On The Potential of Image Moments for Medical Diagnosis
Abstract
1. Introduction
2. Related Works
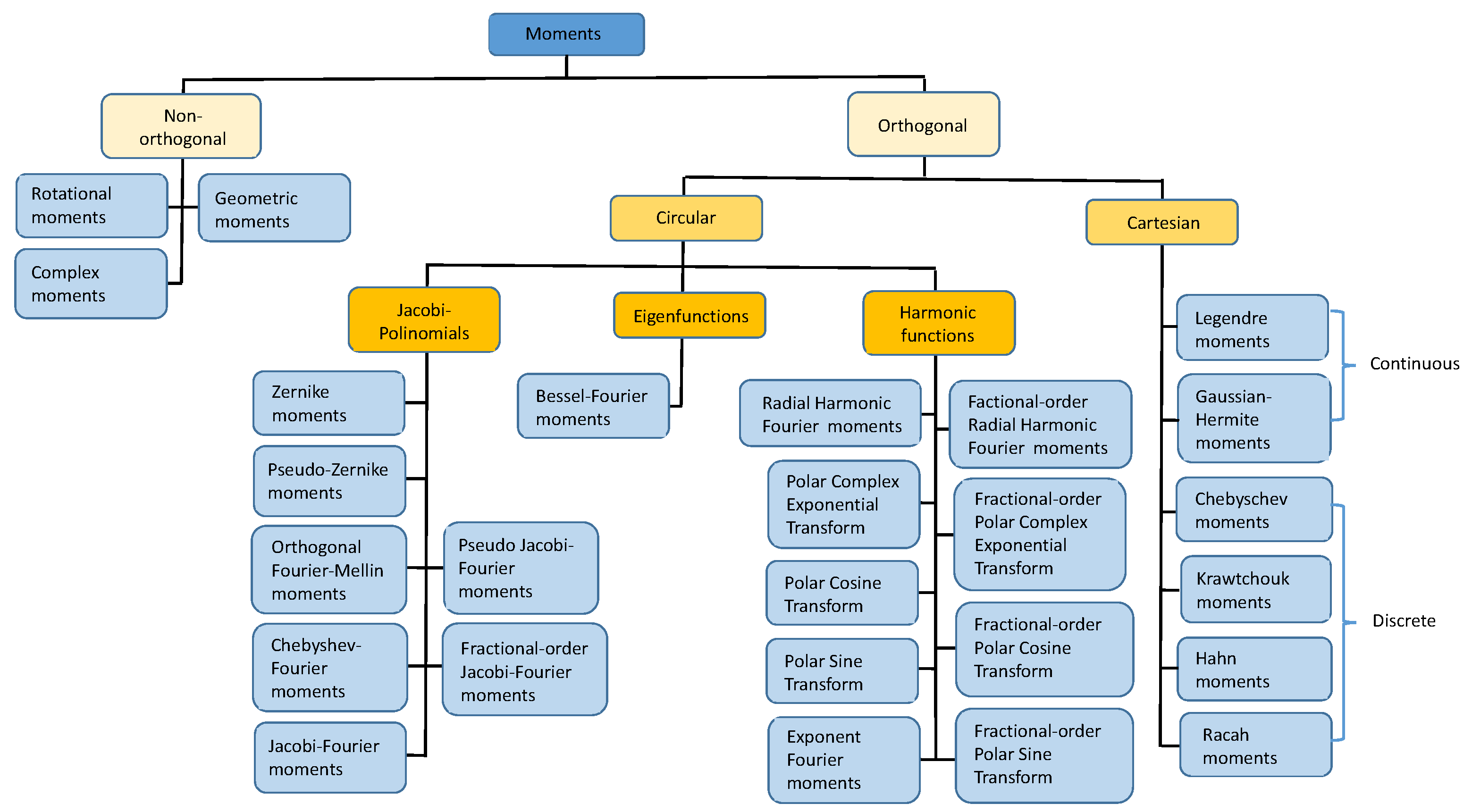
3. Overview of Moments
4. Medical Image Classification
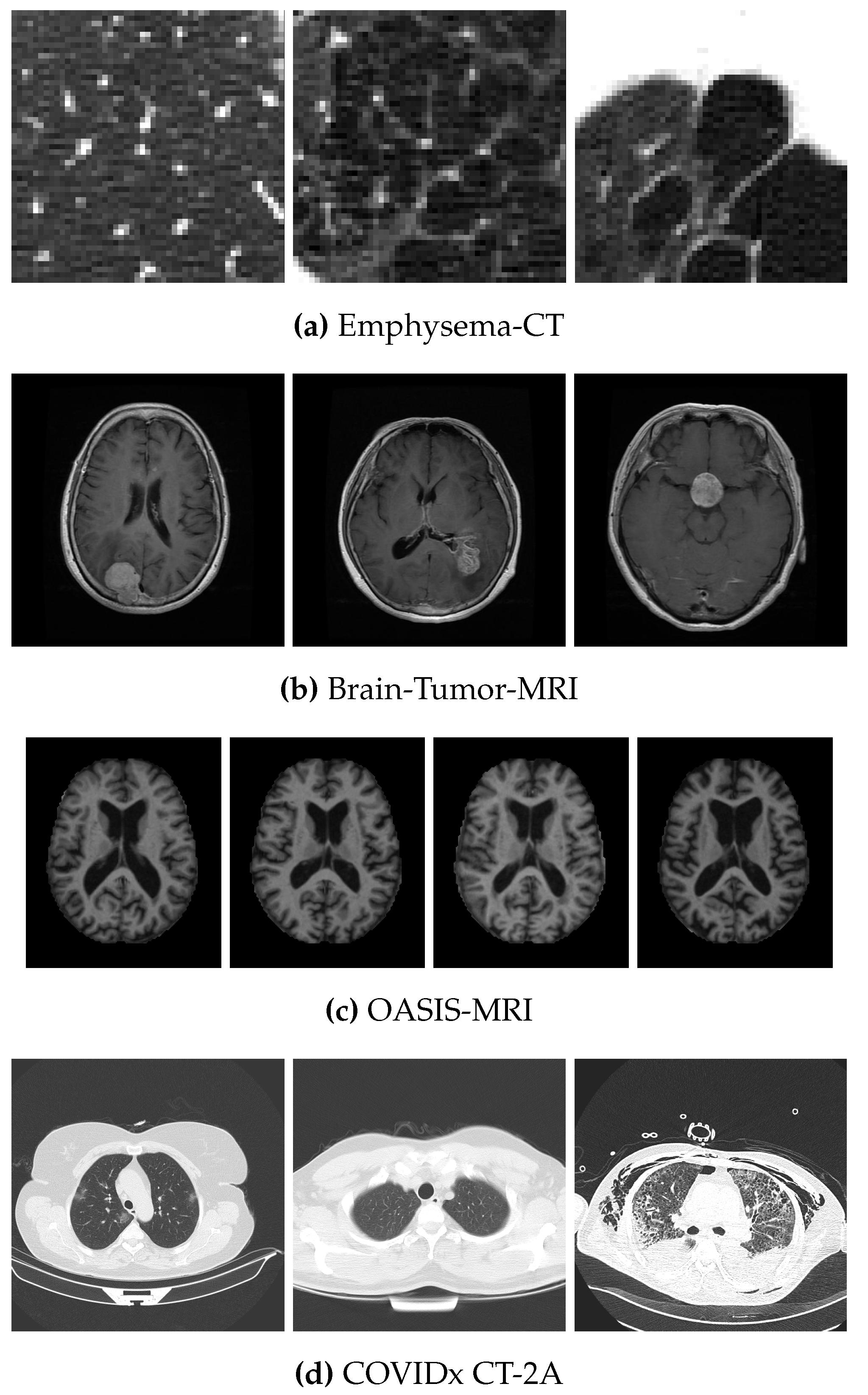
4.1. Data Sets
4.2. Methods
- Cartesian moments: continuous Legendre moments (LM), discrete Chebyshev moments of the first order (CHM), discrete Chebyshev moments of the second order (CH2M).
