Initial Investigations of the Cranial Size and Shape of Adult Eurasian Otters (Lutra lutra) in Great Britain
Abstract
1. Introduction
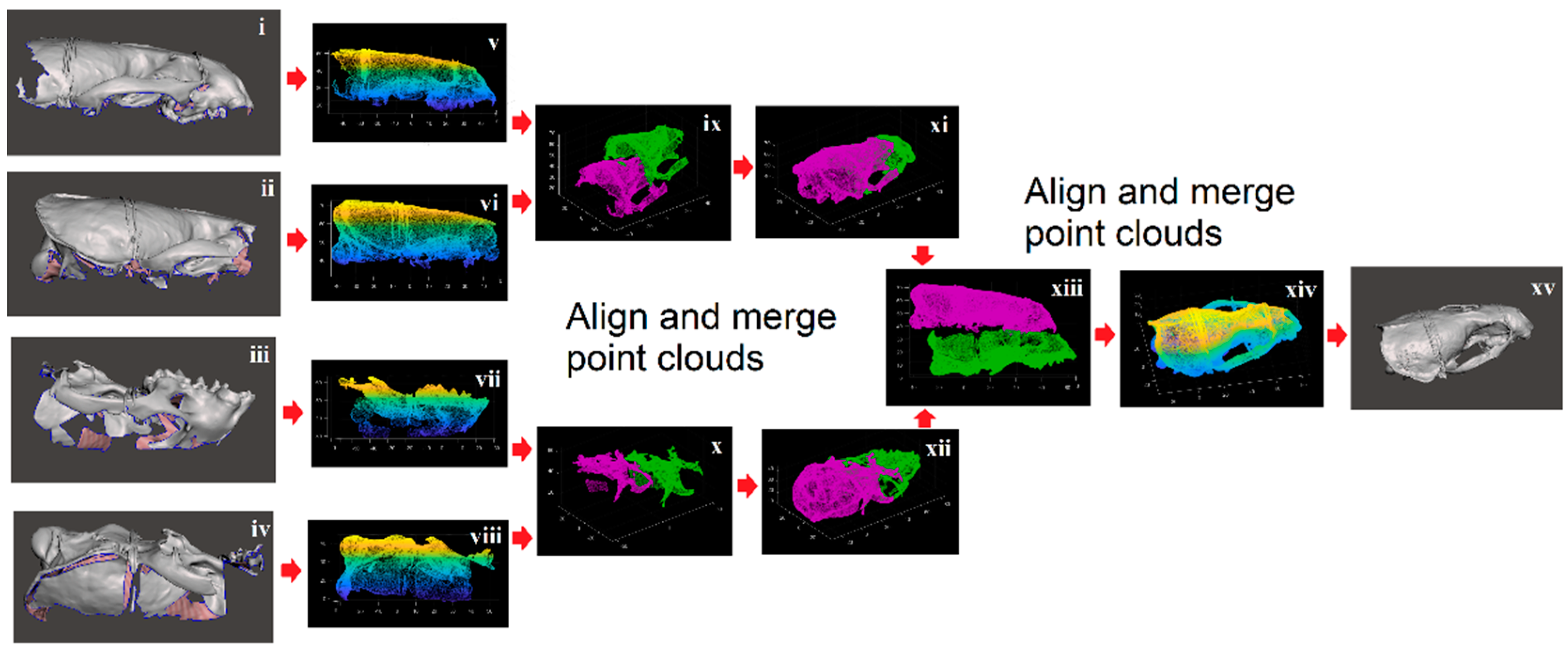
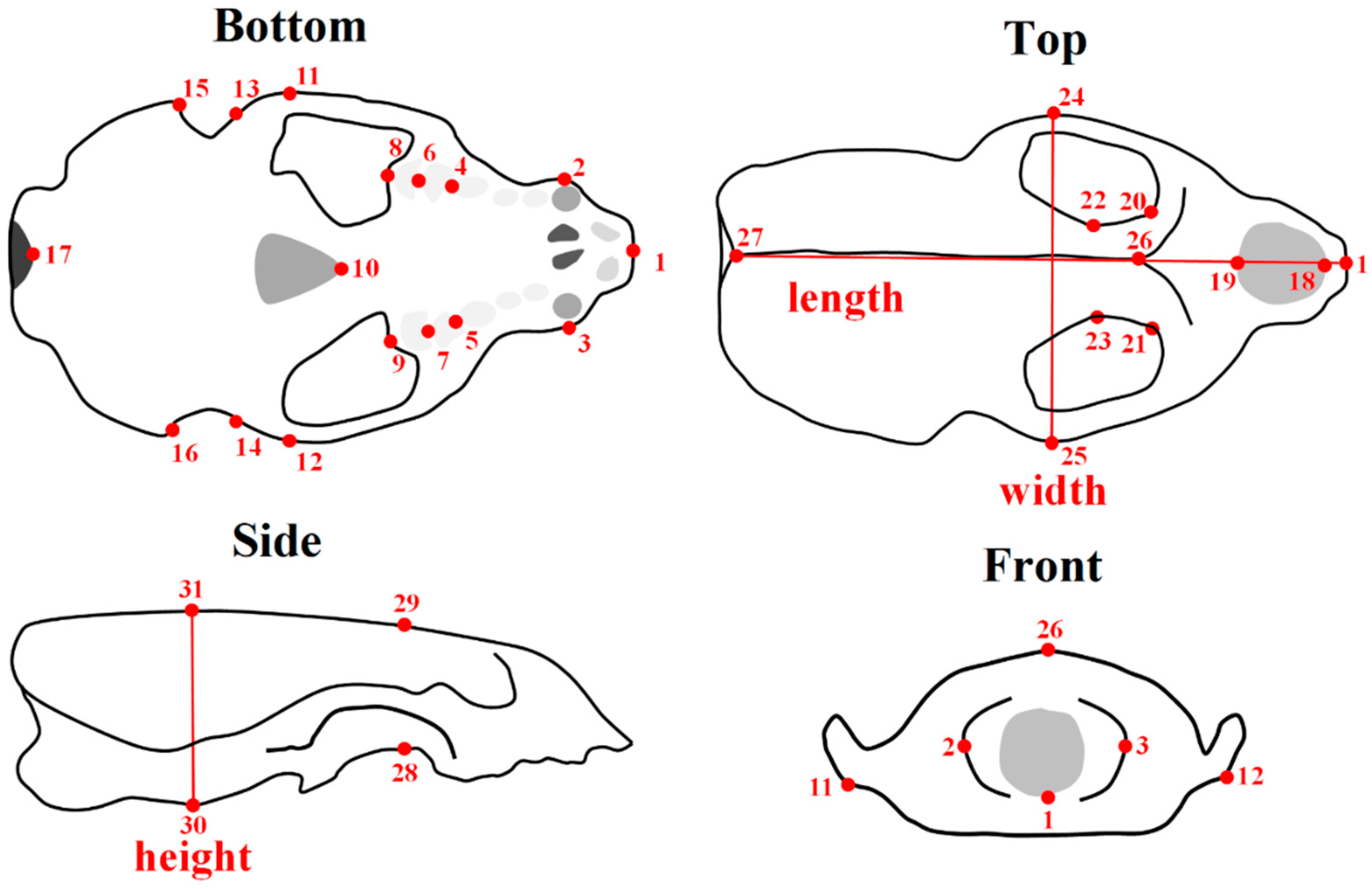
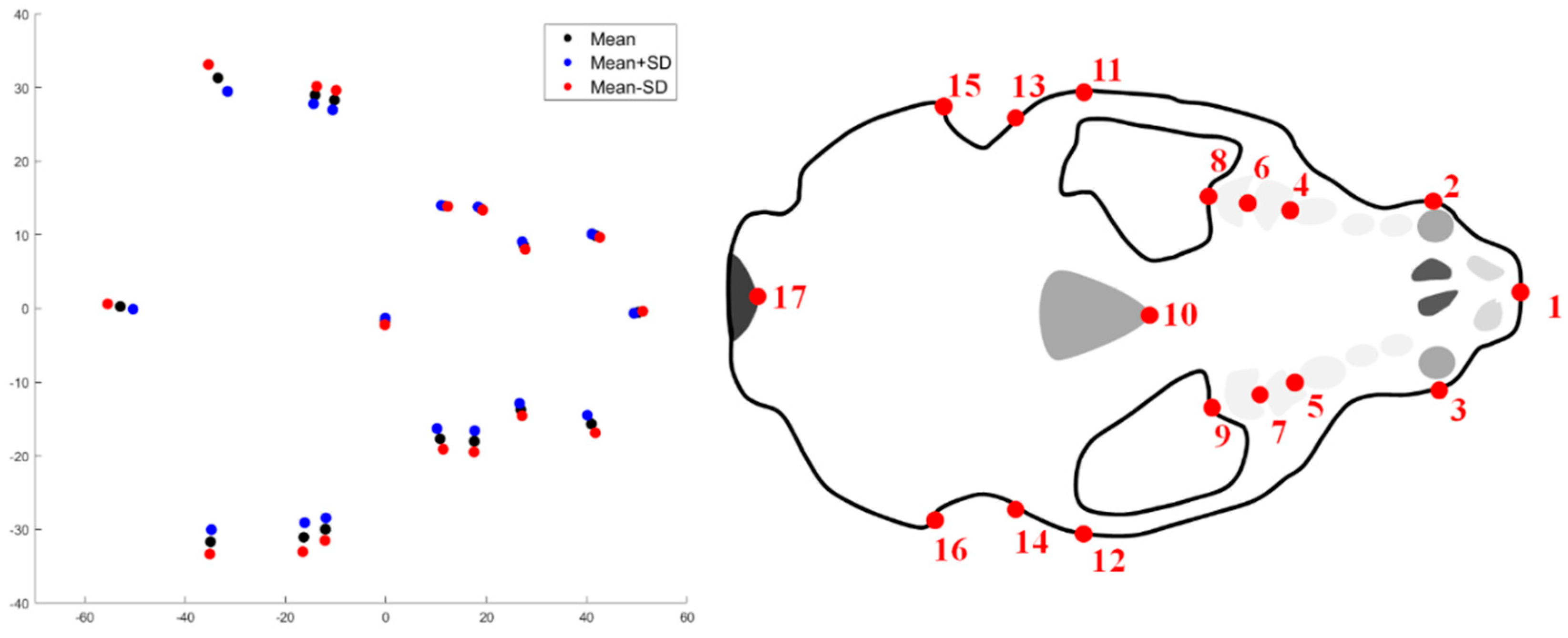
2. Materials and Methods
2.1. Shape Acquisition
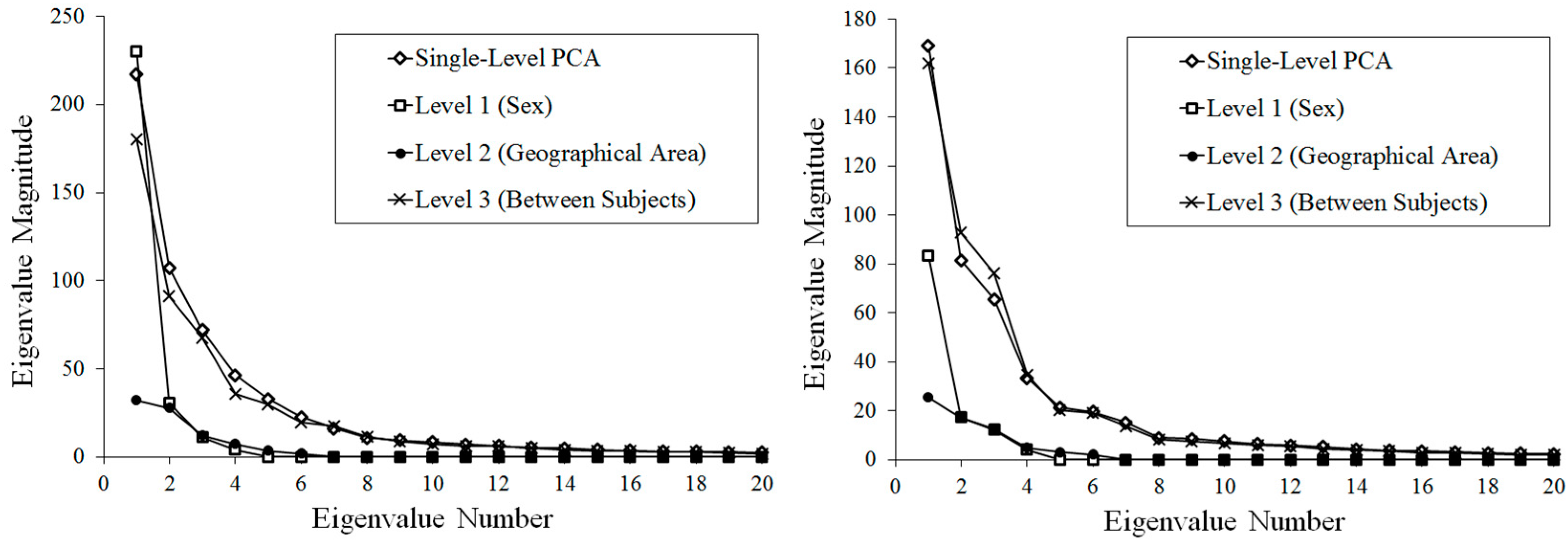
2.2. Statistical Analysis
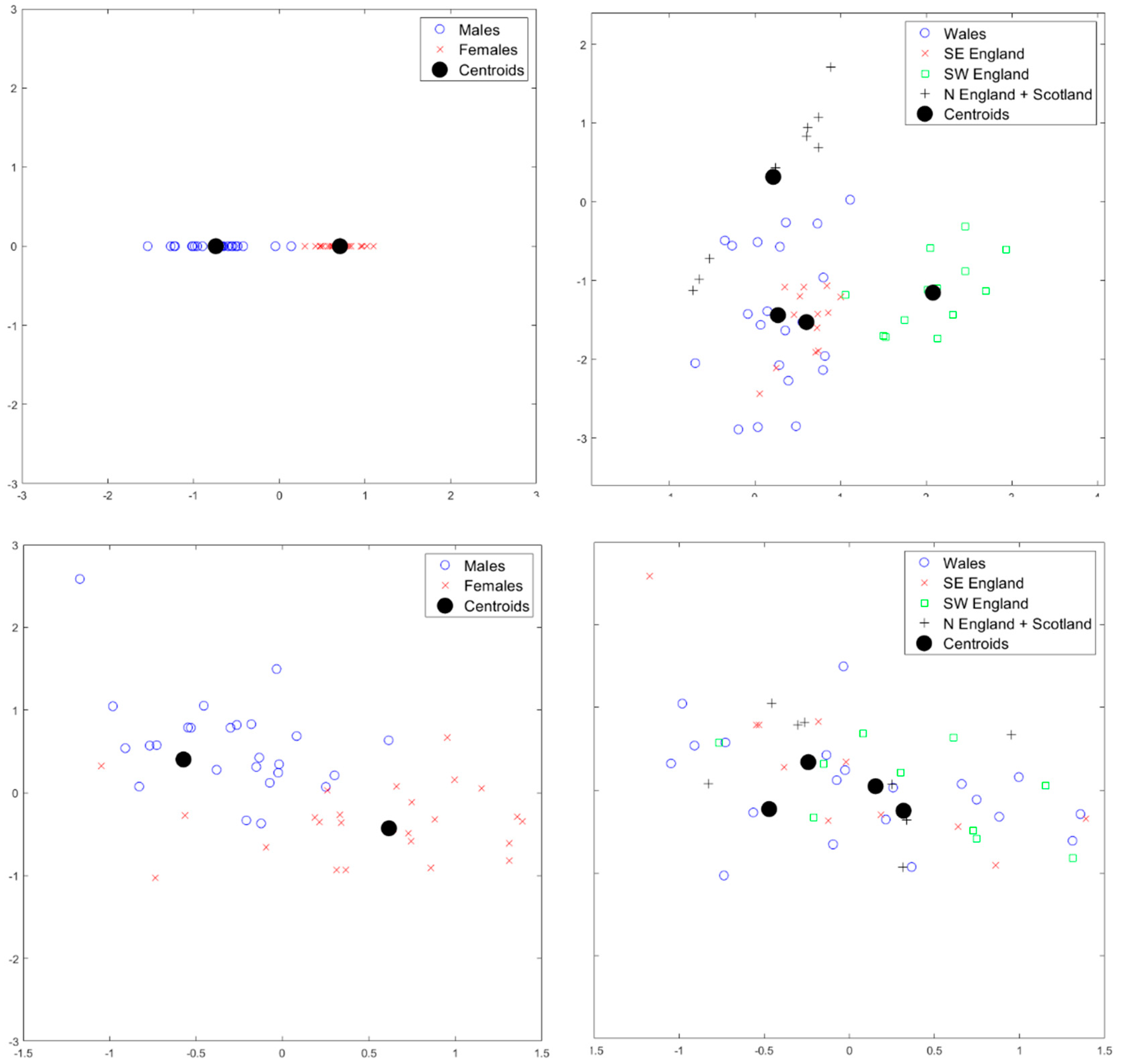
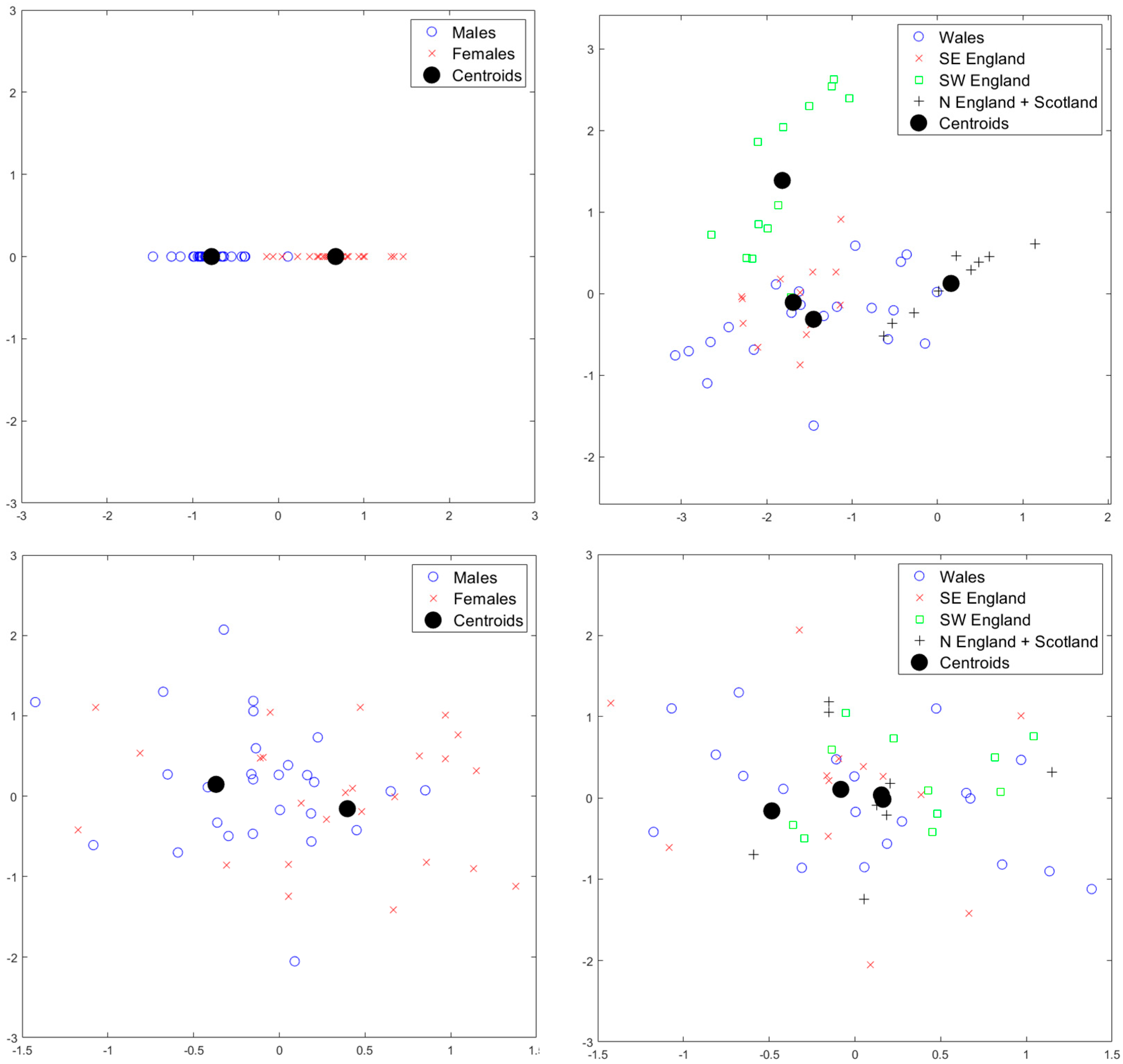
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Appendix A
References
- Zelditch, M.L.; Swiderski, D.L.; Sheets, H.D. Geometric Morphometrics for Biologists: A Primer; Academic Press: London, UK, 2012. [Google Scholar]
- Elewa Ashraf, M.T. (Ed.) Morphometrics: Applications in Biology and Paleontology; Springer Science & Business Media: Berlin, Germany, 2004; Volume 14. [Google Scholar]
- Tatsuta, H.; Takahashi, K.H.; Sakamaki, Y. Geometric morphometrics in entomology: Basics and applications. Entomol. Sci. 2018, 21, 164–184. [Google Scholar] [CrossRef]
- Mitteroecker, P.; Gunz, P. Advances in Geometric Morphometrics. Evol. Biol. 2009, 36, 235–247. [Google Scholar] [CrossRef]
- Klingenberg, C.P. Size, shape, and form: Concepts of allometry in geometric morphometrics. Dev. Genes Evol. 2016, 226, 113–137. [Google Scholar] [CrossRef] [PubMed]
