Accelerating 3D Medical Image Segmentation by Adaptive Small-Scale Target Localization
Abstract
1. Introduction
2. Related Work
2.1. Medical Imaging
2.2. Nonmedical Imaging
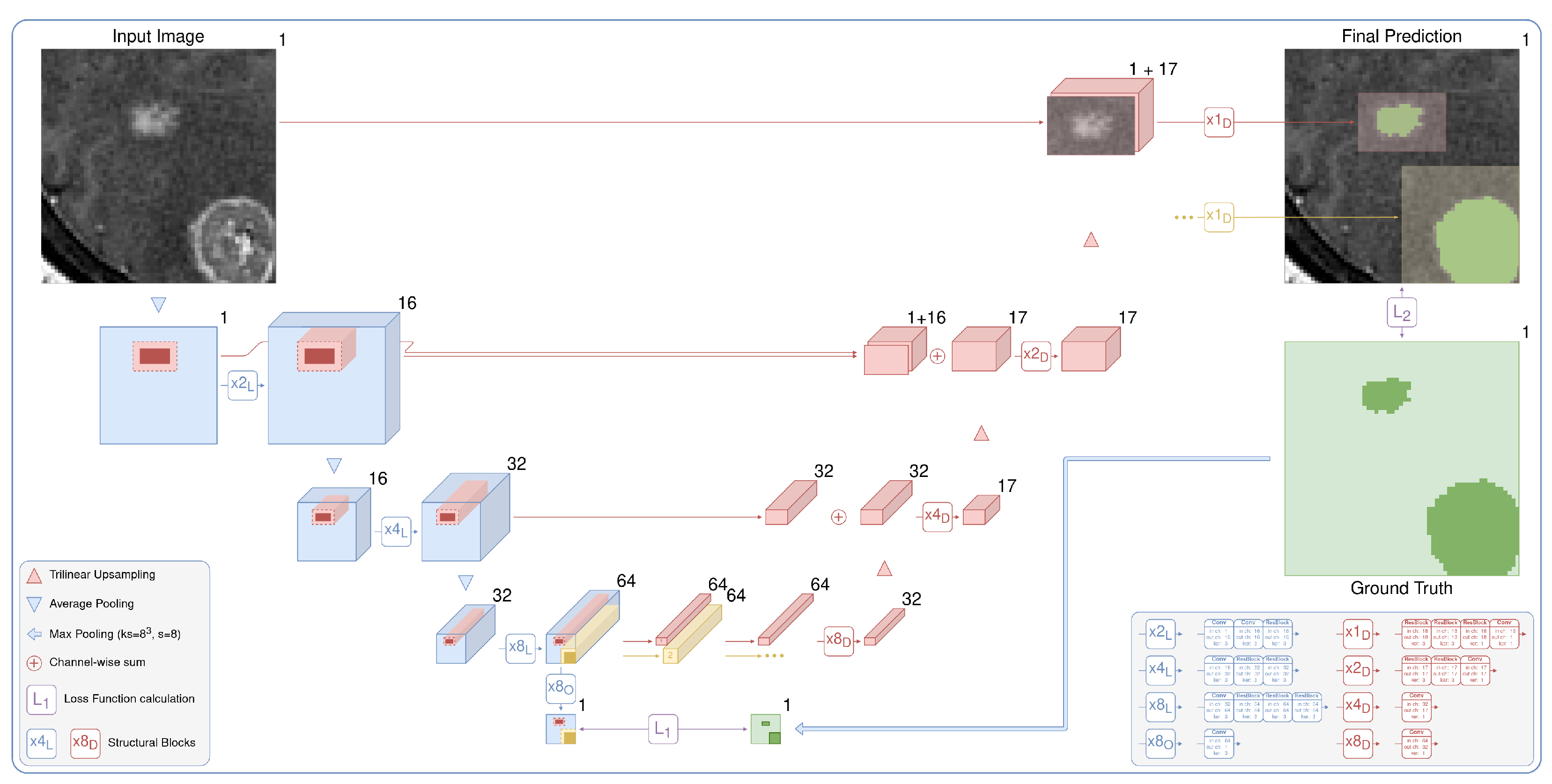
3. Method
3.1. Target Localization
3.2. Detailed Segmentation
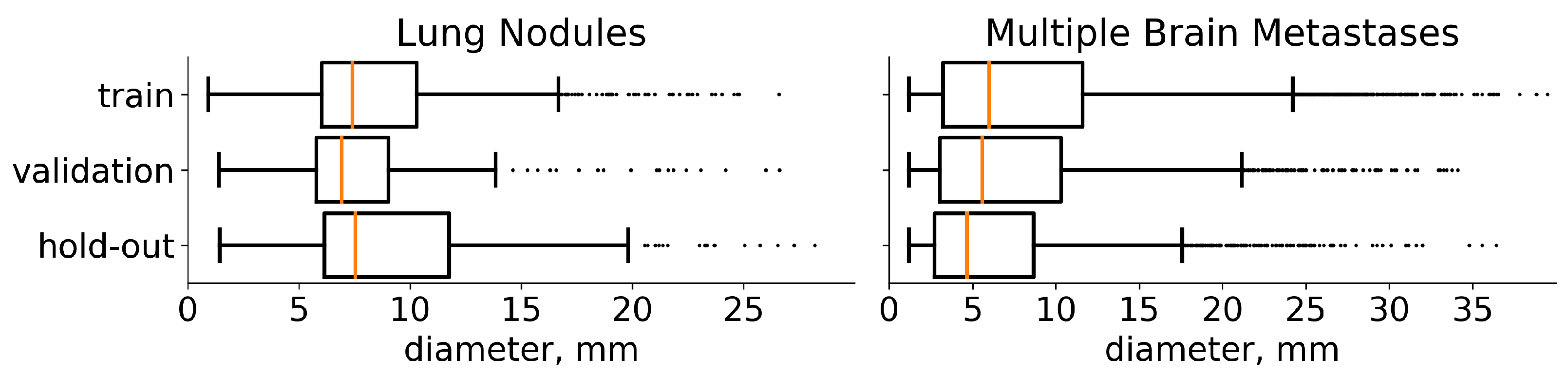
4. Data
5. Experiments and Results
5.1. Training
5.2. Experimental Setup
5.3. Results
6. Discussion
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MDPI | Multidisciplinary Digital Publishing Institute |
| BraTS | Multimodal Brain Tumor Segmentation |
| CNN | Convolutional Neural Network |
| CPU | Central Processing Unit |
| CT | Computed Tomography |
| DSC | Dice Score |
| FP | False Positive |
| FROC | Free-response Receiver Operating Characteristic |
| GPU | Graphics Processing Unit |
| LUNA16 | Lung Nodule Analysis 2016 |
| MRI | Magnetic Resonance Imaging |
| RAM | Random Access Memory |
| ReLU | Rectified Linear Unit |
References
- Pham, D.L.; Xu, C.; Prince, J.L. Current methods in medical image segmentation. Annu. Rev. Biomed. Eng. 2000, 2, 315–337. [Google Scholar] [CrossRef]
- The Lancet Digital Health. Leaving cancer diagnosis to the computers. Lancet Digit. Health 2020, 2, e49. [Google Scholar] [CrossRef]
- Meyer, P.; Noblet, V.; Mazzara, C.; Lallement, A. Survey on deep learning for radiotherapy. Comput. Biol. Med. 2018, 98, 126–146. [Google Scholar] [CrossRef]
- Comelli, A. Fully 3D Active Surface with Machine Learning for PET Image Segmentation. J. Imaging 2020, 6, 113. [Google Scholar] [CrossRef]