- Circular Jacobi polynomials based moments: Zernike moments (ZM), pseudo-Zernike moments (PZM), orthogonal Fourier–Mellin moments (OFMM), Chebyshev–Fourier moments (CHFM), pseudo-Jacobi–Fourier moments (PJFM), Jacobi–Fourier moments (JFM), fractional-order Jacobi–Fourier moments (FrJFM).
- Circular eigenfunction-based moments: Bessel–Fourier moments (BFM).
- Circular harmonic-function-based moments: radial harmonic Fourier moments (RHFM), exponent Fourier moments (EFM), polar complex exponential transform (PCET), polar cosine transform (PCT), polar sine transform (PST), fractional-order radial harmonic Fourier moments (FrRHFM), fractional-order polar complex exponential transform (FrPCET), fractional-order polar cosine transform (FrPCT), fractional-order polar sine transform (FrPST).
5. Experimental Results
5.1. Experimental Setup
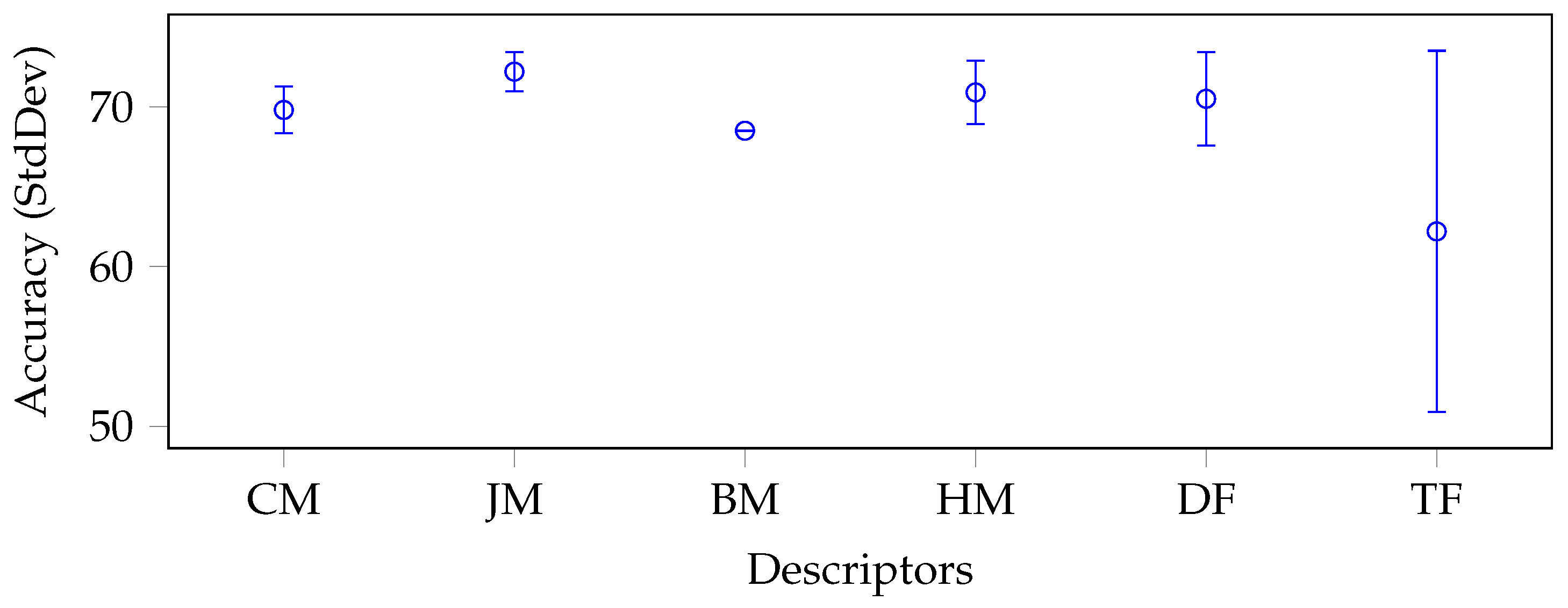
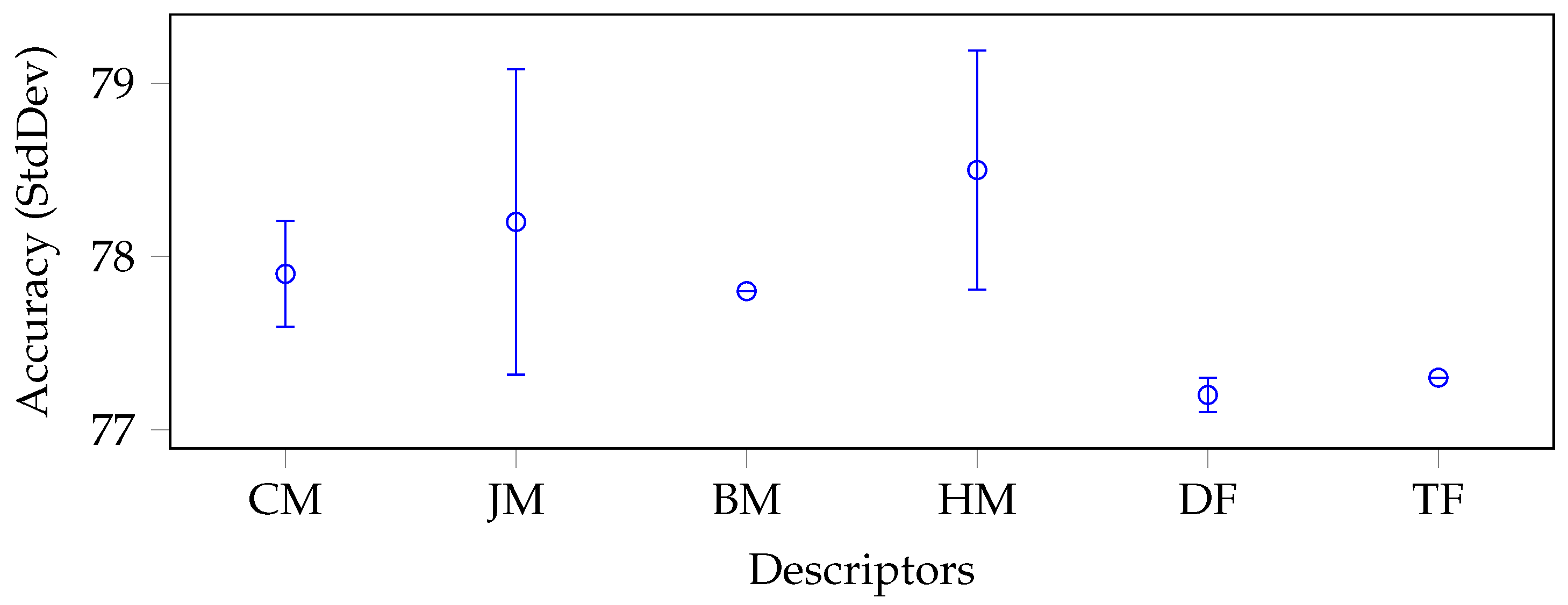
5.2. Results
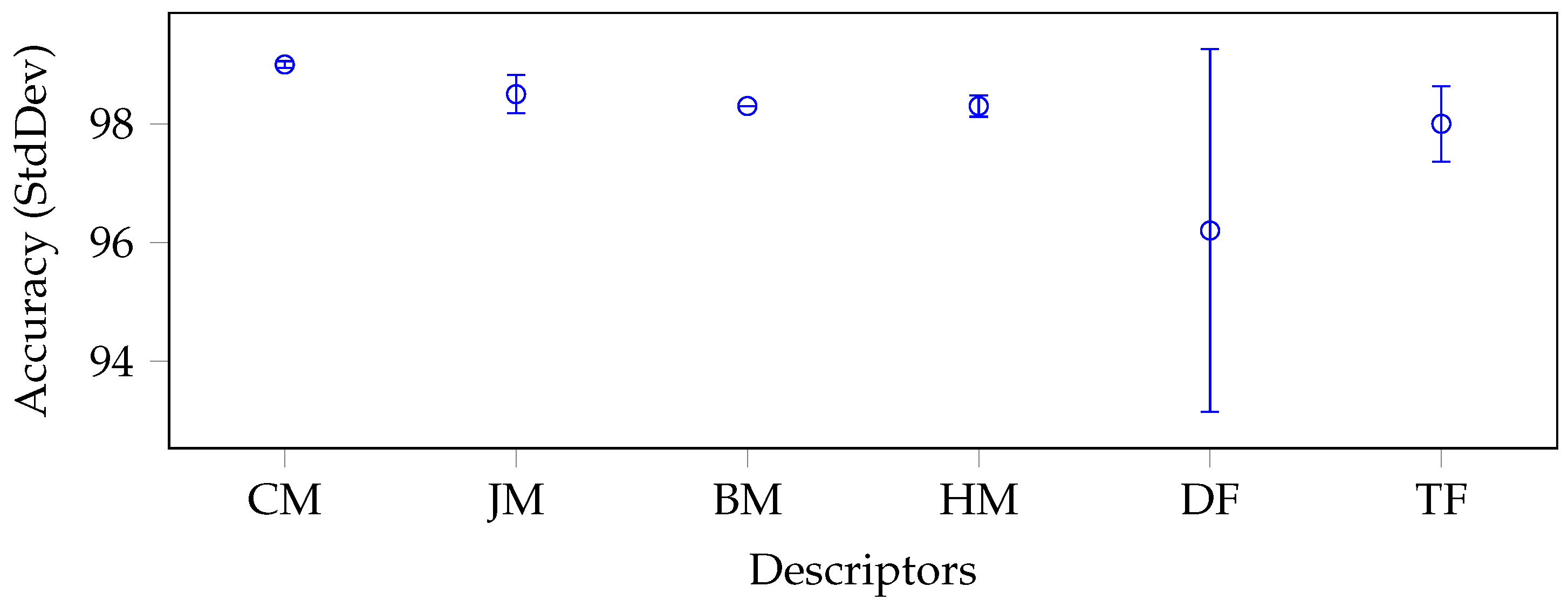
5.3. Discussion
- The moments offered comparable performance to deep features, despite being represented by order of magnitude fewer features than deep features.
- The moments provided extremely robust results, with a reduced standard deviation per category in all the experiments performed.
- Cartesian and harmonic moments were behaved the best, both on average and in terms of the number of absolute best results they offered in the different experiments. They were also the most stable, with low standard deviations.
- There was no tendency for the moments to improve over the years; as mentioned in the previous point, the Cartesian moments performed best among the older ones. Moreover, the use of the fractional domain, one of the most recent innovations in the field, did not bring any significant advantage, as can be seen by comparing against the equivalent moments in the integer domain.
- The naive Bayes classifier was not suitable for the task under consideration, especially when trained by any category of moments, since the assumption that the features were linearly independent was too strong for this type of descriptors.
- Although moments allowed high accuracies for the SVM, k-NN and BT classifiers, the classifier that benefited the most from the moments was the k-NN classifier. It achieved significant absolute best results in all data sets except COVIDx CT-2A, where it had a marginal difference with the absolute top performer, the SVM.
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| CAD | Computer-aided diagnostic |
| CNN | Convolutional neural network |
| VGGNet | Very deep convolutional networks |
| LBP | Local binary pattern |
| CT | Computed tomography |
| MR | Magnetic resonance |
| LM | Continuous Legendre moments |
| CHM | Discrete Chebyshev moments |
| CH2M | Discrete Chebyshev moments of the second order |
| ZM | Zernike moments |
| PZM | Pseudo-Zernike moments |
| OFMM | Orthogonal Fourier–Mellin moments |
| CHFM | Chebyshev–Fourier moments |
| PJFM | Pseudo-Jacobi–Fourier moments |
| JFM | Jacobi–Fourier moments |
| FrJFM | Fractional-order Jacobi–Fourier moments |
| BFM | Bessel–Fourier moments |
| RHFM | Radial harmonic Fourier moments |
| EFM | Exponent Fourier moments |
| PCET | Polar complex exponential transform |
| PCT | Polar cosine transform |
| PST | Polar sine transform |
| FrRHFM | Fractional-order radial harmonic Fourier moments |
| FrPCET | Fractional-order polar complex exponential transform |
| FrPCT | Fractional-order polar cosine transform |
| FrPST | Fractional-order polar sine transform |
| GLCM | Grey-level co-occurrence matrix |
| SVM | Support vector machine |
| k-NN | k-nearest neighbour |
| DT | Decision trees |
| NB | Naive Bayes |
| BT | Bagged trees |
| TP | True positive |
| FP | False positive |
| TN | True negative |
| FN | False negative |
References
- Bhattacharjee, S.; Mukherjee, J.; Nag, S.; Maitra, I.K.; Bandyopadhyay, S.K. Review on Histopathological Slide Analysis using Digital Microscopy. Int. J. Adv. Sci. Technol. 2014, 62, 65–96. [Google Scholar] [CrossRef]
- Kothari, S.; Phan, J.H.; Young, A.N.; Wang, M.D. Histological image classification using biologically interpretable shape-based features. BMC Med. Imaging 2013, 13, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Kowal, M.; Filipczuk, P.; Obuchowicz, A.; Korbicz, J.; Monczak, R. Computer-aided diagnosis of breast cancer based on fine needle biopsy microscopic images. Comput. Biol. Med. 2013, 43, 1563–1572. [Google Scholar] [CrossRef]