- Izenman, A.J. Linear discriminant analysis. In Modern Multivariate Statistical Techniques; Springer: New York, NY, USA, 2013; pp. 237–280. [Google Scholar]
- Huberty, C.J.; Olejnik, S. Applied MANOVA and Discriminant Analysis; John Wiley & Sons: Hoboken, NJ, USA, 2006; Volume 498. [Google Scholar]
- Monteiro, L.R. Multivariate Regression Models and Geometric Morphometrics: The Search for Causal Factors in the Analysis of Shape. Syst. Biol. 1999, 48, 192–199. [Google Scholar] [CrossRef]
- Lecron, F.; Boisvert, J.; Benjelloun, M.; Labelle, H.; Mahmoudi, S. Multilevel statistical shape models: A new framework for modeling hierarchical structures. In Proceedings of the 9th IEEE International Symposium on Biomedical Imaging (ISBI), Barcelona, Spain, 2–5 May 2012; pp. 1284–1287. [Google Scholar]
- Farnell, D.J.J.; Popat, H.; Richmond, S. Multilevel principal component analysis (mPCA) in shape analysis: A feasibility study in medical and dental imaging. Comput. Methods Programs Biomed. 2016, 129, 149–159. [Google Scholar] [CrossRef]
- Farnell, D.J.J.; Galloway, J.; Zhurov, A.I.; Richmond, S.; Perttiniemi, P.; Katic, V. Initial Results of Multilevel Principal Components Analysis of Facial Shape. In Proceedings of the Medical Image Understanding and Analysis, 21st Annual Conference, MIUA 2017, Edinburgh, UK, 11–13 July 2017; Springer: Cham, Switzerland, 2017; pp. 674–685. [Google Scholar]
