Applications of Computational Methods in Biomedical Breast Cancer Imaging Diagnostics: A Review
Abstract
1. Introduction
2. Types of Biomedical Imaging
2.1. Mammography
2.2. Tomosynthesis
2.3. Ultrasound Imaging
2.4. Dedicated Breast Computed Tomography
2.5. Magnetic Resonance Imaging
2.6. Diffusion-Weighted Imaging
2.7. Computed Tomography
2.8. Near-Infrared (NIR) Fluorescence
2.9. Single-Photon Emission Computed Tomography
3. Computational Techniques Used in Breast Cancer Imaging Diagnostics
3.1. Machine Learning Algorithms
3.1.1. Support Vector Machines
3.1.2. Artificial Neural Network
3.2. Deep Learning
3.2.1. Convolutional Neural Network
3.2.2. Generative Adversarial Networks
3.3. Robotics
3.3.1. Reinforcement Learning
3.3.2. Robotic Tools for Breast Cancer Diagnosis
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Adekiya, T.A.; Aruleba, R.T.; Khanyile, S.; Masamba, P.; Oyinloye, B.E.; Kappo, A.P. Structural analysis and epitope prediction of MHC class-1-chain related protein-a for cancer vaccine development. Vaccines 2018, 6, 1. [Google Scholar] [CrossRef]
- Aruleba, R.T.; Adekiya, T.A.; Oyinloye, B.E.; Kappo, A.P. Structural studies of predicted ligand binding sites and molecular docking analysis of Slc2a4 as a therapeutic target for the treatment of cancer. Int. J. Mol. Sci. 2018, 19, 386. [Google Scholar] [CrossRef]
- Oyinloye, B.E.; Adekiya, T.A.; Aruleba, R.T.; Ojo, O.A.; Ajiboye, B.O. Structure-Based Docking Studies of GLUT4 Towards Exploring Selected Phytochemicals from Solanum xanthocarpum as a Therapeutic Target for the Treatment of Cancer. Curr. Drug Discov. Technol. 2019, 16, 406–416. [Google Scholar] [CrossRef]
- Shapiro, C.L. Cancer survivorship. N. Engl. J. Med. 2018, 379, 2438–2450. [Google Scholar] [CrossRef]
- Akram, M.; Iqbal, M.; Daniyal, M.; Khan, A.U. Awareness and current knowledge of breast cancer. Biol. Res. 2017, 50, 33. [Google Scholar] [CrossRef]
- Singh, S.K.; Singh, S.; Lillard, J.W., Jr.; Singh, R. Drug delivery approaches for breast cancer. Int. J. Nanomed. 2017, 12, 6205. [Google Scholar] [CrossRef]
- Wang, H.; Zheng, B.; Yoon, S.W.; Ko, H.S. A support vector machine-based ensemble algorithm for breast cancer diagnosis. Eur. J. Oper. Res. 2018, 267, 687–699. [Google Scholar] [CrossRef]
- Dande, P.; Samant, P. Acquaintance to artificial neural networks and use of artificial intelligence as a diagnostic tool for tuberculosis: A review. Tuberculosis 2018, 108, 1–9. [Google Scholar] [CrossRef]
- Lengauer, T.; Sing, T. Bioinformatics-assisted anti-HIV therapy. Nat. Rev. Genet. 2006, 4, 790–797. [Google Scholar] [CrossRef]
- Vaishya, R.; Javaid, M.; Khan, I.H.; Haleem, A. Artificial Intelligence (AI) applications for COVID-19 pandemic. Diabet. Metab. Syndr. Clin. Res. Rev. 2020, 14, 337–339. [Google Scholar] [CrossRef]
- Nover, A.B.; Jagtap, S.; Anjum, W.; Yegingil, H.; Shih, W.Y.; Shih, W.-H.; Brooks, A.D. Modern breast cancer detection: A technological review. Int. J. Biomed. Imaging 2009, 2009, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Kolb, T.M.; Lichy, J.; Newhouse, J.H. Comparison of the performance of screening mammography, physical examination, and breast US and evaluation of factors that influence them: An analysis of 27825 patient evaluations. Radiology 2002, 225, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Nelson, H.D.; Tyne, K.; Naik, A.; Bougatsos, C.; Chan, B.K.; Humphrey, L. Screening for breast cancer: An update for the US Preventive Services Task Force. Ann. Intern. Med. 2009, 151, 727–737. [Google Scholar] [CrossRef] [PubMed]
- Bagni, B.; Franceschetto, A.; Casolo, A.; De Santis, M.; Bagni, I.; Pansini, F.; Di Leo, C. Scintimammography with 99mTc-MIBI and magnetic resonance imaging in the evaluation of breast cancer. Eur. J. Nucl. Med. Mol. Imaging 2003, 30, 1383–1388. [Google Scholar] [CrossRef]
- Lladó, X.; Oliver, A.; Freixenet, J.; Martí, R.; Martí, J. A textural approach for mass false positive reduction in mammography. Comput. Med. Imaging Graph. 2009, 33, 415–422. [Google Scholar] [CrossRef] [PubMed]
- Aiken, L.S.; West, S.G.; Woodward, C.K.; Reno, R.R. Health beliefs and compliance with mammography-screening recommendations in asymptomatic women. Health Psychol. 1994, 13, 122. [Google Scholar] [CrossRef]
