code: UEC23/94 on 5 February 2024).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The datasets presented in this article are not readily available because the genetic data contains private variants which could be used to identify participants. It is essential that participant identity remains anonymous for privacy reasons.
Acknowledgments
We gratefully acknowledge all sample donors who participated in this study. Thanks to Michael Travers, David Jones, Alan Hay, and the three well-documented Hay test takers for their support. Thank you to Tunde Huszar and Graham S Holton for their support and expert input.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| CE | Common Era |
| Y-SNP | Y-chromosome Single Nucleotide Polymorphism |
| Y-DNA | Y-chromosome Deoxyribonucleic Acid |
| NPE | Non-Paternity Event |
| FPE | False Paternal Event |
| EPP | Extra Pair Paternity |
| FTDNA | Family Tree DNA |
| IBD | Identity-By-Descent |
| MultiGen_SNP | Multigenerational Single Nucleotide Polymorphism |
| SNP_Progen | Single Nucleotide Polymorphism Progenitor |
| T2T | Telomere-to-Telomere Genome |
References
- Avni, Chen, Dana Sinai, Uri Blasbalg, and Paz Toren. 2023. Discovering your presumed father is not your biological father: Psychiatric ramifications of independently uncovered non-paternity events resulting from direct-to-consumer DNA testing. Psychiatry Research 323: 115142. [Google Scholar] [CrossRef] [PubMed]
- Barron, Evan Macleod. 1997. The Scottish War of Independence. New York: Barnes & Noble Books. [Google Scholar]
- Barrow, Geoffrey, and Wallis Steuart. 1973. The Kingdom of the Scots: Government, Church and Society from the Eleventh to the Fourteenth Century. London: Edward Arnold. [Google Scholar]
- Barrow, Geoffrey, and Wallis Steuart. 1985. David I of Scotland (1124–1153): The Balance of New and Old. Reading: University of Reading. [Google Scholar]
- Black, George F. 1946. The Surnames of Scotland: Their Origin, Meaning, and History. New York: New York Public Library. [Google Scholar]
- Bonito, Maria, Eugenia D’Atanasio, Francesco Ravasini, Selene Cariati, Andrea Finocchio, Andrea Novelletto, Beniamino Trombetta, and Fulvio Cruciani. 2021. New insights into the evolution of human Y chromosome palindromes through mutation and gene conversion. Human Molecular Genetics 30: 2272–85. [Google Scholar] [CrossRef] [PubMed]
- Broun, Dauvit. 1997. The Birth of Scottish History. The Scottish Historical Review 76: 4–22. [Google Scholar] [CrossRef]
- Broun, Dauvit. 2015. Britain and the beginning of Scotland. Journal of the British Academy 3: 107–37. [Google Scholar] [CrossRef]
- Broun, Dauvit, Alice Taylor, Matthew Hammond, Roibeard Ó. Maolalaigh, Keith J. Stringer, Bradley John, David Carpenter, Amanda Beam, John Reuben, Michele Pasin, and et al. 2007a. “David Hay, lord of Errol. Document 3/276/4 (C.A. Rent., 337, no. 48).” People of Medieval Scotland 1093–1371. Available online: https://poms.ac.uk/record/factoid/53657/ (accessed on 11 February 2025).
- Broun, Dauvit, Alice Taylor, Matthew Hammond, Roibeard Ó. Maolalaigh, Keith J. Stringer, Bradley John, David Carpenter, Amanda Beam, John Reuben, Michele Pasin, and et al. 2007b. “Document 1/5/32 (RRS, i, no. 141).” People of Medieval Scotland 1093–1371. Available online: https://poms.ac.uk/record/factoid/18122/ (accessed on 11 February 2025).
- Broun, Dauvit, Alice Taylor, Matthew Hammond, Roibeard Ó. Maolalaigh, Keith J. Stringer, Bradley John, David Carpenter, Amanda Beam, John Reuben, Michele Pasin, and et al. 2007c. “Document 3/276/1 (St A. Lib., 313).” People of Medieval Scotland 1093–1371. Available online: https://poms.ac.uk/record/source/4610/ (accessed on 11 February 2025).