- Farnell, D.J.J.; Galloway, J.; Zhurov, A.I.; Richmond, S.; Perttiniemi, P.; Lähdesmäki, R. What’s in a Smile? Initial Results of Multilevel Principal Components Analysis of Facial Shape and Image Texture. Medical Image Understanding and Analysis. MIUA 2018. Commun. Comput. Inf. Sci. 2018, 894, 177–188. [Google Scholar]
- Farnell, D.J.J.; Galloway, J.; Zhurov, A.I.; Richmond, S.; Marshall, D.; Rosin, P.L.; Al-Meyah, K.; Perttiniemi, P.; Lähdesmäki, R. What’s in a Smile? Initial Analyses of Dynamic Changes in Facial Shape and Appearance. J. Imaging 2019, 5, 2. [Google Scholar] [CrossRef]
- Farnell, D.J.J.; Galloway, J.; Zhurov, A.I.; Richmond, S. Multilevel Models of Age-Related Changes in Facial Shape in Adolescents. Medical Image Understanding and Analysis. MIUA 2019. Commun. Comput. Inf. Sci. 2020, 1065, 101–113. [Google Scholar]
- Farnell, D.J.J.; Richmond, S.; Galloway, J.; Zhurov, A.I.; Pirttiniemi, P.; Heikkinen, T.; Harila, V.; Matthews, H.; Claes, P. Multilevel Principal Components Analysis of Three-Dimensional Facial Growth in Adolescents. Comput. Methods Programs Biomed. 2019, 188, 105272. [Google Scholar] [CrossRef]