- Kennedy, D.A.; Lee, T.; Seely, D. A comparative review of thermography as a breast cancer screening technique. Integr. Cancer Ther. 2009, 8, 9–16. [Google Scholar] [CrossRef]
- Schillaci, O.; Buscombe, J.R. Breast scintigraphy today: Indications and limitations. Eur. J. Nucl. Med. Mol. Imaging 2004, 31, S35–S45. [Google Scholar] [CrossRef]
- Cherel, P.; Hagay, C.; Benaim, B.; De Maulmont, C.; Engerand, S.; Langer, A.; Talma, V. Mammographic evaluation of dense breasts: Techniques and limits. J. Radiol. 2008, 89, 1156. [Google Scholar] [CrossRef]
- Mori, M.; Akashi-Tanaka, S.; Suzuki, S.; Daniels, M.I.; Watanabe, C.; Hirose, M.; Nakamura, S. Diagnostic accuracy of contrast-enhanced spectral mammography in comparison to conventional full-field digital mammography in a population of women with dense breasts. Breast Cancer 2017, 24, 104–110. [Google Scholar] [CrossRef]
- Jung, J.I.; Kim, H.H.; Park, S.H.; Song, S.W.; Chung, M.H.; Kim, H.S.; Kim, K.J.; Ahn, M.I.; Seo, S.B.; Hahn, S.T. Thoracic manifestations of breast cancer and its therapy. Radiographics 2004, 24, 1269–1285. [Google Scholar] [CrossRef] [PubMed]
- Savaridas, S.L.; Spratt, J.D.; Cox, J. Incidence and potential significance of internal mammary lymphadenopathy on computed tomography in patients with a diagnosis of primary breast cancer. Breast Cancer Basic Clin. Res. 2015, 9. [Google Scholar] [CrossRef] [PubMed]
- Dobbins, J.T. Tomosynthesis imaging: At a translational crossroads. Med. Phys. 2009, 36, 1956–1967. [Google Scholar] [CrossRef] [PubMed]
- Friedewald, S.M.; Rafferty, E.A.; Rose, S.L.; Durand, M.A.; Plecha, D.M.; Greenberg, J.S.; Hayes, M.K.; Copit, D.S.; Carlson, K.L.; Cink, T.M. Breast cancer screening using tomosynthesis in combination with digital mammography. JAMA 2014, 311, 2499–2507. [Google Scholar] [CrossRef] [PubMed]
- Niklason, L.T.; Christian, B.T.; Niklason, L.E.; Kopans, D.B.; Castleberry, D.E.; Opsahl-Ong, B.; Landberg, C.E.; Slanetz, P.J.; Giardino, A.A.; Moore, R. Digital tomosynthesis in breast imaging. Radiology 1997, 205, 399–406. [Google Scholar] [CrossRef] [PubMed]
- Poplack, S.P.; Tosteson, T.D.; Kogel, C.A.; Nagy, H.M. Digital breast tomosynthesis: Initial experience in 98 women with abnormal digital screening mammography. Am. J. Roentgenol. 2007, 189, 616–623. [Google Scholar] [CrossRef] [PubMed]
- Lång, K.; Andersson, I.; Rosso, A.; Tingberg, A.; Timberg, P.; Zackrisson, S. Performance of one-view breast tomosynthesis as a stand-alone breast cancer screening modality: Results from the Malmö Breast Tomosynthesis Screening Trial, a population-based study. Eur. Radiol. 2016, 26, 184–190. [Google Scholar] [CrossRef]
- Sechopoulos, I. A review of breast tomosynthesis. Part I. The image acquisition process. Med. Phys. 2013, 40, 014301. [Google Scholar] [CrossRef]
- Van de Sompel, D.; Brady, M.; Boone, J. Task-based performance analysis of FBP, SART and ML for digital breast tomosynthesis using signal CNR and Channelised Hotelling Observers. Med. Image Anal. 2011, 15, 53–70. [Google Scholar] [CrossRef]
- O’Brien, W.D., Jr. Ultrasound–biophysics mechanisms. Prog. Biophys. Mol. Biol. 2007, 93, 212–255. [Google Scholar]
- Mason, T.J. Therapeutic ultrasound an overview. Ultrason. Sonochem. 2011, 18, 847–852. [Google Scholar] [CrossRef]
- Dewall, R.J. Ultrasound elastography: Principles, techniques, and clinical applications. Crit. Rev. Biomed. Eng. 2013, 41, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Lu, G.; Qin, B.; Fei, B. Ultrasound Imaging Technologies for Breast Cancer Detection and Management: A Review. Ultrasound Med. Biol. 2018, 44, 37–70. [Google Scholar] [CrossRef]
- Thornton, G.D.; McPhail, M.J.W.; Nayagam, S.; Hewitt, M.J.; Vlavianos, P.; Monahan, K.J. Endoscopic ultrasound guided fine needle aspiration for the diagnosis of pancreatic cystic neoplasms: A meta-analysis. Pancreatology 2013, 13, 48–57. [Google Scholar] [CrossRef]
- Liu, R.; Adler, D.G. Duplication cysts: Diagnosis, management, and the role of endoscopic ultrasound. Endosc. Ultrasound 2014, 3, 152–160. [Google Scholar] [CrossRef]