- Greenspan, H.; Van Ginneken, B.; Summers, R.M. Guest editorial deep learning in medical imaging: Overview and future promise of an exciting new technique. IEEE Trans. Med. Imaging 2016, 35, 1153–1159. [Google Scholar] [CrossRef]
- Setio, A.A.A.; Traverso, A.; De Bel, T.; Berens, M.S.; van den Bogaard, C.; Cerello, P.; Chen, H.; Dou, Q.; Fantacci, M.E.; Geurts, B.; et al. Validation, comparison, and combination of algorithms for automatic detection of pulmonary nodules in computed tomography images: The LUNA16 challenge. Med. Image Anal. 2017, 42, 1–13. [Google Scholar] [CrossRef]
- Bakas, S.; Reyes, M.; Jakab, A.; Bauer, S.; Rempfler, M.; Crimi, A.; Shinohara, R.T.; Berger, C.; Ha, S.M.; Rozycki, M.; et al. Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BRATS challenge. arXiv 2018, arXiv:1811.02629. [Google Scholar]
- Liu, X.; Faes, L.; Kale, A.U.; Wagner, S.K.; Fu, D.J.; Bruynseels, A.; Mahendiran, T.; Moraes, G.; Shamdas, M.; Kern, C.; et al. A comparison of deep learning performance against health-care professionals in detecting diseases from medical imaging: A systematic review and meta-analysis. Lancet Digit. Health 2019, 1, e271–e297. [Google Scholar] [CrossRef]
- Ardila, D.; Kiraly, A.P.; Bharadwaj, S.; Choi, B.; Reicher, J.J.; Peng, L.; Tse, D.; Etemadi, M.; Ye, W.; Corrado, G.; et al. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat. Med. 2019, 25, 954–961. [Google Scholar] [CrossRef]