- Rodríguez, J.H.; Javier, F.; Fraile, C.; Conde, M.J.R.; Llorente, P.L.G. Computer aided detection and diagnosis in medical imaging: A review of clinical and educational applications. In Proceedings of the TEEM ’16: Fourth International Conference on Technological Ecosystems for Enhancing Multiculturality, Salamanca, Spain, 2–4 November 2016; pp. 517–524. [Google Scholar]
- Yanase, J.; Triantaphyllou, E. A systematic survey of computer-aided diagnosis in medicine: Past and present development. Expert Syst. Appl. 2019, 138, 112821. [Google Scholar] [CrossRef]
- Di Ruberto, C.; Fodde, G.; Putzu, L. Comparison of Statistical Features for Medical Colour Image Classification. In International Conference ICVS on Computer Vision Systems; Lecture Notes in Computer Science; Springer International Publishing: Berlin/Heidelberg, Germany, 2015; Volume 9163, pp. 3–13. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. In Proceedings of the 25th International Conference on Neural Information Processing Systems, Lake Tahoe, Nevada, 3–6 December 2012; Volume 1, pp. 1097–1105. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. In Proceedings of the 3rd International Conference on Learning Representations, ICLR 2015, Conference Track Proceedings, San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- Yandex, A.B.; Lempitsky, V. Aggregating Local Deep Features for Image Retrieval. In Proceedings of the 2015 IEEE International Conference on Computer Vision (ICCV), Washington, DC, USA, 7–13 December 2015; pp. 1269–1277. [Google Scholar]
- Kim, W.; Goyal, B.; Chawla, K.; Lee, J.; Kwon, K. Attention-Based Ensemble for Deep Metric Learning. In Proceedings of the Computer Vision—ECCV 2018, Munich, Germany, 8–14 September 2018; pp. 760–777. [Google Scholar]
- Loddo, A.; Putzu, L. On the Reliability of CNNs in Clinical Practice: A Computer-Aided Diagnosis System Case Study. Appl. Sci. 2022, 12, 3269. [Google Scholar] [CrossRef]
- Gelzinis, A.; Verikas, A.; Bacauskiene, M. Increasing the discrimination power of the co-occurrence matrix-based features. Pattern Recognit. 2007, 40, 2367–2372. [Google Scholar] [CrossRef]
- Gong, D.; Zhang, Y.; Jiang, J.; Liu, J. Steganalysis for GIF images based on colors-gradient co-occurrence matrix. Opt. Commun. 2012, 285, 4961–4965. [Google Scholar] [CrossRef]
- Peckinpaugh, S.H. An improved method for computing gray-level co-occurrence matrix based texture measures. CVGIP Graph. Model. Image Process. 1991, 53, 574–580. [Google Scholar] [CrossRef]
- Ojala, T.; Pietikäinen, M.; Harwood, D. A comparative study of texture measures with classification based on featured distributions. Pattern Recognit. 1996, 29, 51–59. [Google Scholar] [CrossRef]
- Singh, C.; Singh, J. A survey on rotation invariance of orthogonal moments and transforms. Signal Process. 2021, 185, 108086. [Google Scholar] [CrossRef]
- Yang, B.; Kostková, J.; Flusser, J.; Suk, T.; Bujack, R. Rotation invariants of vector fields from orthogonal moments. Pattern Recognit. 2018, 74, 110–121. [Google Scholar] [CrossRef]
- Wang, X.; Yang, T.; Guo, F. Image analysis by circularly semi-orthogonal moments. Pattern Recognit. 2016, 49, 226–236. [Google Scholar] [CrossRef]
- Li, X.; Song, A. A new edge detection method using Gaussian-Zernike moment operator. In Proceedings of the IEEE 2nd International Asia Conference on Informatics in Control, Automation and Robotics (CAR), Wuhan, China, 6–7 March 2010; Volume 1, pp. 276–279. [Google Scholar]
- Obulakonda, R.R.; Eswara, R.B.; Keshava, R.E. Ternary patterns and moment invariants for texture classification. ICTACT J. Image Video Process. 2016, 7, 1295–1298. [Google Scholar]
- Tahmasbi, A.; Saki, F.; Shokouhi, S.B. Classification of benign and malignant masses based on Zernike moments. Comput Biol Med. 2011, 41, 726–735. [Google Scholar] [CrossRef] [PubMed]
- Vijayalakshmi, B.; Bharathi, V.S. Classification of CT liver images using local binary pattern with Legendre moments. Curr. Sci. 2016, 110, 687–691. [Google Scholar] [CrossRef]
- Wu, K.; Garnier, C.; Coatrieux, J.L.; Shu, H. A preliminary study of moment-based texture analysis for medical images. In Proceedings of the 2010 Annual International Conference of the IEEE Engineering in Medicine and Biology, San Francisco, CA, USA, 31 August–4 September 2010; pp. 5581–5584. [Google Scholar]
- Hu, M.K. Visual pattern recognition by moment invariants. IRE Trans. Inf. Theory 1962, 8, 179–187. [Google Scholar]