- Park, M.K.; Jo, J.; Kwon, H.; Cho, J.H.; Oh, J.Y.; Noh, M.H.; Nam, K.J. Usefulness of acoustic radiation force impulse elastography in the differential diagnosis of benign and malignant solid pancreatic lesions. Ultrasonography 2014, 33, 26. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.-H.; Xu, H.-X.; Xie, X.-Y.; Lu, M.-D.; Kuang, M.; Xu, Z.-F.; Liu, G.-J.; Wang, Z.; Liang, J.-Y.; Chen, L.-D. Differential diagnosis between benign and malignant gallbladder diseases with real-time contrast-enhanced ultrasound. Eur. Radiol. 2010, 20, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Masroor, I.; Afzal, S.; Suffian, S.N. Imaging guided breast interventions. J. Coll. Physicians Surg. Pak. 2016, 26, 521–526. [Google Scholar]
- Giuliano, V.; Giuliano, C. Improved breast cancer detection in asymptomatic women using 3D-automated breast ultrasound in mammographically dense breasts. Clin. Imaging 2013, 37, 480–486. [Google Scholar] [CrossRef]
- Bachawal, S.V.; Jensen, K.C.; Lutz, A.M.; Gambhir, S.S.; Tranquart, F.; Tian, L.; Willmann, J.K. Earlier detection of breast cancer with ultrasound molecular imaging in a transgenic mouse model. Cancer Res. 2013, 73, 1689–1698. [Google Scholar] [CrossRef]
- Lindfors, K.K.; Boone, J.M.; Nelson, T.R.; Yang, K.; Kwan, A.L.; Miller, D.F. Dedicated breast CT: Initial clinical experience. Radiology 2008, 246, 725–733. [Google Scholar] [CrossRef]
- Kuzmiak, C.M.; Cole, E.B.; Zeng, D.; Tuttle, L.A.; Steed, D.; Pisano, E.D. Dedicated three-dimensional breast computed tomography: Lesion characteristic perception by radiologists. J. Clin. Imaging Sci. 2016, 6, 14. [Google Scholar] [CrossRef]
- Shah, J.P.; Mann, S.D.; McKinley, R.L.; Tornai, M.P. Characterization of CT Hounsfield units for 3D acquisition trajectories on a dedicated breast CT system. J. X-ray Sci. Technol. 2018, 26, 535–551. [Google Scholar] [CrossRef]
- Boone, J.M.; Nelson, T.R.; Lindfors, K.K.; Seibert, J.A. Dedicated breast CT: Radiation dose and image quality evaluation. Radiology 2001, 221, 657–667. [Google Scholar] [CrossRef]
- Sarno, A.; Mettivier, G.; Russo, P. Dedicated breast computed tomography: Basic aspects. Med. Phys. 2015, 42, 2786–2804. [Google Scholar] [CrossRef]
- Radhakrishna, S.; Agarwal, S.; Parikh, P.M.; Kaur, K.; Panwar, S.; Sharma, S.; Dey, A.; Saxena, K.; Chandra, M.; Sud, S. Role of magnetic resonance imaging in breast cancer management. South Asian J. Cancer 2018, 7, 69–71. [Google Scholar] [CrossRef]
- Sardanelli, F.; Giuseppetti, G.M.; Panizza, P.; Bazzocchi, M.; Fausto, A.; Simonetti, G.; Lattanzio, V.; Del Maschio, A. Sensitivity of MRI versus mammography for detecting foci of multifocal, multicentric breast cancer in fatty and dense breasts using the whole-breast pathologic examination as a gold standard. Am. J. Roentgenol. 2004, 183, 1149–1157. [Google Scholar] [CrossRef]
- Lee, C.H.; Dershaw, D.D.; Kopans, D.; Evans, P.; Monsees, B.; Monticciolo, D.; Brenner, R.J.; Bassett, L.; Berg, W.; Feig, S. Breast cancer screening with imaging: Recommendations from the Society of Breast Imaging and the ACR on the use of mammography, breast MRI, breast ultrasound, and other technologies for the detection of clinically occult breast cancer. J. Am. Coll. Radiol. 2010, 7, 18–27. [Google Scholar] [CrossRef]
- Morrow, M.; Waters, J.; Morris, E. MRI for breast cancer screening, diagnosis, and treatment. Lancet 2011, 378, 1804–1811. [Google Scholar] [CrossRef]
- Gareth, E.D.; Nisha, K.; Yit, L.; Soujanye, G.; Emma, H.; Massat, N.J.; Maxwell, A.J.; Sarah, I.; Rosalind, E.; Leach, M.O. MRI breast screening in high-risk women: Cancer detection and survival analysis. Breast Cancer Res. Treat. 2014, 145, 663–672. [Google Scholar] [CrossRef]
- Lehman, C.D.; Gatsonis, C.; Kuhl, C.K.; Hendrick, R.E.; Pisano, E.D.; Hanna, L.; Peacock, S.; Smazal, S.F.; Maki, D.D.; Julian, T.B. MRI evaluation of the contralateral breast in women with recently diagnosed breast cancer. N. Engl. J. Med. 2007, 356, 1295–1303. [Google Scholar] [CrossRef] [PubMed]