- Galloway, J.; Farnell, D.J.J.; Richmond, S.; Zhurov, A.I. Multilevel Analysis of the Influence of Maternal Smoking and Alcohol Consumption on the Facial Shape of English Adolescents. J. Imaging 2020, 6, 34. [Google Scholar] [CrossRef]
- Chanin, P.R.F. The Handbook of British Mammals, 3rd ed.; Blackwell: Oxford, UK, 1991. [Google Scholar]
- Mucci, N.; Arrendal, J.; Ansorge, H.; Bailey, M.; Bodner, M.; Delibes, M.; Ferrando, A.; Fournier, P.; Fournier, C.; Godoy, J.A.; et al. Genetic diversity and landscape genetic structure of otter (Lutra lutra) populations in Europe. Conserv. Genet. 2010, 11, 583–599. [Google Scholar] [CrossRef]
- Stanton, D.W.; Hobbs, G.I.; McCafferty, D.J.; Chadwick, E.A.; Philbey, A.W.; Saccheri, I.J.; Slater, F.M.; Bruford, M.W. Contrasting genetic structure of the Eurasian otter (Lutra lutra) across a latitudinal divide. J. Mammal. 2014, 95, 814–823. [Google Scholar] [CrossRef]
- Stanton, D.W.G.; Hobbs, G.I.; Chadwick, E.A.; Slater, F.M.; Bruford, M.W. Mitochondrial genetic diversity and structure of the European otter (Lutra lutra) in Britain. Conserv. Genet. 2009, 10, 733–737. [Google Scholar] [CrossRef]