- Broun, Dauvit, Alice Taylor, Matthew Hammond, Roibeard Ó. Maolalaigh, Keith J. Stringer, Bradley John, David Carpenter, Amanda Beam, John Reuben, Michele Pasin, and et al. 2007d. “Document 3/540/1 (Reid, de Soulis, no. 1).” People of Medieval Scotland 1093–1371. Available online: https://poms.ac.uk/record/factoid/67772/ (accessed on 11 February 2025).
- Chen, Hao, Yan Lu, Dongsheng Lu, and Shuhua Xu. 2021. Y-LineageTracker: A high-throughput analysis framework for Y-chromosomal next-generation sequencing data. BMC Bioinformatics 22: 114. [Google Scholar] [CrossRef]
- Davis, Caleb, Michael Sager, Göran Runfeldt, Elliott Greenspan, Arjan Bormans, Bennett Greenspan, and Connie Bormans. 2019. The Science Behind FamilyTreeDNA’s Big Y-700 Test. Family Tree DNA Gene by Gene, Ltd. Available online: https://blog.familytreedna.com/wp-content/uploads/2018/06/big_y_700_white_paper_compressed.pdf (accessed on 11 February 2025).
- Denise Syndercombe Court. 2021. The Y chromosome and its use in forensic DNA analysis. Emerging Topics in Life Sciences 5: 427–41. [Google Scholar] [CrossRef]
- DePew, Kyle, Maurice Gleeson, and Bart Jaski. 2024. Tracing the Sons of Brión. The R1b-A259 Y-DNA Subclade and the Uí Briúin Dynasty of Connacht. Peritia 34: 9–45. [Google Scholar] [CrossRef]
- Douglas, Robert, and John Philip Wood. 1813. The Peerage of Scotland: Containing an Historical and Genealogical Account of the Nobility of That Kingdom, from Their Origin to the Present Generation. Collected from the Public Records. Edinburgh: G. Ramsay. [Google Scholar]
- Durie, Bruce. 2022. Clans, Families and Kinship Structures in Scotland—An Essay. Genealogy 6: 88. [Google Scholar] [CrossRef]
- Erzurumluoglu, A. Mesut, Denis Baird, Tom G. Richardson, Nicholas J. Timpson, and Santiago Rodriguez. 2018. Using Y-Chromosomal Haplogroups in Genetic Association Studies and Suggested Implications. Genes 9: 45. [Google Scholar] [CrossRef]
- Fournier, Romain, Zoi Tsangalidou, David Reich, and Pier Francesco Palamara. 2023. Haplotype-based inference of recent effective population size in modern and ancient DNA samples. Nature Communications 14: 7945. [Google Scholar] [CrossRef]
- FTDNA. 2024. Family Tree DNA Privacy Statement. Available online: https://www.familytreedna.com/legal/privacy-statement (accessed on 11 February 2025).
- FTDNA. 2025a. Discover Haplogroup R-YP4138. Available online: https://discover.familytreedna.com/y-dna/R-YP4138/story (accessed on 11 February 2025).
- FTDNA. 2025b. Hay FTDNA Project. Available online: https://www.familytreedna.com/public/hay?iframe=ydna-results-overview (accessed on 11 February 2025).
- FTDNA. 2025c. Public Haplotrees. Available online: https://www.familytreedna.com/public/y-dna-haplotree/A?srsltid=AfmBOorjw6jphtFCpF_WtYBi3xm3pDOnNjYYrfPjy5Qe0GlHrrZssNdd (accessed on 11 February 2025).
- Gentula, Milos Cetkovic, and Aco Nevski. 2015. NEVGEN Y-DNA Haplogroup Predictor. Available online: https://www.nevgen.org/ (accessed on 11 February 2025).