- Passaperuma, K.; Warner, E.; Causer, P.; Hill, K.; Messner, S.; Wong, J.; Jong, R.; Wright, F.; Yaffe, M.; Ramsay, E. Long-term results of screening with magnetic resonance imaging in women with BRCA mutations. Br. J. Cancer 2012, 107, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Bang, O.Y.; Li, W. Applications of diffusion-weighted imaging in diagnosis, evaluation, and treatment of acute ischemic stroke. Precis. Future Med. 2019, 3, 69–76. [Google Scholar] [CrossRef]
- Chung, J.W.; Park, S.H.; Kim, N.; Kim, W.J.; Park, J.H.; Ko, Y.; Yang, M.H.; Jang, M.S.; Han, M.K.; Jung, C. Trial of ORG 10172 in Acute Stroke Treatment (TOAST) classification and vascular territory of ischemic stroke lesions diagnosed by diffusion-weighted imaging. J. Am. Heart Assoc. 2014, 3, e001119. [Google Scholar] [CrossRef] [PubMed]
- Malayeri, A.A.; El Khouli, R.H.; Zaheer, A.; Jacobs, M.A.; Corona-Villalobos, C.P.; Kamel, I.R.; Macura, K.J. Principles and applications of diffusion-weighted imaging in cancer detection, staging, and treatment follow-up. Radiographics 2011, 31, 1773–1791. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.; Lucas-Quesada, F.A.; Sinha, U.; De Bruhl, N.; Bassett, L.W. In vivo diffusion-weighted MRI of the breast: Potential for lesion characterization. J. Magn. Reson. Imaging 2002, 15, 693–704. [Google Scholar] [CrossRef]
- Menezes, G.L.; Knuttel, F.M.; Stehouwer, B.L.; Pijnappel, R.M.; van den Bosch, M.A. Magnetic resonance imaging in breast cancer: A literature review and future perspectives. World J. Clin. Oncol. 2014, 5, 61. [Google Scholar] [CrossRef]
- Costantini, M.; Belli, P.; Rinaldi, P.; Bufi, E.; Giardina, G.; Franceschini, G.; Petrone, G.; Bonomo, L. Diffusion-weighted imaging in breast cancer: Relationship between apparent diffusion coefficient and tumour aggressiveness. Clin. Radiol. 2010, 65, 1005–1012. [Google Scholar] [CrossRef]
- Tan, S.; Rahmat, K.; Rozalli, F.; Mohd-Shah, M.; Aziz, Y.; Yip, C.; Vijayananthan, A.; Ng, K. Differentiation between benign and malignant breast lesions using quantitative diffusion-weighted sequence on 3 T MRI. Clin. Radiol. 2014, 69, 63–71. [Google Scholar] [CrossRef]
- Baltzer, P.A.; Bickel, H.; Spick, C.; Wengert, G.; Woitek, R.; Kapetas, P.; Clauser, P.; Helbich, T.H.; Pinker, K. Potential of noncontrast magnetic resonance imaging with diffusion-weighted imaging in characterization of breast lesions: Intraindividual comparison with dynamic contrast-enhanced magnetic resonance imaging. Investig. Radiol. 2018, 53, 229–235. [Google Scholar] [CrossRef]
- Chilla, G.S.; Tan, C.H.; Xu, C.; Poh, C.L. Diffusion weighted magnetic resonance imaging and its recent trend—A survey. Quant. Imaging Med. Surg. 2015, 5, 407. [Google Scholar] [PubMed]
- Baliyan, V.; Das, C.J.; Sharma, R.; Gupta, A.K. Diffusion weighted imaging: Technique and applications. World J. Radiol. 2016, 8, 785. [Google Scholar] [CrossRef] [PubMed]
- Scarfe, W.C.; Li, Z.; Aboelmaaty, W.; Scott, S.; Farman, A. Maxillofacial cone beam computed tomography: Essence, elements and steps to interpretation. Aust. Dent. J. 2012, 57, 46–60. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Lal, P. Recent development on computer aided tissue engineering—A review. Comput. Methods Programs Biomed. 2002, 67, 85–103. [Google Scholar] [CrossRef]
- Suga, K.; Yuan, Y.; Ogasawara, N.; Okada, M.; Matsunaga, N. Localization of breast sentinel lymph nodes by MR lymphography with a conventional gadolinium contrast agent: Preliminary observations in dongs and humans. Acta Radiol. 2003, 44, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Foo, S.Y.; Gray, K. Computed tomography (CT) staging in breast cancer. Clin. Radiol. 2015, 70, S13. [Google Scholar] [CrossRef]
- Okamura, Y.; Yoshizawa, N.; Yamaguchi, M.; Kashiwakura, I. Application of dual-energy computed tomography for breast cancer diagnosis. Int. J. Med. Phys. Clin. Eng. Radiat. Oncol. 2016, 5, 288–297. [Google Scholar] [CrossRef]
- Park, E.K.; Seo, B.K.; Kwon, M.; Cho, K.R.; Woo, O.H.; Song, S.E.; Cha, J.; Lee, H.Y. Low-dose perfusion computed tomography for breast cancer to quantify tumor vascularity: Correlation with prognostic biomarkers. Investig. Radiol. 2019, 54, 273–281. [Google Scholar] [CrossRef] [PubMed]