- Kamnitsas, K.; Ledig, C.; Newcombe, V.F.; Simpson, J.P.; Kane, A.D.; Menon, D.K.; Rueckert, D.; Glocker, B. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med. Image Anal. 2017, 36, 61–78. [Google Scholar] [CrossRef]
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning dense volumetric segmentation from sparse annotation. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Berlin, Germany, 2016; pp. 424–432. [Google Scholar]
- Chen, D.; Liu, S.; Kingsbury, P.; Sohn, S.; Storlie, C.B.; Habermann, E.B.; Naessens, J.M.; Larson, D.W.; Liu, H. Deep learning and alternative learning strategies for retrospective real-world clinical data. NPJ Digit. Med. 2019, 2, 1–5. [Google Scholar] [CrossRef]
- European Society of Radiology. Renewal of radiological equipment. Insights Imaging 2014, 5, 543–546. [Google Scholar] [CrossRef] [PubMed]
- McDonald, R.J.; Schwartz, K.M.; Eckel, L.J.; Diehn, F.E.; Hunt, C.H.; Bartholmai, B.J.; Erickson, B.J.; Kallmes, D.F. The effects of changes in utilization and technological advancements of cross-sectional imaging on radiologist workload. Acad. Radiol. 2015, 22, 1191–1198. [Google Scholar] [CrossRef] [PubMed]
- Lindfors, K.K.; O’Connor, J.; Parker, R.A. False-positive screening mammograms: Effect of immediate versus later work-up on patient stress. Radiology 2001, 218, 247–253. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Jacobs, C.; Setio, A.A.A.; Traverso, A.; van Ginneken, B. LUng Nodule Analysis 2016. Available online: https://luna16.grand-challenge.org (accessed on 9 December 2019).