- Hobbs, G.I.; Chadwick, E.A.; Bruford, M.W.; Slater, F.M. Bayesian Clustering Techniques and Progressive Partitioning to Identify Population Structuring Within a Recovering Otter Population in the UK. J. Appl. Ecol. 2011, 48, 1206–1217. [Google Scholar] [CrossRef]
- Lynch, J.M.; O’Sullivan, W.M. Cranial form and sexual dimorphism in the Irish otter Lutra L. In Biology and Environment: Proceedings of the Royal Irish Academy; Royal Irish Academy: Dublin, Ireland, 1993; pp. 97–105. [Google Scholar]
- Moorhouse-Gann, R.J.; Kean, E.F.; Parry, G.; Valladares, S.; Chadwick, E.A. Dietary complexity and hidden costs of prey switching in a generalist top predator. Ecol. Evol. 2020, 10, 6395–6408. [Google Scholar] [CrossRef]
- Lau, A.C.C.; Asahara, M.; Han, S.Y.; Kimura, J. Sexual dimorphism of the Eurasian otter (Lutra lutra) in South Korea: Craniodental geometric morphometry. J. Vet. Med. Sci. 2016, 16, 0018. [Google Scholar] [CrossRef][Green Version]
- Lynch, J.M.; Conroy, J.W.H.; Kitchener, A.C.; Jefferies, D.J.; Hayden, T.J. Variation in cranial form and sexual dimorphism among five European populations of the otter Lutra. J. Zool. 1996, 238, 81–96. [Google Scholar] [CrossRef]
- Wiig, Ø. Sexual dimorphism in the skull of minks Mustela vison, badgers Meles and otters Lutra. Zool. J. Linn. Soc. 1986, 87, 163–179. [Google Scholar] [CrossRef]