- Shah, J.P.; Mann, S.D.; McKinley, R.L.; Tornai, M.P. Three dimensional dose distribution comparison of simple and complex acquisition trajectories in dedicated breast CT. Med. Phys. 2015, 42, 4497–4510. [Google Scholar] [CrossRef]
- Hawrysz, D.J.; Sevick-Muraca, E.M. Developments toward diagnostic breast cancer imaging using near-infrared optical measurements and fluorescent contrast agents1. Neoplasia 2000, 2, 388–417. [Google Scholar] [CrossRef]
- Troyan, S.L.; Kianzad, V.; Gibbs-Strauss, S.L.; Gioux, S.; Matsui, A.; Oketokoun, R.; Ngo, L.; Khamene, A.; Azar, F.; Frangioni, J.V. The FLARE™ intraoperative near-infrared fluorescence imaging system: A first-in-human clinical trial in breast cancer sentinel lymph node mapping. Ann. Surg. Oncol. 2009, 16, 2943–2952. [Google Scholar] [CrossRef] [PubMed]
- Tagaya, N.; Yamazaki, R.; Nakagawa, A.; Abe, A.; Hamada, K.; Kubota, K.; Oyama, T. Intraoperative identification of sentinel lymph nodes by near-infrared fluorescence imaging in patients with breast cancer. Am. J. Surg. 2008, 195, 850–853. [Google Scholar] [CrossRef] [PubMed]
- Verbeek, F.P.; Troyan, S.L.; Mieog, J.S.D.; Liefers, G.-J.; Moffitt, L.A.; Rosenberg, M.; Hirshfield-Bartek, J.; Gioux, S.; van de Velde, C.J.; Vahrmeijer, A.L. Near-infrared fluorescence sentinel lymph node mapping in breast cancer: A multicenter experience. Breast Cancer Res. Treat. 2014, 143, 333–342. [Google Scholar] [CrossRef] [PubMed]
- Mieog, J.S.D.; Troyan, S.L.; Hutteman, M.; Donohoe, K.J.; Van Der Vorst, J.R.; Stockdale, A.; Liefers, G.-J.; Choi, H.S.; Gibbs-Strauss, S.L.; Putter, H. Toward optimization of imaging system and lymphatic tracer for near-infrared fluorescent sentinel lymph node mapping in breast cancer. Ann. Surg. Oncol. 2011, 18, 2483–2491. [Google Scholar] [CrossRef] [PubMed]
- Sevick-Muraca, E.M.; Sharma, R.; Rasmussen, J.C.; Marshall, M.V.; Wendt, J.A.; Pham, H.Q.; Bonefas, E.; Houston, J.P.; Sampath, L.; Adams, K.E. Imaging of lymph flow in breast cancer patients after microdose administration of a near-infrared fluorophore: Feasibility study. Radiology 2008, 246, 734–741. [Google Scholar] [CrossRef]
- Altınoǧlu, E.I.; Russin, T.J.; Kaiser, J.M.; Barth, B.M.; Eklund, P.C.; Kester, M.; Adair, J.H. Near-infrared emitting fluorophore-doped calcium phosphate nanoparticles for in vivo imaging of human breast cancer. ACS Nano 2008, 2, 2075–2084. [Google Scholar] [CrossRef] [PubMed]
- Poellinger, A.; Burock, S.; Grosenick, D.; Hagen, A.; Lüdemann, L.; Diekmann, F.; Engelken, F.; Macdonald, R.; Rinneberg, H.; Schlag, P.-M. Breast cancer: Early-and late-fluorescence near-infrared imaging with indocyanine green—A preliminary study. Radiology 2011, 258, 409–416. [Google Scholar] [CrossRef]
- Ke, S.; Wen, X.; Gurfinkel, M.; Charnsangavej, C.; Wallace, S.; Sevick-Muraca, E.M.; Li, C. Near-infrared optical imaging of epidermal growth factor receptor in breast cancer xenografts. Cancer Res. 2003, 63, 7870–7875. [Google Scholar]
- Vallabhajosula, S.; Polack, B.D.; Babich, J.W. Molecular Imaging of Prostate Cancer: Radiopharmaceuticals for Positron Emission Tomography (PET) and Single-Photon Emission Computed Tomography (SPECT). In Precision Molecular Pathology of Prostate Cancer; Springer: Berlin/Heidelberg, Germany, 2018; pp. 475–501. [Google Scholar]
- Pellikka, P.A.; She, L.; Holly, T.A.; Lin, G.; Varadarajan, P.; Pai, R.G.; Bonow, R.O.; Pohost, G.M.; Panza, J.A.; Berman, D.S. Variability in ejection fraction measured by echocardiography, gated single-photon emission computed tomography, and cardiac magnetic resonance in patients with coronary artery disease and left ventricular dysfunction. JAMA Netw. Open 2018, 1, e181456. [Google Scholar] [CrossRef]
- Noyce, A.J.; Dickson, J.; Rees, R.N.; Bestwick, J.P.; Isaias, I.U.; Politis, M.; Giovannoni, G.; Warner, T.T.; Lees, A.J.; Schrag, A. Dopamine reuptake transporter-single-photon emission computed tomography and transcranial sonography as imaging markers of prediagnostic Parkinson’s disease. Mov. Disord. 2018, 33, 478–482. [Google Scholar] [CrossRef]
- Frangioni, J.V. New technologies for human cancer imaging. J. Clin. Oncol. 2008, 26, 4012. [Google Scholar] [CrossRef] [PubMed]