- Di Ruberto, C.; Putzu, L.; Rodriguez, G. Fast and accurate computation of orthogonal moments for texture analysis. Pattern Recognit. 2018, 83, 498–510. [Google Scholar] [CrossRef]
- Ryabchykov, O.; Ramoji, A.; Bocklitz, T.; Foerster, M.; Hagel, S.; Kroegel, C.; Bauer, M.; Neugebauer, U.; Popp, J. Leukocyte subtypes classification by means of image processing. In Proceedings of the 2016 Federated Conference on Computer Science and Information Systems (FedCSIS), Gdańsk, Poland, 11–14 September 2016; Volume 8, pp. 309–316. [Google Scholar]
- Elaziz, M.; Hosny, K.; Hemedan, A.; Darwish, M. Improved recognition of bacterial species using novel fractional-order orthogonal descriptors. Appl. Soft Comput. 2020, 95, 106504. [Google Scholar] [CrossRef]
- Mukundan, R. Local Tchebichef Moments for Texture Analysis. In Moments and Moment Invariants—Theory and Applications; Science Gate Publishing: Thrace, Greece, 2014; pp. 127–142. [Google Scholar]
- Xu, Z.; Xilin, L.; Yang, C.; Huazhong, S. Medical Image Blind Integrity Verification with Krawtchouk Moments. Int. J. Biomed. Imaging 2018, 2018, 2572431. [Google Scholar]
- Batioua, I.; Benouini, R.; Zenkouar, K.; Zahi, A.; Hakim, E.F. 3D image analysis by separable discrete orthogonal moments based on Krawtchouk and Tchebichef polynomials. Pattern Recognit. 2017, 71, 264–277. [Google Scholar] [CrossRef]
- Moung, E.; Hou, C.; Sufian, M.; Hijazi, M.; Dargham, J.; Omatu, S. Fusion of Moment Invariant Method and Deep Learning Algorithm for COVID-19 Classification. Big Data Cogn. Comput. 2021, 5, 74. [Google Scholar] [CrossRef]
- Lao, H.; Zhang, X. Diagnose Alzheimer’s disease by combining 3D discrete wavelet transform and 3D moment invariants. IET Image Process. 2022, 16, 3948–3964. [Google Scholar] [CrossRef]
- El ogri, O.; Daoui, A.; Yamni, M.; Karmouni, H.; Sayyouri, M.; Qjidaa, H. 2D and 3D Medical Image Analysis by Discrete Orthogonal Moments. Procedia Comput. Sci. 2019, 148, 428–437. [Google Scholar] [CrossRef]
- Mukundan, R.; Ong, S.H.; Lee, P.A. Image analysis by Tchebichef moments. IEEE Trans. Image Process. 2001, 10, 1357–1364. [Google Scholar] [CrossRef] [PubMed]
- Yap, P.T.; Paramesran, R.; Ong, S.H. Image analysis by Krawtchouk moments. IEEE Trans. Image Process. 2003, 12, 1367–1377. [Google Scholar] [PubMed]
- Yap, P.T.; Paramesran, R. An Efficient Method for the Computation of Legendre Moments. IEEE Trans. Pattern Anal. Mach. Intell. 2005, 27, 1996–2002. [Google Scholar]
- Teague, M.R. Image analysis via the general theory of moments. J. Opt. Soc. Am. 1980, 70, 920–930. [Google Scholar] [CrossRef]
- Qi, S.; Zhang, Y.; Wang, C.; Zhou, J.; Cao, X. A Survey of Orthogonal Moments for Image Representation: Theory, Implementation, and Evaluation. ACM Comput. Surv. 2021, 55, 1–35. [Google Scholar] [CrossRef]
- Varga, D. No-Reference Quality Assessment of Authentically Distorted Images Based on Local and Global Features. J. Imaging 2022, 8, 173. [Google Scholar] [CrossRef]
- Flusser, J.; Suk, T. Rotation Moment Invariants for Recognition of Symmetric Objects. IEEE Trans. Image Process. 2006, 15, 3784–3790. [Google Scholar] [CrossRef] [PubMed]
- Rahmalan, H.; Abu, N.A.; Wong, S.L. Using tchebichef moment for fast and efficient image compression. Pattern Recognit. Image Anal. 2010, 20, 505–512. [Google Scholar] [CrossRef]
- Tsougenis, E.D.; Papakostas, G.A.; Koulouriotis, D.E. Image watermarking via separable moments. Multim. Tools Appl. 2015, 74, 3985–4012. [Google Scholar] [CrossRef]
- Hosny, K.; Papakostas, G.; Koulouriotis, D. Accurate reconstruction of noisy medical images using orthogonal moments. In Proceedings of the 2013 18th International Conference on Digital Signal Processing (DSP), Santorini, Greece, 1–3 July 2013; pp. 1–6. [Google Scholar]
- Zhang, Y.; Wang, S.; Sun, P.; Phillips, P. Pathological brain detection based on wavelet entropy and Hu moment invariants. Bio-Med. Mater. Eng. 2015, 26 (Suppl. 1), S1283–S1290. [Google Scholar] [CrossRef] [PubMed]
- Iscan, Z.; Dokur, Z.; Ölmez, T. Tumor detection by using Zernike moments on segmented magnetic resonance brain images. Expert Syst. Appl. 2010, 37, 2540–2549. [Google Scholar] [CrossRef]
- Dogantekin, E.; Yilmaz, M.; Dogantekin, A.; Avci, E.; Sengur, A. A robust technique based on invariant moments—ANFIS for recognition of human parasite eggs in microscopic images. Expert Syst. Appl. 2008, 35, 728–738. [Google Scholar] [CrossRef]