- Cootes, T.F.; Hill, A.; Taylor, C.J.; Haslam, J. Use of Active Shape Models for Locating Structure in Medical Images. Image Vis. Comput. 1994, 12, 355–365. [Google Scholar] [CrossRef]
- Cootes, T.F.; Taylor, C.J.; Cooper, D.H.; Graham, J. Active Shape Models—Their Training and Application. Comp. Vis. Image Und. 1995, 61, 38–59. [Google Scholar] [CrossRef]
- Hill, A.; Cootes, T.F.; Taylor, C.J. Active shape models and the shape approximation problem. Image Vis. Comput. 1996, 14, 601–607. [Google Scholar] [CrossRef]
- Taylor, C.J.; Cootes, T.F.; Lanitis, A.; Edwards, G.; Smyth, P.; Kotcheff, A.C.W. Model-based interpretation of complex and variable images. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1997, 352, 1267–1274. [Google Scholar] [CrossRef] [PubMed]
- Cootes, T.F.; Taylor, C.J. A mixture model for representing shape variation. Image Vis. Comput. 1999, 17, 567–573. [Google Scholar] [CrossRef]
- Cootes, T.F.; Edwards, G.J.; Taylor, C.J. Active appearance models. IEEE Trans. Pattern Anal. Mach. Intell. 2001, 23, 681–685. [Google Scholar] [CrossRef]
- Cootes, T.F.; Taylor, C.J. Anatomical statistical models and their role in feature extraction. Br. J. Radiol. 2004, 77, S133–S139. [Google Scholar] [CrossRef]