- Armato, S.G., III; McLennan, G.; Bidaut, L.; McNitt-Gray, M.F.; Meyer, C.R.; Reeves, A.P.; Zhao, B.; Aberle, D.R.; Henschke, C.I.; Hoffman, E.A.; et al. The lung image database consortium (LIDC) and image database resource initiative (IDRI): A completed reference database of lung nodules on CT scans. Med. Phys. 2011, 38, 915–931. [Google Scholar] [CrossRef] [PubMed]
- Isensee, F.; Kickingereder, P.; Wick, W.; Bendszus, M.; Maier-Hein, K.H. No new-net. In International MICCAI Brainlesion Workshop; Springer: Berlin, Germany, 2018; pp. 234–244. [Google Scholar]
- Imai, H.; Matzek, S.; Le, T.D.; Negishi, Y.; Kawachiya, K. Fast and accurate 3d medical image segmentation with data-swapping method. arXiv 2018, arXiv:1812.07816. [Google Scholar]
- Mehta, R.; Sivaswamy, J. M-net: A convolutional neural network for deep brain structure segmentation. In Proceedings of the 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017), Melbourne, VIC, Australia, 18–21 April 2017; pp. 437–440. [Google Scholar]
- Heinrich, M.P.; Blendowski, M.; Oktay, O. TernaryNet: Faster deep model inference without GPUs for medical 3D segmentation using sparse and binary convolutions. Int. J. Comput. Assist. Radiol. Surg. 2018, 13, 1311–1320. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Berlin, Germany, 2015; pp. 234–241. [Google Scholar]
- Lai, M. Deep learning for medical image segmentation. arXiv 2015, arXiv:1505.02000. [Google Scholar]
- Zhao, N.; Tong, N.; Ruan, D.; Sheng, K. Fully Automated Pancreas Segmentation with Two-Stage 3D Convolutional Neural Networks. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Berlin, Germany, 2019; pp. 201–209. [Google Scholar]
- Wang, C.; MacGillivray, T.; Macnaught, G.; Yang, G.; Newby, D. A two-stage 3D Unet framework for multi-class segmentation on full resolution image. arXiv 2018, arXiv:1804.04341. [Google Scholar]
- Gerard, S.E.; Herrmann, J.; Kaczka, D.W.; Musch, G.; Fernandez-Bustamante, A.; Reinhardt, J.M. Multi-resolution convolutional neural networks for fully automated segmentation of acutely injured lungs in multiple species. Med. Image Anal. 2020, 60, 101592. [Google Scholar] [CrossRef]
- Paszke, A.; Chaurasia, A.; Kim, S.; Culurciello, E. Enet: A deep neural network architecture for real-time semantic segmentation. arXiv 2016, arXiv:1606.02147. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.C. Mobilenetv2: Inverted residuals and linear bottlenecks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 4510–4520. [Google Scholar]