- Liyun, W.; Hefei, L.; Fuhao, Z.; Zhengding, L.; Zhendi, W. Spermatogonium image recognition using Zernike moments. Comput. Methods Programs Biomed. 2009, 95, 10–22. [Google Scholar] [CrossRef]
- Alegre, E.; González-Castro, V.; Alaiz-Rodríguez, R.; García-Ordás, M. Texture and moments-based classification of the acrosome integrity of boar spermatozoa images. Comput. Methods Programs Biomed. 2012, 108, 873–881. [Google Scholar] [CrossRef] [PubMed]
- Di Ruberto, C.; Loddo, A.; Putzu, L. Histological image analysis by invariant descriptors. In Proceedings of the International Conference ICIAP on Image Analysis and Processing, Catania, Italy, 11–15 September 2017; Lecture Notes in Computer Science. Springer International Publishing: Berlin/Heidelberg, Germany, 2017; Volume 10484, pp. 345–356. [Google Scholar]
- Siti, S.; Shaharuddin, S.; Rozi, M. Haralick texture and invariant moments features for breast cancer classification. In Proceedings of the 2016 AIP Conference Proceedings, Bandung, Indonesia, 24–26 October 2016; Volume 1750, pp. 020022-1–020022-6. [Google Scholar]
- Laine, R.; Goodfellow, G.; Young, L.; Travers, J.; Carroll, D.; Dibben, O.; Bright, H.; Kaminski, C. Structured illumination microscopy combined with machine learning enables the high throughput analysis and classification of virus structure. eLife 2018, 7, e40183. [Google Scholar] [CrossRef] [PubMed]
- Balakrishnan, S.; Joseph, P. Stratification of risk of atherosclerotic plaque using Hu’s moment invariants of segmented ultrasonic images. Biomed. Tech. 2022, 67, 391–402. [Google Scholar] [CrossRef]
- Mukundan, R.; Ramakrishnan, K.R. Moment Functions in Image Analysis–Theory and Applications; World Scientific: Singapore, 1998. [Google Scholar] [CrossRef]
- Flusser, J.; Zitova, B.; Suk, T. Moments and Moment Invariants in Pattern Recognition; John Wiley & Sons: Hoboken, NJ, USA, 2009. [Google Scholar]
- Teh, C.H.; Chin, R.T. On image analysis by the methods of moments. IEEE Trans. Pattern Anal. Mach. Intell. 1988, 10, 496–513. [Google Scholar] [CrossRef]
- Abu-Mostafa, Y.S.; Psaltis, D. Recognitive aspects of moment invariants. IEEE Trans. Pattern Anal. Mach. Intell. 1984, 6, 698–706. [Google Scholar] [CrossRef]
- Sheng, Y.; Shen, L. Orthogonal Fourier-Mellin moments for invariant pattern recognition. J. Opt. Soc. Am. 1994, 11, 1748–1757. [Google Scholar] [CrossRef]
- Ping, Z.; Wu, R.; Sheng, Y. Image description with Chebyshev- Fourier moments. J. Opt. Soc. Am. 2002, 19, 1748–1754. [Google Scholar] [CrossRef]
- Ping, Z.; Ren, H.; Zou, J.; Sheng, Y.; Bo, W. Generic orthogonal moments: Jacobi-Fourier moments for invariant image description. Pattern Recognit. 2007, 40, 1245–1254. [Google Scholar] [CrossRef]
- Amu, G.; Hasi, S.; Yang, X.; Ping, Z. Image analysis by pseudo-Jacobi (p = 4, q = 3)-Fourier moments. Appl. Opt. 2004, 43, 2093–2101. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Ping, Z.; Bo, W.; Wu, W.; Sheng, Y. Multidistortion-invariant image recognition with radial harmonic Fourier moments. J. Opt. Soc. Am. 2003, 20, 631–637. [Google Scholar] [CrossRef]
- Hu, H.; Zhang, Y.; Shao, C.; Ju, Q. Orthogonal moments based on exponent functions: Exponent-Fourier moments. Pattern Recognit. 2014, 47, 2596–2606. [Google Scholar] [CrossRef]
- Yap, P.T.; Jiang, X.; Kot, A.C. Two-dimensional polar harmonic transforms for invariant image representation. IEEE Trans. Pattern Anal. Mach. Intell. 2010, 32, 1259–12707. [Google Scholar]
- Xiao, B.; Ma, J.; Wang, X. Image analysis by Bessel-Fourier moments. Pattern Recognit. 2010, 43, 2620–2629. [Google Scholar] [CrossRef]
- Yang, B.; Flusser, J.; Suk, T. 3D rotation invariants of Gaussian–Hermite moments. Pattern Recognit. Lett. 2015, 54, 18–26. [Google Scholar] [CrossRef]
- Yap, P.T.; Paramesran, R.; Ong, S.H. Image Analysis Using Hahn Moments. IEEE Trans. Pattern Anal. Mach. Intellig. 2007, 29, 2057–2062. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Shu, H.; Liang, J.; Luo, L.; Coatrieux, J.L. Image analysis by discrete orthogonal Racah moments. Signal Process. 2007, 87, 687–708. [Google Scholar] [CrossRef]
- Hoang, T.V.; Tabbone, S. Generic polar harmonic transforms for invariant image representation. Image Vis. Comput. 2014, 32, 497–509. [Google Scholar] [CrossRef]
- Hoang, T.V.; Tabbone, S. Generic polar harmonic transforms for invariant image description. In Proceedings of the 2011 18th IEEE International Conference on Image Processing, Brussels, Belgium, 11–14 September 2011; pp. 829–832. [Google Scholar]