- Glynn, Claire L. 2022. Bridging Disciplines to Form a New One: The Emergence of Forensic Genetic Genealogy. Genes 13: 1381. [Google Scholar] [CrossRef]
- Gretzinger, Joscha, Duncan Sayer, Pierre Justeau, Eveline Altena, Maria Pala, Katharina Dulias, Ceiridwen J. Edwards, Susanne Jodoin, Laura Lacher, Susanna Sabin, and et al. 2022. The Anglo-Saxon migration and the formation of the early English gene pool. Nature 610: 112–19. [Google Scholar] [CrossRef]
- Greytak, Ellen M., CeCe Moore, and Steven L. Armentrout. 2019. Genetic genealogy for cold case and active investigations. Forensic Science International 299: 103–13. [Google Scholar] [CrossRef] [PubMed]
- Guan, Yongtao. 2014. Detecting structure of haplotypes and local ancestry. Genetics 196: 625–42. [Google Scholar] [CrossRef] [PubMed]
- Hallast, Pille, Chiara Batini, Daniel Zadik, Pierpaolo Maisano Delser, Jon H. Wetton, Eduardo Arroyo-Pardo, Gianpiero L. Cavalleri, Peter de Knijff, Giovanni Destro Bisol, Berit M. Dupuy, and et al. 2015. The Y-chromosome tree bursts into leaf: 13,000 high-confidence SNPs covering the majority of known clades. Molecular Biology and Evolution 32: 661–73. [Google Scholar] [CrossRef]
- Hallast, Pille, Peter Ebert, Mark Loftus, Feyza Yilmaz, Peter A. Audano, Glennis A. Logsdon, Marc J. Bonder, Weichen Zhou, Wolfram Höps, Kwondo Kim, and et al. 2023. Assembly of 43 human Y chromosomes reveals extensive complexity and variation. Nature 621: 355–64. [Google Scholar] [CrossRef]
- Hammond, Matthew. 2006. Ethnicity and the Writing of Medieval Scottish History. The Scottish Historical Review 85: 1–27. [Google Scholar] [CrossRef][Green Version]
- Hanks, Patrick. 2003. Dictionary of American Family Names: 3-Volume Set. New York: Oxford University Press, p. 146. [Google Scholar]
- Holton, Graham S. 2023. John Roy Stewart and his genealogical legacy. Journal of Genealogy and Family History 7: 5. [Google Scholar] [CrossRef]
- Holton, Graham S., and Alasdair F. Macdonald. 2020. Declaration of Arbroath Family History Project. Foundation for Medieval Genealogy. Available online: https://fmg.ac/publications/arbroath-bg (accessed on 11 February 2025).
- ISOGG. 2024. FTT SNP Index. Available online: https://isogg.org/wiki/FTT_SNP_index (accessed on 11 February 2025).
- Jain, Chirag, Arang Rhie, Nancy Hansen, Sergey Koren, and Adam M. Phillippy. 2022. Long-read mapping to repetitive reference sequences using Winnowmap2. Nature Methods 19: 705–10. [Google Scholar] [CrossRef]
- Jain, Miten, Sergey Koren, Karen H. Miga, Josh Quick, Arthur C. Rand, Thomas A. Sasani, John R. Tyson, Andrew D. Beggs, Alexander T. Dilthey, Ian T. Fiddes, and et al. 2018. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nature Biotechnology 36: 338–45. [Google Scholar] [CrossRef]
- Jobling, Mark A., and Chris Tyler-Smith. 2003. The human Y chromosome: An evolutionary marker comes of age. Nature Reviews Genetics 4: 598–612. [Google Scholar] [CrossRef] [PubMed]
- King, Henry S. 1874. The Norman People and Their Existing Descendants in the British Dominions and the United States of America. London: Henry S. King & Company. [Google Scholar]
- King, Turi E., and Mark A. Jobling. 2009a. Founders, drift, and infidelity: The relationship between Y chromosome diversity and patrilineal surnames. Molecular Biology and Evolution 26: 1093–102. [Google Scholar] [CrossRef]
- King, Turi E., and Mark A. Jobling. 2009b. What’s in a name? Y chromosomes, surnames and the genetic genealogy revolution. Trends in Genetics 25: 351–60. [Google Scholar] [CrossRef] [PubMed]
- King, Turi E., Gloria Gonzalez Fortes, Patricia Balaresque, Mark G. Thomas, David Balding, Pierpaolo Maisano Delser, Rita Neumann, Walther Parson, Michael Knapp, Susan Walsh, and et al. 2014. Identification of the remains of King Richard III. Nature Communications 5: 5631. [Google Scholar] [CrossRef] [PubMed]