- Vesal, S.; Maier, A.; Ravikumar, N. Fully Automated 3D Cardiac MRI Localisation and Segmentation Using Deep Neural Networks. J. Imaging 2020, 6, 65. [Google Scholar] [CrossRef]
- Lin, N.U.; Lee, E.Q.; Aoyama, H.; Barani, I.J.; Barboriak, D.P.; Baumert, B.G.; Bendszus, M.; Brown, P.D.; Camidge, D.R.; Chang, S.M.; et al. Response assessment criteria for brain metastases: Proposal from the RANO group. Lancet Oncol. 2015, 16, e270–e278. [Google Scholar] [CrossRef]
- Bankier, A.A.; MacMahon, H.; Goo, J.M.; Rubin, G.D.; Schaefer-Prokop, C.M.; Naidich, D.P. Recommendations for measuring pulmonary nodules at CT: A statement from the Fleischner Society. Radiology 2017, 285, 584–600. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Nair, V.; Hinton, G.E. Rectified linear units improve restricted boltzmann machines. In Proceedings of the 27th International Conference on Machine Learning (ICML-10), Haifa, Israel, 21–24 June 2010; pp. 807–814. [Google Scholar]
- Zhao, Z.Q.; Zheng, P.; Xu, S.t.; Wu, X. Object detection with deep learning: A review. IEEE Trans. Neural Netw. Learn. Syst. 2019, 30, 3212–3232. [Google Scholar] [CrossRef]
- Milletari, F.; Navab, N.; Ahmadi, S.A. V-net: Fully convolutional neural networks for volumetric medical image segmentation. In Proceedings of the 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; pp. 565–571. [Google Scholar]
- Van Ginneken, B.; Armato, S.G., III; de Hoop, B.; van Amelsvoort-van de Vorst, S.; Duindam, T.; Niemeijer, M.; Murphy, K.; Schilham, A.; Retico, A.; Fantacci, M.E.; et al. Comparing and combining algorithms for computer-aided detection of pulmonary nodules in computed tomography scans: The ANODE09 study. Med. Image Anal. 2010, 14, 707–722. [Google Scholar] [CrossRef] [PubMed]
- Quekel, L.G.; Kessels, A.G.; Goei, R.; van Engelshoven, J.M. Miss rate of lung cancer on the chest radiograph in clinical practice. Chest 1999, 115, 720–724. [Google Scholar] [CrossRef] [PubMed]
- Azad, R.; Asadi-Aghbolaghi, M.; Fathy, M.; Escalera, S. Bi-directional convlstm u-net with densley connected convolutions. In Proceedings of the IEEE/CVF International Conference on Computer Vision Workshops, Seoul, Korea, 27–28 October 2019. [Google Scholar]
- Seo, H.; Huang, C.; Bassenne, M.; Xiao, R.; Xing, L. Modified U-Net (mU-Net) with incorporation of object-dependent high level features for improved liver and liver-tumor segmentation in CT images. IEEE Trans. Med. Imaging 2019, 39, 1316–1325. [Google Scholar] [CrossRef] [PubMed]