- Koizumi, M.; Koyama, M. Comparison between single photon emission computed tomography with computed tomography and planar scintigraphy in sentinel node biopsy in breast cancer patients. Ann. Nucl. Med. 2019, 33, 160–168. [Google Scholar] [CrossRef] [PubMed]
- Van der Ploeg, I.M.; Nieweg, O.E.; Kroon, B.B.; Rutgers, E.J.; Baas-Vrancken Peeters, M.J.; Vogel, W.V.; Hoefnagel, C.A.; Olmos, R.A. The yield of SPECT/CT for anatomical lymphatic mapping in patients with breast cancer. Eur. J. Nucl. Med. Mol. Imaging 2009, 36, 903–909. [Google Scholar] [CrossRef] [PubMed]
- Maza, S.; Valencia, R.; Geworski, L.; Zander, A.; Guski, H.; Winzer, K.J.; Munz, D.L. Peritumoural versus subareolar administration of technetium-99m nanocolloid for sentinel lymph node detection in breast cancer: Preliminary results of a prospective intra-individual comparative study. Eur. J. Nucl. Med. Mol. Imaging 2003, 30, 651–656. [Google Scholar] [CrossRef] [PubMed]
- Lerman, H.; Lievshitz, G.; Zak, O.; Metser, U.; Schneebaum, S.; Even-Sapir, E. Improved sentinel node identification by SPECT/CT in overweight patients with breast cancer. J. Nucl. Med. 2007, 48, 201–206. [Google Scholar]
- Pecking, A.P.; Wartski, W.; Cluzan, R.; Bellet, D.; Albérini, J. SPECT–CT fusion imaging radionuclide lymphoscintigraphy: Potential for limb lymphedema assessment and sentinel node detection in breast cancer. In Cancer Metastasis and the Lymphovascular System: Basis for Rational Therapy; Springer: Berlin/Heidelberg, Germany, 2007; pp. 79–84. [Google Scholar]
- Mann, S.D.; Perez, K.L.; McCracken, E.K.; Shah, J.P.; Wong, T.Z.; Tornai, M.P. Initial in vivo quantification of Tc-99m sestamibi uptake as a function of tissue type in healthy breasts using dedicated breast SPECT-CT. J. Oncol. 2012, 2012, 1–7. [Google Scholar] [CrossRef]
- Bowen, S.L.; Wu, Y.; Chaudhari, A.J.; Fu, L.; Packard, N.J.; Burkett, G.W.; Yang, K.; Lindfors, K.K.; Shelton, D.K.; Hagge, R. Initial characterization of a dedicated breast PET/CT scanner during human imaging. J. Nucl. Med. 2009, 50, 1401–1408. [Google Scholar] [CrossRef][Green Version]
- Tornai, M.P.; Shah, J.P.; Mann, S.D.; McKinley, R.L. Development of Fully-3D CT in a Hybrid SPECT-CT Breast Imaging System. In Proceedings of the 13th International Workshop on Breast Imaging, Malmo, Sweden, 19–22 June 2016; pp. 567–575. [Google Scholar]
- Crotty, D.J.; Brady, S.L.; Jackson, D.V.C.; Toncheva, G.I.; Anderson, C.E.; Yoshizumi, T.T.; Tornai, M.P. Evaluation of the absorbed dose to the breast using radiochromic film in a dedicated CT mammotomography system employing a quasi-monochromatic X-ray beam. Med. Phys. 2011, 38, 3232–3245. [Google Scholar] [CrossRef]
- Suthaharan, S. Machine learning models and algorithms for big data classification. Integr. Ser. Inf. Syst. 2016, 36, 1–12. [Google Scholar]
- Tsai, H.-H.; Chang, Y.-C. Facial expression recognition using a combination of multiple facial features and support vector machine. Soft Comput. 2018, 22, 4389–4405. [Google Scholar] [CrossRef]
- Manavalan, B.; Shin, T.H.; Lee, G. PVP-SVM: Sequence-based prediction of phage virion proteins using a support vector machine. Front. Microbiol. 2018, 9, 476. [Google Scholar] [CrossRef]
- Chen, S.; Wu, X.; Zhang, R. A novel twin support vector machine for binary classification problems. Neural Process. Lett. 2016, 44, 795–811. [Google Scholar] [CrossRef]
- Acharya, U.R.; Ng, E.Y.-K.; Tan, J.-H.; Sree, S.V. Thermography based breast cancer detection using texture features and support vector machine. J. Med. Syst. 2012, 36, 1503–1510. [Google Scholar] [CrossRef]
- Maglogiannis, I.; Zafiropoulos, E.; Anagnostopoulos, I. An intelligent system for automated breast cancer diagnosis and prognosis using SVM based classifiers. Appl. Intell. 2009, 30, 24–36. [Google Scholar] [CrossRef]
- Huang, Y.-L.; Wang, K.-L.; Chen, D.-R. Diagnosis of breast tumors with ultrasonic texture analysis using support vector machines. Neural Comput. Appl. 2006, 15, 164–169. [Google Scholar] [CrossRef]
- Abu-Elanien, A.E.; Salama, M.; Ibrahim, M. Determination of transformer health condition using artificial neural networks. In Proceedings of the 19th International Symposium on Innovations in Intelligent Systems and Applications, Warsaw, Poland, 28–30 June 2011; pp. 1–5. [Google Scholar]