- Xiao, B.; Li, L.; Li, Y.; Li, W.; Wang, G. Image analysis by fractional-order orthogonal moments. Inf. Sci. 2017, 382, 135–149. [Google Scholar] [CrossRef]
- Hosny, K.M.; Darwish, M.M.; Eltoukhy, M.M. Novel Multi-Channel Fractional-Order Radial Harmonic Fourier Moments for Color Image Analysis. IEEE Access 2020, 8, 40732–40743. [Google Scholar] [CrossRef]
- He, B.; Liu, J.; Yang, T.; Xiao, B.; Peng, Y. Quaternion fractional-order color orthogonal moment-based image representation and recognition. EURASIP J. Image Video Process. 2021, 2021, 1–35. [Google Scholar] [CrossRef]
- Yang, H.; Qi, S.; Tian, J.; Niu, P.; Wang, X. Robust and discriminative image representation: Fractional-order Jacobi-Fourier moments. Pattern Recognit. 2021, 115, 107898. [Google Scholar] [CrossRef]
- Sørensen, L.; Shaker, S.B.; de Bruijne, M. Quantitative Analysis of Pulmonary Emphysema Using Local Binary Patterns. IEEE Trans. Med Imaging 2010, 29, 559–569. [Google Scholar] [CrossRef]
- Cheng, J.; Huang, W.; Cao, S.; Yang, R.; Yang, W.; Yun, Z.; Wang, Z.; Feng, Q. Enhanced Performance of Brain Tumor Classification via Tumor Region Augmentation and Partition. PLoS ONE 2015, 10, e0140381. [Google Scholar] [CrossRef]
- Marcus, D.S.; Wang, T.H.; Parker, J.; Csernansky, J.G.; Morris, J.C.; Buckner, R.L. Open Access Series of Imaging Studies (OASIS): Cross-sectional MRI Data in Young, Middle Aged, Nondemented, and Demented Older Adults. J. Cogn. Neurosci. 2007, 19, 1498–1507. [Google Scholar] [CrossRef]
- Gunraj, H.; Wang, L.; Wong, A. COVIDNet-CT: A Tailored Deep Convolutional Neural Network Design for Detection of COVID-19 Cases From Chest CT Images. Front. Med. 2020, 7, 1025. [Google Scholar] [CrossRef] [PubMed]
- Putzu, L.; Di Ruberto, C. Rotation Invariant Co-occurrence Matrix Features. In Proceedings of the 19th International Conference ICIAP on Image Analysis and Processing, Catania, Italy, 11–15 September 2017; Lecture Notes in Computer Science. Springer International Publishing: Berlin/Heidelberg, Germany, 2017; Volume 10484, pp. 391–401. [Google Scholar]
- Ojala, T.; Pietikainen, M.; Maenpaa, T. Multiresolution gray-scale and rotation invariant texture classification with local binary pattern. IEEE Trans. Pattern Anal. Mach. Intell. 2002, 24, 971–987. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. Imagenet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE conference on computer vision and pattern recognition, New Orleans, LA, USA, 18–24 June 2009; pp. 248–255. [Google Scholar]
- Putzu, L.; Piras, L.; Giacinto, G. Convolutional neural networks for relevance feedback in content based image retrieval. Multimed Tools Appl. 2020, 79, 26995–27021. [Google Scholar] [CrossRef]
- Donahue, J.; Jia, Y.; Vinyals, O.; Hoffman, J.; Zhang, N.; Tzeng, E.; Darrell, T. DeCAF: A Deep Convolutional Activation Feature for Generic Visual Recognition. In Proceedings of the 31st International Conference on International Conference on Machine Learning, Beijing, China, 22–24 June 2014; Volume 32, pp. I-647–I-655. [Google Scholar]
- Lin, Y.; Lv, F.; Zhu, S.; Yang, M.; Cour, T.; Yu, K.; Cao, L.; Huang, T.S. Large-scale image classification: Fast feature extraction and SVM training. In Proceedings of the 24th IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2011, Colorado Springs, CO, USA, 20–25 June 2011; IEEE Computer Society: Washington, DC, USA, 2011; pp. 1689–1696. [Google Scholar]
- Cover, T.; Hart, P. Nearest neighbor pattern classification. IEEE Trans. Inf. Theory 1967, 13, 21–27. [Google Scholar] [CrossRef]
- Coppersmith, D.; Hong, S.; Hosking, J. Partitioning nominal attributes in decision trees. Data Min. Knowl. Discov. 1999, 3, 197–217. [Google Scholar] [CrossRef]
- Hastie, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction; Springer Series in Statistics; Springer: Berlin/Heidelberg, Germany, 2009. [Google Scholar]
- Putzu, L.; Loddo, A.; Di Ruberto, C. Invariant Moments, Textural and Deep Features for Diagnostic MR and CT Image Retrieval. In Proceedings of the Computer Analysis of Images and Patterns; Springer International Publishing: Cham, Switzerland, 2021; pp. 287–297. [Google Scholar]

| Ref. | Descriptors | Year |
|---|---|---|
| [37] | Legendre moments | 1980 |
| [34] | Chebyshev moments of the first order | 2001 |
| [34] | Chebyshev moments of the second order | 2001 |
| [37] | Zernike moments | 1980 |
| [55] | Pseudo-Zernike moments | 1988 |
| [57] | Orthogonal Fourier–Mellin moments | 1994 |
| [58] | Chebyshev–Fourier moments | 2002 |