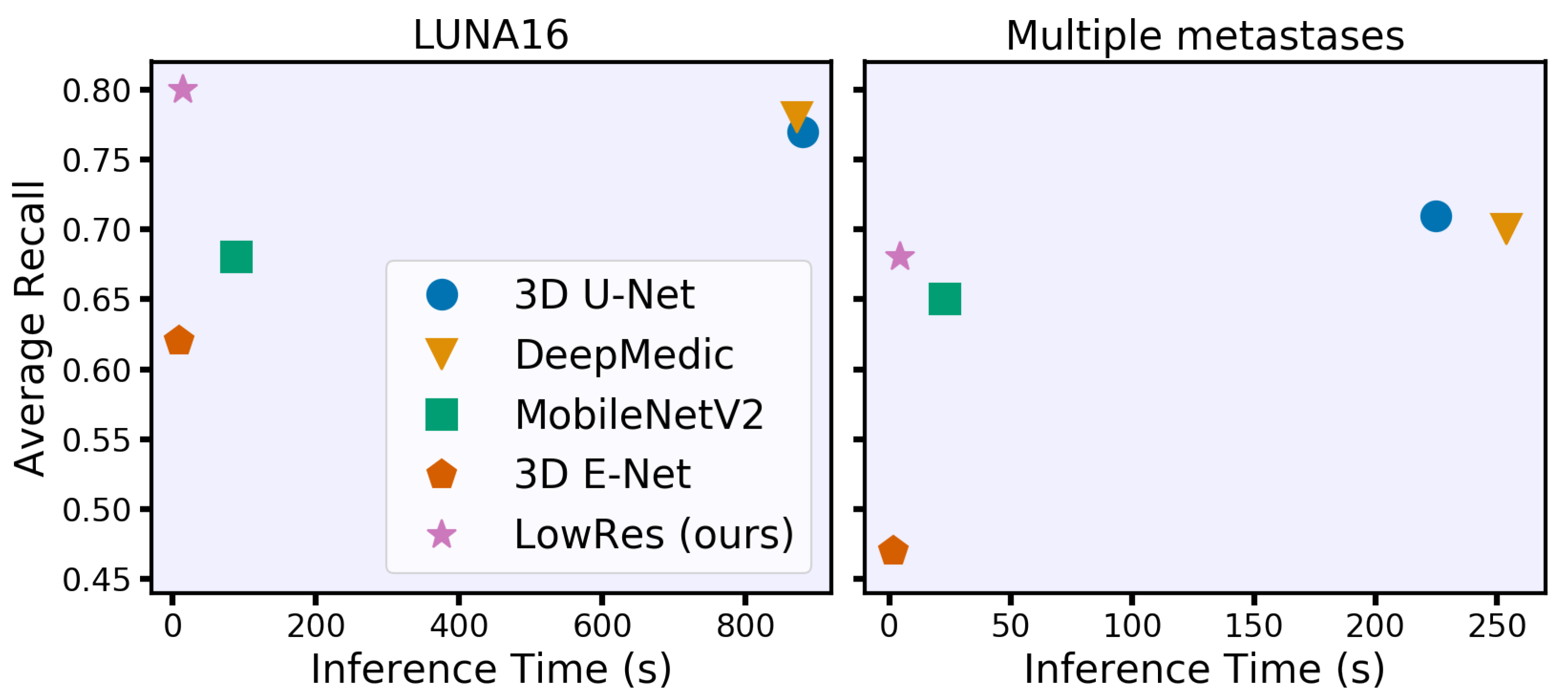
| Inference Time * (CPU Threads) | Quality Metrics | ||||
|---|---|---|---|---|---|
| 4 Threads | 8 Threads | 16 Threads | Avg Recall | Obj DSC | |
| 3D U-Net | 1293 (100) | 880 (61) | 828 (62) | 0.77 (0.02) | 0.82 (0.16) |
| DeepMedic | 1139 (162) | 872 (138) | 840 (127) | 0.78 (0.02) | 0.78 (0.20) |
| 3D MobileNetV2 | 108 (14) | 89 (12) | 74 (10) | 0.68 (0.02) | 0.75 (0.22) |
| 3D E-Net | 11 (1.2) | 9.5 (1.2) | 9.2 (1.0) | 0.62 (0.02) | 0.70 (0.22) |
| LowRes | 23 (2.9) | 15 (1.9) | 13 (1.7) | 0.80 (0.02) | 0.75 (.18) |
| Inference Time * (CPU Threads) | Quality Metrics | ||||
|---|---|---|---|---|---|
| 4 Threads | 8 Threads | 16 Threads | Avg Recall | Obj DSC | |
| 3D U-Net | 342 (36) | 225 (20) | 202 (16) | 0.71 (0.01) | 0.72 (0.21) |
| DeepMedic | 381 (74) | 254 (50) | 226 (46) | 0.70 (0.02) | 0.69 (0.23) |
| 3D MobileNetV2 | 33 (4.6) | 23 (3.0) | 21 (2.4) | 0.65 (0.01) | 0.69 (0.22) |
| 3D E-Net | 3.2 (0.3) | 1.7 (0.2) | 1.9 (0.3) | 0.47 (0.01) | 0.59 (0.23) |
| LowRes | 6.1 (0.7) | 4.3 (0.6) | 3.7 (0.6) | 0.68 (0.01) | 0.64 (0.22) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shirokikh, B.; Shevtsov, A.; Dalechina, A.; Krivov, E.; Kostjuchenko, V.; Golanov, A.; Gombolevskiy, V.; Morozov, S.; Belyaev, M. Accelerating 3D Medical Image Segmentation by Adaptive Small-Scale Target Localization. J. Imaging 2021, 7, 35. https://doi.org/10.3390/jimaging7020035
Shirokikh B, Shevtsov A, Dalechina A, Krivov E, Kostjuchenko V, Golanov A, Gombolevskiy V, Morozov S, Belyaev M. Accelerating 3D Medical Image Segmentation by Adaptive Small-Scale Target Localization. Journal of Imaging. 2021; 7(2):35. https://doi.org/10.3390/jimaging7020035
Chicago/Turabian StyleShirokikh, Boris, Alexey Shevtsov, Alexandra Dalechina, Egor Krivov, Valery Kostjuchenko, Andrey Golanov, Victor Gombolevskiy, Sergey Morozov, and Mikhail Belyaev. 2021. "Accelerating 3D Medical Image Segmentation by Adaptive Small-Scale Target Localization" Journal of Imaging 7, no. 2: 35. https://doi.org/10.3390/jimaging7020035
APA StyleShirokikh, B., Shevtsov, A., Dalechina, A., Krivov, E., Kostjuchenko, V., Golanov, A., Gombolevskiy, V., Morozov, S., & Belyaev, M. (2021). Accelerating 3D Medical Image Segmentation by Adaptive Small-Scale Target Localization. Journal of Imaging, 7(2), 35. https://doi.org/10.3390/jimaging7020035