- Allen, P.D.; Graham, J.; Farnell, D.J.J.; Harrison, E.J.; Jacobs, R.; Nicopolou-Karayianni, K.; Lindh, C.; van der Stelt, P.F.; Horner, K.; Devlin, H. Detecting reduced bone mineral density from dental radiographs using statistical shape models. IEEE Trans. Inf. Technol. Biomed. 2007, 11, 601–610. [Google Scholar] [CrossRef]
- Bookstein, F.L. Pathologies of Between-Groups Principal Components Analysis in Geometric Morphometrics. Evol. Biol. 2019, 46, 271–302. [Google Scholar] [CrossRef]



| Sex (Measurement Type) | Length | Width | Height | |
|---|---|---|---|---|
| Male (Physical) | Mean (mm) | 100.53 | 70.53 | 41.30 |
| SD (mm) | 3.96 | 2.89 | 1.62 | |
| Female (Physical) | Mean (mm) | 93.68 | 65.09 | 39.64 |
| SD (mm) | 3.26 | 2.14 | 1.36 | |
| Male (GUI) | Mean (mm) | 100.38 | 70.10 | 41.84 |
| SD (mm) | 3.72 | 3.57 | 1.36 | |
| Female (GUI) | Mean (mm) | 93.44 | 63.92 | 39.14 |
| SD (mm) | 3.25 | 2.42 | 1.22 | |
| Region | Length | Width | Height | |
|---|---|---|---|---|
| Wales | Mean (mm) | 97.23 | 67.00 | 40.76 |
| SD (mm) | 4.08 | 3.10 | 1.45 | |
| SE England | Mean (mm) | 99.03 | 69.21 | 41.14 |
| SD (mm) | 6.57 | 4.98 | 1.95 | |
| SW England | Mean (mm) | 94.63 | 66.57 | 39.42 |
| SD (mm) | 4.34 | 2.90 | 1.45 | |
| North England/Scotland | Mean (mm) | 99.08 | 70.75 | 40.81 |