- Lisboa, P.J.; Taktak, A.F. The use of artificial neural networks in decision support in cancer: A systematic review. Neural Netw. 2006, 19, 408–415. [Google Scholar] [CrossRef] [PubMed]
- Abbass, H.A. An evolutionary artificial neural networks approach for breast cancer diagnosis. Artif. Intell. Med. 2002, 25, 265–281. [Google Scholar] [CrossRef]
- Tourassi, G.D.; Markey, M.K.; Lo, J.Y.; Floyd, C.E., Jr. A neural network approach to breast cancer diagnosis as a constraint satisfaction problem. Med. Phys. 2001, 28, 804–811. [Google Scholar] [CrossRef] [PubMed]
- Janghel, R.; Shukla, A.; Tiwari, R.; Kala, R. Breast cancer diagnosis using artificial neural network models. In Proceedings of the 3rd International Conference on Information Sciences and Interaction Sciences, Chengdu, China, 23–25 June 2010; pp. 89–94. [Google Scholar]
- Delen, D.; Walker, G.; Kadam, A. Predicting breast cancer survivability: A comparison of three data mining methods. Artif. Intell. Med. 2005, 34, 113–127. [Google Scholar] [CrossRef]
- Sarvestani, A.S.; Safavi, A.; Parandeh, N.; Salehi, M. Predicting breast cancer survivability using data mining techniques. In Proceedings of the 2nd International Conference on Software Technology and Engineering, San Juan, PR, USA, 3–5 October 2010; p. V2-227. [Google Scholar]
- Çakır, A.; Demirel, B. A software tool for determination of breast cancer treatment methods using data mining approach. J. Med. Syst. 2011, 35, 1503–1511. [Google Scholar] [CrossRef]
- Şahan, S.; Polat, K.; Kodaz, H.; Güneş, S. A new hybrid method based on fuzzy-artificial immune system and k-nn algorithm for breast cancer diagnosis. Comput. Biol. Med. 2007, 37, 415–423. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, A.; Tiwari, A. Breast cancer diagnosis using genetically optimized neural network model. Expert Syst. Appl. 2015, 42, 4611–4620. [Google Scholar] [CrossRef]
- Karabatak, M. A new classifier for breast cancer detection based on Naïve Bayesian. Measurement 2015, 72, 32–36. [Google Scholar] [CrossRef]
- Bagui, S.C.; Bagui, S.; Pal, K.; Pal, N.R. Breast cancer detection using rank nearest neighbor classification rules. Pattern Recognit. 2003, 36, 25–34. [Google Scholar] [CrossRef]
- Chen, H.-L.; Yang, B.; Liu, J.; Liu, D.-Y. A support vector machine classifier with rough set-based feature selection for breast cancer diagnosis. Expert Syst. Appl. 2011, 38, 9014–9022. [Google Scholar] [CrossRef]
- Polat, K.; Güneş, S. Breast cancer diagnosis using least square support vector machine. Digit. Signal Process. 2007, 17, 694–701. [Google Scholar] [CrossRef]
- Le Cun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Zhao, R.; Yan, R.; Chen, Z.; Mao, K.; Wang, P.; Gao, R.X. Deep learning and its applications to machine health monitoring. Mech. Syst. Signal Process. 2019, 115, 213–237. [Google Scholar] [CrossRef]
- Zou, J.; Huss, M.; Abid, A.; Mohammadi, P.; Torkamani, A.; Telenti, A. A primer on deep learning in genomics. Nat. Genet. 2019, 51, 12–18. [Google Scholar] [CrossRef]
- Lee, S.M.; Seo, J.B.; Yun, J.; Cho, Y.-H.; Vogel-Claussen, J.; Schiebler, M.L.; Gefter, W.B.; Van Beek, E.J.; Goo, J.M.; Lee, K.S. Deep Learning Applications in Chest Radiography and Computed Tomography. J. Thorac. Imaging 2019, 34, 75–85. [Google Scholar] [CrossRef]
- Mishra, S.; Glaws, A.; Palanisamy, P. Predictive Analytics in Future Power Systems: A Panorama and State-Of-The-Art of Deep Learning Applications. In Optimization, Learning, and Control for Interdependent Complex Networks; Springer: Berlin/Heidelberg, Germany, 2020; pp. 147–182. [Google Scholar]
- Kalchbrenner, N.; Grefenstette, E.; Blunsom, P. A convolutional neural network for modelling sentences. arXiv 2014, arXiv:1404.2188. [Google Scholar]
- Cong, I.; Choi, S.; Lukin, M.D. Quantum convolutional neural networks. Nat. Phys. 2019, 15, 1273–1278. [Google Scholar] [CrossRef]
- Coley, C.W.; Jin, W.; Rogers, L.; Jamison, T.F.; Jaakkola, T.S.; Green, W.H.; Barzilay, R.; Jensen, K.F. A graph-convolutional neural network model for the prediction of chemical reactivity. Chem. Sci. 2019, 10, 370–377. [Google Scholar] [CrossRef] [PubMed]