| [60] | Pseudo-Jacobi–Fourier moments | 2004 |
| [59] | Jacobi–Fourier moments | 2007 |
| [73] | Fractional-order Jacobi–Fourier moments | 2021 |
| [64] | Bessel–Fourier moments | 2010 |
| [61] | Radial harmonic Fourier moments | 2003 |
| [62] | Exponent Fourier moments | 2014 |
| [63] | Polar complex exponential transform | 2010 |
| [63] | Polar cosine transform | 2010 |
| [63] | Polar sine transform | 2010 |
| [71] | Fractional-order radial harmonic Fourier moments | 2020 |
| [68] | Fractional-order polar complex exponential transform | 2014 |
| [68] | Fractional-order polar cosine transform | 2014 |
| [68] | Fractional-order polar sine transform | 2014 |
| Data Set | No. of Images | Size | Classes | No. of Images per Class |
|---|---|---|---|---|
| Emphysema-CT | 168 | 3 | 59–50–59 | |
| Brain-Tumor-MRI | 870 | 3 | 143–436–291 | |
| OASIS-MRI | 436 | 4 | 336–70–28–2 | |
| COVIDx CT-2A | 4942 | 3 | 3253–873–816 |
| Category | Descriptor | Size | Classifiers | AVG | AVG Cat. | ||||
|---|---|---|---|---|---|---|---|---|---|
| SVM | k-NN | DT | NB | BT | |||||
| CM | LM | 28 | 71.4 | 68.5 | 64.9 | 63.7 | 64.3 | 66.6 | 68.4 |
| CHM | 21 | 69.6 | 69.6 | 70.2 | 63.7 | 69.0 | 68.4 | ||
| CH2M | 21 | 69.6 | 71.4 | 70.8 | 67.3 | 72.0 | 70.2 | ||
| JM | ZM | 21 | 70.2 | 73.8 | 68.5 | 70.2 | 72.0 | 70.9 | 68.0 |
| PZM | 36 | 71.4 | 70.8 | 66.7 | 64.9 | 67.3 | 68.2 | ||
| OFMM | 66 | 70.8 | 72.0 | 68.5 | 61.9 | 66.7 | 68.0 | ||
| CHFM | 66 | 64.3 | 72.0 | 72.0 | 61.3 | 63.7 | 66.7 | ||
| PJFM | 66 | 69.0 | 72.0 | 70.2 | 59.5 | 69.6 | 68.1 | ||
| JFM | 66 | 69.6 | 70.8 | 69.6 | 61.9 | 67.9 | 68.0 | ||
| FrJFM | 66 | 66.1 | 73.8 | 69.6 | 61.3 | 61.3 | 66.4 | ||
| BM | BFM | 66 | 69.6 | 68.5 | 67.3 | 60.1 | 69.0 | 66.9 | 66.9 |
| HM | RHFM | 66 | 68.5 | 72.0 | 69.6 | 64.3 | 73.2 | 69.5 | 67.7 |
| EFM | 121 | 67.3 | 70.2 | 67.3 | 63.7 | 67.9 | 67.3 | ||
| PCET | 121 | 65.5 | 67.9 | 70.8 | 58.9 | 69.0 | 66.4 | ||
| PCT | 66 | 65.6 | 72.0 | 69.0 | 60.7 | 66.7 | 66.8 | ||
| PST | 55 | 69.6 | 69.6 | 67.9 | 61.9 | 64.3 | 66.7 | ||
| FrRHFM | 66 | 70.2 | 70.8 | 73.2 | 66.7 | 75.0 | 71.2 | ||
| FrPCET | 121 | 66.7 | 75.0 | 70.8 | 58.9 | 69.0 | 68.1 | ||
| FrPCT | 66 | 64.9 | 70.2 | 67.9 | 61.9 | 63.1 | 65.6 | ||
| FrPST | 55 | 69.6 | 70.2 | 69.0 | 64.3 | 66.7 | 68.0 | ||
| DF | AlexNet | 4096 | 78.9 | 73.8 | 63.1 | 72.6 | 76.8 | 73.0 | 72.0 |
| VGG19 | 4096 | 76.8 | 70.8 | 63.1 | 70.2 | 79.2 | 72.0 | ||
| ResNet50 | 1000 | 78.6 | 70.8 | 64.3 | 75.0 | 77.4 | 73.2 | ||
| GoogLeNet | 1000 | 76.8 | 66.7 | 61.9 | 72.0 | 71.4 | 69.8 | ||
| TF | LBP18 | 36 | 53.0 | 54.2 | 56.5 | 54.2 | 56.5 | 54.9 | 63.3 |
| HARri | 52 | 76.8 | 70.2 | 64.3 | 69.6 | 77.4 | 71.7 | ||
| - AVG | - | 69.6 | 70.3 | 67.6 | 64.3 | 69.1 | - | ||
| Category | Descriptor | Size | Classifiers | AVG | AVG Cat. | ||||
|---|---|---|---|---|---|---|---|---|---|
| SVM | k-NN | DT | NB | BT | |||||
| CM | LM | 28 | 95.9 | 97.7 | 90.7 | 91.0 | 95.2 | 94.1 | 93.7 |
| CHM | 21 | 94.6 | 97.9 | 89.7 | 91.0 | 94.7 | 93.6 | ||
| CH2M | 21 | 94.1 | 97.4 | 89.9 | 90.5 | 94.9 | 93.4 | ||
| JM | ZM | 21 | 93.4 | 93.3 | 90.2 | 89.7 | 93.4 | 92.0 | 91.8 |
| PZM | 36 | 94.1 | 94.9 | 90.5 | 89.9 | 94.8 | 92.8 | ||
| OFMM | 66 | 93.8 | 95.5 | 88.6 | 88.5 | 92.9 | 91.9 | ||
| CHFM | 66 | 93.7 | 96.2 | 87.1 | 87.7 | 93.7 | 91.7 | ||
| PJFM | 66 | 92.6 | 95.3 | 90.5 | 88.0 | 91.6 | 91.6 | ||
| JFM | 66 | 94.5 | 93.0 | 89.1 | 88.0 | 92.2 | 91.4 | ||
| FrJFM | 66 | 90.8 | 95.5 | 89.9 | 87.8 | 92.6 | 91.3 | ||
| BM | BFM | 66 | 94.3 | 95.9 | 89.1 | 86.9 | 94.0 | 92.0 | 92.0 |
| HM | RHFM | 66 | 93.6 | 95.4 | 88.4 | 87.6 | 92.9 | 91.6 | 91.7 |
| EFM | 121 | 94.9 | 98.2 | 87.1 | 86.7 | 92.6 | 91.9 | ||
| PCET | 121 | 96.7 | 97.7 | 87.4 | 86.7 | 92.0 | 92.1 | ||
| PCT | 66 | 92.2 | 94.4 | 87.4 | 88.3 | 91.4 | 90.7 | ||
| PST | 55 | 93.4 | 95.3 | 89.7 | 87.0 | 93.4 | 91.8 | ||
| FrRHFM | 66 | 94.1 | 95.2 | 89.2 | 88.3 | 92.0 | 91.8 | ||
| FrPCET | 121 | 95.9 | 98.2 | 89.4 | 87.0 | 94.6 | 93.0 | ||
| FrPCT | 66 | 94.0 | 94.5 | 88.4 | 87.6 | 91.5 | 91.2 | ||
| FrPST | 55 | 91.7 | 93.6 | 89.8 | 87.5 | 92.4 | 91.0 | ||
| DF | AlexNet | 4096 | 94.6 | 94.7 | 88.6 | 83.9 | 93.1 | 91.0 | 90.8 |
| VGG19 | 4096 | 95.4 | 94.9 | 84.4 | 87.7 | 93.0 | 91.1 | ||
| ResNet50 | 1000 | 95.7 | 95.9 | 88.5 | 87.4 | 93.8 | 92.3 | ||
| GoogLeNet | 1000 | 94.4 | 91.1 | 83.7 | 83.1 | 91.8 | 88.8 | ||
| TF | LBP18 | 36 | 92.3 | 93.6 | 89.4 | 77.1 | 90.3 | 88.5 | 90.3 |
| HARri | 52 | 93.4 | 92.3 | 88.5 | 92.3 | 93.9 | 92.1 | ||