| SD (mm) | 4.17 | 2.39 | 1.75 | |
| ANOVA | F = 2.563; df = 3, 55; P = 0.064 | F = 3.991; df = 3, 55; P = 0.012 | F = 5.386; df = 3, 55; P = 0.03 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Farnell, D.J.J.; Khor, C.; Ayre, W.N.; Doyle, Z.; Chadwick, E.A. Initial Investigations of the Cranial Size and Shape of Adult Eurasian Otters (Lutra lutra) in Great Britain. J. Imaging 2020, 6, 106. https://doi.org/10.3390/jimaging6100106
Farnell DJJ, Khor C, Ayre WN, Doyle Z, Chadwick EA. Initial Investigations of the Cranial Size and Shape of Adult Eurasian Otters (Lutra lutra) in Great Britain. Journal of Imaging. 2020; 6(10):106. https://doi.org/10.3390/jimaging6100106
Chicago/Turabian StyleFarnell, Damian J. J., Chern Khor, Wayne Nishio Ayre, Zoe Doyle, and Elizabeth A. Chadwick. 2020. "Initial Investigations of the Cranial Size and Shape of Adult Eurasian Otters (Lutra lutra) in Great Britain" Journal of Imaging 6, no. 10: 106. https://doi.org/10.3390/jimaging6100106
APA StyleFarnell, D. J. J., Khor, C., Ayre, W. N., Doyle, Z., & Chadwick, E. A. (2020). Initial Investigations of the Cranial Size and Shape of Adult Eurasian Otters (Lutra lutra) in Great Britain. Journal of Imaging, 6(10), 106. https://doi.org/10.3390/jimaging6100106