- Kivisild, Toomas. 2017. The study of human Y chromosome variation through ancient DNA. Human Genetics 136: 529–46. [Google Scholar] [CrossRef]
- Klyosov, Anatole, and Igor L. Rozhanskii. 2012. Haplogroup R1a as the Proto Indo-Europeans and the Legendary Aryans as Witnessed by the DNA of Their Current Descendants. Advances in Anthropology 2: 1–13. [Google Scholar] [CrossRef]
- Lall, Gurdeep Matharu, Marteen H. D. Larmuseau, Jon H. Wetton, Chiara Batini, Pille Hallast, Tunde I. Huszar, Daniel Zadik, Sigurd Aase, Tina Baker, Patricia Balaresque, and et al. 2021. Subdividing Y-chromosome haplogroup R1a1 reveals Norse Viking dispersal lineages in Britain. European Journal of Human Genetics 29: 512–23. [Google Scholar] [CrossRef]
- Larmuseau, Marteen H. D., Philippe Delorme, Patrick Germain, Nancy Vanderheyden, Anja Gilissen, Anneleen Van Geystelen, Jean-Jacques Cassiman, and Ronny Decorte. 2014. Genetic genealogy reveals true Y haplogroup of House of Bourbon contradicting recent identification of the presumed remains of two French Kings. European Journal of Human Genetics 22: 681–87. [Google Scholar] [CrossRef]
- Larmuseau, Marteen H. D., Sofie Claerhout, Leen Gruyters, Kelly Nivelle, Michiel Vandenbosch, Anke Peeters, Pieter van den Berg, Tom Wenseleers, and Ronny Decorte. 2017. Genetic-genealogy approach reveals low rate of extrapair paternity in historical Dutch populations. American Journal of Human Biology 29: e23046. [Google Scholar] [CrossRef]
- Lemos, Bernardo, Alan T. Branco, and Daniel L. Hartl. 2010. Epigenetic effects of polymorphic Y chromosomes modulate chromatin components, immune response, and sexual conflict. Proceedings of the National Academy of Sciences USA 107: 15826–31. [Google Scholar] [CrossRef]
- Lovett, S. T., P. T. Drapkin, V. A. Sutera, Jr., and T. J. Gluckman-Peskind. 1993. A sister-strand exchange mechanism for recA-independent deletion of repeated DNA sequences in Escherichia coli. Genetics 135: 631–42. [Google Scholar] [CrossRef]
- Lundy, Darryl. 2019a. The Peerage.com. Major Malcolm Vivian Hay of Seaton. Available online: https://www.thepeerage.com/p3702.htm#i37015 (accessed on 11 February 2025).
- Lundy, Darryl. 2019b. The Peerage.com. Sir Robert Travers. Available online: https://www.thepeerage.com/p15106.htm#i151060 (accessed on 11 February 2025).
- Lynch, Michael. 1992. Scotland: A New History. London: Pimlico. [Google Scholar]
- McClure, Peter. 2015. English topographic surnames with fused Anglo Norman preposition and article: Myth or reality? Nomina 38: 33–69. [Google Scholar]
- McDonald, Iain. 2021. Improved Models of Coalescence Ages of Y-DNA Haplogroups. Genes 12: 862. [Google Scholar] [CrossRef] [PubMed]
- Moncreiffe of that Ilk Iain, and Jackson W. Armstrong. 2010. The Law of Succession: Origins and Background of the Law of Succession to Arms and Dignitaries in Scotland. Edinburgh: Edinburgh University Press. [Google Scholar]
- Morez, A., K. Britton, G. Noble, T. Gunther, A. Gotherstrom, R. Rodriguez-Varela, N. Kashuba, R. Martiniano, S. Talamo, N. J. Evans, and et al. 2023. Imputed genomes and haplotype-based analyses of the Picts of early medieval Scotland reveal fine-scale relatedness between Iron Age, early medieval and the modern people of the UK. PLoS Genetics 19: e1010360. [Google Scholar] [CrossRef] [PubMed]
- Nait Saada, Juba, Georgios Kalantzis, Derek Shyr, Fergus Cooper, Martin Robinson, Alexander Gusev, and Pier Francesco Palamara. 2020. Identity-by-descent detection across 487,409 British samples reveals fine scale population structure and ultra-rare variant associations. Nature Communications 11: 6130. [Google Scholar] [CrossRef]
- NBCI. 2025. Homo Sapiens Isolate NA24385 Chromosome Y GenBank: CP086569.2. Available online: https://www.ncbi.nlm.nih.gov/nuccore/CP086569.2?report=genbank (accessed on 11 February 2025).