| - AVG | - | 94.0 | 95.3 | 88.7 | 87.6 | 93.0 | - | - | |
| Category | Descriptor | Size | Classifiers | AVG | AVG Cat. | ||||
|---|---|---|---|---|---|---|---|---|---|
| SVM | k-NN | DT | NB | BT | |||||
| CM | LM | 28 | 77.8 | 78.2 | 77.1 | 71.8 | 77.5 | 76.5 | 76.5 |
| CHM | 21 | 78.0 | 77.6 | 77.1 | 72.9 | 77.1 | 76.5 | ||
| CH2M | 21 | 77.5 | 78.0 | 77.1 | 72.5 | 77.1 | 76.4 | ||
| JM | ZM | 21 | 77.3 | 78.2 | 77.1 | 70.6 | 77.3 | 76.1 | 76.3 |
| PZM | 36 | 77.1 | 78.4 | 77.1 | 72.0 | 78.2 | 76.6 | ||
| OFMM | 66 | 77.1 | 78.2 | 77.1 | 72.5 | 77.1 | 76.4 | ||
| CHFM | 66 | 77.1 | 80.0 | 77.1 | 71.3 | 77.1 | 76.5 | ||
| PJFM | 66 | 77.1 | 77.8 | 77.1 | 70.6 | 75.7 | 75.7 | ||
| JFM | 66 | 77.1 | 78.0 | 78.0 | 71.8 | 77.8 | 76.5 | ||
| FrJFM | 66 | 78.2 | 77.1 | 77.3 | 72.5 | 77.1 | 76.4 | ||
| BM | BFM | 66 | 77.1 | 77.8 | 77.1 | 74.1 | 76.6 | 76.5 | 76.5 |
| HM | RHFM | 66 | 77.1 | 78.4 | 78.4 | 72.0 | 76.6 | 76.5 | 76.9 |
| EFM | 121 | 77.1 | 78.0 | 77.1 | 73.9 | 76.6 | 76.5 | ||
| PCET | 121 | 77.3 | 78.4 | 77.1 | 74.8 | 78.0 | 77.1 | ||
| PCT | 66 | 77.1 | 78.9 | 77.1 | 72.7 | 77.1 | 76.6 | ||
| PST | 55 | 77.1 | 79.4 | 79.6 | 72.7 | 79.1 | 77.6 | ||
| FrRHFM | 66 | 77.1 | 78.9 | 77.1 | 72.0 | 78.4 | 76.7 | ||
| FrPCET | 121 | 77.5 | 77.1 | 77.8 | 75.5 | 77.1 | 77.0 | ||
| FrPCT | 66 | 77.1 | 79.1 | 77.1 | 72.5 | 77.1 | 76.9 | ||
| FrPST | 55 | 77.3 | 78.2 | 79.1 | 72.2 | 77.1 | 76.8 | ||
| DF | AlexNet | 4096 | 77.1 | 77.1 | 76.8 | 77.3 | 77.1 | 77.1 | 76.9 |
| VGG19 | 4096 | 78.7 | 77.1 | 75.7 | 77.5 | 78.9 | 77.6 | ||
| ResNet50 | 1000 | 77.1 | 77.3 | 75.7 | 75.5 | 77.1 | 76.5 | ||
| GoogLeNet | 1000 | 78.4 | 77.1 | 77.1 | 72.9 | 76.4 | 76.4 | ||
| TF | LBP18 | 36 | 77.1 | 77.3 | 77.1 | 72.0 | 75.5 | 75.8 | 75.7 |
| HARri | 52 | 77.1 | 77.3 | 77.1 | 69.7 | 77.1 | 75.7 | ||
| - AVG | - | 77.4 | 78.0 | 77.3 | 72.9 | 77.2 | - | - | |
| Category | Descriptor | Size | Classifiers | AVG | AVG Cat. | ||||
|---|---|---|---|---|---|---|---|---|---|
| SVM | k-NN | DT | NB | BT | |||||
| CM | LM | 28 | 99.0 | 97.2 | 94.3 | 76.2 | 98.8 | 93.1 | 92.0 |
| CHM | 21 | 99.0 | 96.9 | 93.9 | 72.0 | 98.7 | 92.1 | ||
| CH2M | 21 | 99.1 | 95.3 | 91.3 | 71.5 | 97.5 | 90.9 | ||
| JM | ZM | 21 | 98.9 | 95.4 | 92.2 | 72.2 | 97.7 | 91.3 | 91.7 |
| PZM | 36 | 99.0 | 96.7 | 93.4 | 75.8 | 98.4 | 92.7 | ||
| OFMM | 66 | 98.4 | 97.0 | 93.0 | 73.5 | 98.2 | 92.0 | ||
| CHFM | 66 | 98.3 | 97.0 | 92.8 | 72.5 | 98.2 | 91.8 | ||
| PJFM | 66 | 98.3 | 97.1 | 92.2 | 71.8 | 98.3 | 91.5 | ||
| JFM | 66 | 98.3 | 96.9 | 92.4 | 71.8 | 98.1 | 91.5 | ||
| FrJFM | 66 | 98.2 | 97.1 | 92.2 | 71.8 | 98.1 | 91.5 | ||
| BM | BFM | 66 | 98.3 | 97.0 | 92.3 | 73.2 | 98.1 | 91.8 | 91.8 |
| HM | RHFM | 66 | 98.3 | 97.1 | 92.6 | 71.4 | 98.1 | 91.5 | 91.6 |
| EFM | 121 | 98.3 | 97.6 | 91.5 | 70.3 | 98.2 | 91.2 | ||
| PCET | 121 | 98.0 | 97.7 | 92.5 | 72.1 | 98.4 | 91.7 | ||
| PCT | 66 | 98.4 | 97.2 | 93.1 | 72.2 | 98.5 | 91.9 | ||
| PST | 55 | 98.5 | 97.0 | 92.2 | 71.9 | 98.4 | 91.6 | ||
| FrRHFM | 66 | 98.3 | 97.1 | 92.6 | 71.4 | 98.2 | 91.5 | ||
| FrPCET | 121 | 98.1 | 97.6 | 92.2 | 72.5 | 98.3 | 91.7 | ||
| FrPCT | 66 | 98.5 | 97.3 | 93.0 | 72.3 | 98.4 | 91.9 | ||
| FrPST | 55 | 98.5 | 97.0 | 92.2 | 71.8 | 98.3 | 91.6 | ||
| DF | AlexNet | 4096 | 97.5 | 97.5 | 97.4 | 97.4 | 97.5 | 97.5 | 96.0 |
| VGG19 | 4096 | 99.4 | 99.4 | 99.4 | 99.1 | 99.4 | 99.3 | ||
| ResNet50 | 1000 | 95.8 | 95.8 | 95.6 | 95.0 | 96.5 | 95.7 | ||
| GoogLeNet | 1000 | 92.2 | 91.4 | 91.5 | 90.9 | 90.6 | 91.3 | ||
| TF | LBP18 | 36 | 98.4 | 98.0 | 95.0 | 74.1 | 97.9 | 92.7 | 92.4 |
| HARri | 52 | 97.5 | 96.5 | 94.7 | 73.9 | 97.5 | 92.0 | ||
| - AVG | - | 98.1 | 96.8 | 93.3 | 76.1 | 97.9 | - | - | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Ruberto, C.; Loddo, A.; Putzu, L. On The Potential of Image Moments for Medical Diagnosis. J. Imaging 2023, 9, 70. https://doi.org/10.3390/jimaging9030070
Di Ruberto C, Loddo A, Putzu L. On The Potential of Image Moments for Medical Diagnosis. Journal of Imaging. 2023; 9(3):70. https://doi.org/10.3390/jimaging9030070
Chicago/Turabian StyleDi Ruberto, Cecilia, Andrea Loddo, and Lorenzo Putzu. 2023. "On The Potential of Image Moments for Medical Diagnosis" Journal of Imaging 9, no. 3: 70. https://doi.org/10.3390/jimaging9030070
APA StyleDi Ruberto, C., Loddo, A., & Putzu, L. (2023). On The Potential of Image Moments for Medical Diagnosis. Journal of Imaging, 9(3), 70. https://doi.org/10.3390/jimaging9030070