- Ormrod, W. Mark, Joanna Story, and Elizabeth M. Tyler. 2020. Migrants in Medieval England, C. 500-c. 1500. Liverpool: Liverpool University Press. [Google Scholar]
- Paul, J. B. 1904. The Scots Peerage: Founded on Wood’s Edition of Sir Robert Douglas’s Peerage of Scotland; Containing an Historical and Genealogical Account of the Nobility of that Kingdom. Edinburgh: David Douglas. [Google Scholar]
- Petersen, Jakob, Jens Kandt, and Paul A. Longley. 2022. British surname origins, population structure and health outcomes-an observational study of hospital admissions. Scientific Reports 12: 2156. [Google Scholar] [CrossRef]
- Rhie, Arang, Sergey Nurk, Monika Cechova, Savannah J. Hoyt, Dylan J. Taylor, Nicolas Altemose, Paul W. Hook, Sergey Koren, Mikko Rautiainen, Ivan A. Alexandrov, and et al. 2023. The complete sequence of a human Y chromosome. Nature 621: 344–54. [Google Scholar] [CrossRef]
- Ritchie, Robert Lindsaey Graeme. 1954. The Normans in Scotland. Edinburgh: Edinburgh University Press. [Google Scholar]
- Shaw, Emma L., Debra J. Donnelly, Gideon Boadu, Rachel Burke, and Robert J. Parkes. 2024. DNA Testing and Identities in Family History Research. Genealogy 8: 75. [Google Scholar] [CrossRef]
- Smith, Albert Hugh. 1928. The Place-Names of the North Riding of Yorkshire. English Place-Name Society. London: Cambridge University Press, vol. V. [Google Scholar]
- Snow, Dean R. 2001. Scotland’s Irish Origins. Archaeology 54: 46–51. [Google Scholar]
- Sole-Morata, Neus., Jaume Bertranpetit, David Comas, and Francesc Calafell. 2015. Y-chromosome diversity in Catalan surname samples: Insights into surname origin and frequency. European Journal of Human Genetics 23: 1549–57. [Google Scholar] [CrossRef]
- Stead, Philip. 2023. Clan Forbes Family History Project. History Scotland 28: 29. [Google Scholar]
- The Institute for Name-Studies. 2025a. Key to English Place-Names. Available online: http://kepn.nottingham.ac.uk/map/place/Yorkshire%20NR/Richmond (accessed on 11 February 2025).
- The Institute for Name-Studies. 2025b. Key to English Place-Names: Barnard Castle. Available online: http://kepn.nottingham.ac.uk/map/county/Durham (accessed on 11 February 2025).
- Travers, Samuel Smith. 1864. A Collection of Pedigrees of the Family of Travers, Abstracts of Documents. Oxford: J. H. and J. Parker. [Google Scholar]
- Underhill, Peter A., G. David Poznik, Siiri Rootsi, Mari Jarve, Alice A. Lin, Jianbin Wang, Ben Passarelli, Jad Kanbar, Natalie M. Myres, Roy J. King, and et al. 2015. The phylogenetic and geographic structure of Y-chromosome haplogroup R1a. European Journal of Human Genetics 23: 124–31. [Google Scholar] [CrossRef]
- Viguera, Enrique, Danielle Canceill, and S. Dusko Ehrlich. 2001. Replication slippage involves DNA polymerase pausing and dissociation. Embo Journal 20: 2587–95. [Google Scholar] [CrossRef]
- Woolf, Alex. 2007. From Pictland to Alba, 789–1070. Edinburgh: Edinburgh University Press. [Google Scholar]
- YFull. 2025. Haplogroup YTree v13.02.00, R-Y515442(FTT162). Available online: https://www.yfull.com/tree/R-Y515442/ (accessed on 11 February 2025).
- Zhabagin, Maxat, Alizhan Bukayev, Zhanargul Dyussenova, Altyn Zhuraliyeva, Assel Tashkarayeva, Aigul Zhunussova, Baglan Aidarov, Akynkali Darmenov, Ainur Akilzhanova, Uli Schamiloglu, and et al. 2024. Y-Chromosomal insights into the paternal genealogy of the Kerey tribe have called into question their descent from the Stepfather of Genghis Khan. PLoS ONE 19: e0309080. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).