- Indolia, S.; Goswami, A.K.; Mishra, S.; Asopa, P. Conceptual understanding of convolutional neural network-a deep learning approach. Procedia Comput. Sci. 2018, 132, 679–688. [Google Scholar] [CrossRef]
- Amit, G.; Ben-Ari, R.; Hadad, O.; Monovich, E.; Granot, N.; Hashoul, S. Classification of breast MRI lesions using small-size training sets: Comparison of deep learning approaches. In Proceedings of the Medical Imaging: Computer-Aided Diagnosis Conference, Orlando, FL, USA, 11–16 February 2017; p. 101341H. [Google Scholar]
- Nahid, A.-A.; Mehrabi, M.A.; Kong, Y. Histopathological breast cancer image classification by deep neural network techniques guided by local clustering. BioMed Res. Int. 2018. [Google Scholar] [CrossRef] [PubMed]
- Murtaza, G.; Shuib, L.; Mujtaba, G.; Raza, G. Breast cancer multi-classification through deep neural network and hierarchical classification approach. Multimed. Tools Appl. 2019. [Google Scholar] [CrossRef]
- Dabeer, S.; Khan, M.M.; Islam, S. Cancer diagnosis in histopathological image: CNN based approach. Inform. Med. Unlocked 2019, 16, 100231. [Google Scholar] [CrossRef]
- Tan, Y.; Sim, K.; Ting, F. Breast cancer detection using convolutional neural networks for mammogram imaging system. In Proceedings of the 27th International Conference on Robotics, Automation and Sciences (ICORAS), Melaka, Malaysia, 27–29 November 2017; pp. 1–5. [Google Scholar]
- Byra, M.; Piotrzkowska-Wróblewska, H.; Dobruch-Sobczak, K.; Nowicki, A. Combining Nakagami imaging and convolutional neural network for breast lesion classification. In Proceedings of the 2017 IEEE International Ultrasonics Symposium (IUS), Washington, DC, USA, 6–9 September 2017; pp. 1–4. [Google Scholar]
- Gao, F.; Wu, T.; Li, J.; Zheng, B.; Ruan, L.; Shang, D.; Patel, B. SD-CNN: A shallow-deep CNN for improved breast cancer diagnosis. Comput. Med. Imaging Graph. 2018, 70, 53–62. [Google Scholar] [CrossRef]
- Wang, Z.; Li, M.; Wang, H.; Jiang, H.; Yao, Y.; Zhang, H.; Xin, J. Breast cancer detection using extreme learning machine based on feature fusion with CNN deep features. IEEE Access 2019, 7, 105146–105158. [Google Scholar] [CrossRef]
- Litjens, G.; Sánchez, C.I.; Timofeeva, N.; Hermsen, M.; Nagtegaal, I.; Kovacs, I.; Hulsbergen-Van De Kaa, C.; Bult, P.; Van Ginneken, B.; Van Der Laak, J. Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis. Sci. Rep. 2016, 6, 26286. [Google Scholar] [CrossRef]
- Sun, W.; Tseng, T.-L.B.; Zhang, J.; Qian, W. Enhancing deep convolutional neural network scheme for breast cancer diagnosis with unlabeled data. Comput. Med. Imaging Graph. 2017, 57, 4–9. [Google Scholar] [CrossRef] [PubMed]
- Araújo, T.; Aresta, G.; Castro, E.; Rouco, J.; Aguiar, P.; Eloy, C.; Polónia, A.; Campilho, A. Classification of breast cancer histology images using convolutional neural networks. PLoS ONE 2017, 12, e0177544. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Julio, Y.F.; Prieto-Guevara, M.J.; Nieto-Bernal, W.; Meriño-Fuentes, I.; Guerrero-Avendaño, A. Framework for the development of data-driven Mamdani-type fuzzy clinical decision support systems. Diagnostics 2019, 9, 52. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Liu, L.; Wang, Z.; Dai, G.; Xie, Y. Transferring deep neural networks for the differentiation of mammographic breast lesions. Sci. China Technol. Sci. 2019, 62, 441–447. [Google Scholar] [CrossRef]
- Ragab, D.A.; Sharkas, M.; Marshall, S.; Ren, J. Breast cancer detection using deep convolutional neural networks and support vector machines. PeerJ 2019, 7, e6201. [Google Scholar] [CrossRef]
- Acharya, S.; Alsadoon, A.; Prasad, P.; Abdullah, S.; Deva, A. Deep convolutional network for breast cancer classification: Enhanced loss function (ELF). J. Supercomput. 2020. [Google Scholar] [CrossRef]
- Goodfellow, I.J. On distinguishability criteria for estimating generative models. arXiv 2014, arXiv:1412.6515. [Google Scholar]
- Kazeminia, S.; Baur, C.; Kuijper, A.; van Ginneken, B.; Navab, N.; Albarqouni, S.; Mukhopadhyay, A. GANs for medical image analysis. Artif. Intell. Med. 2020. [Google Scholar] [CrossRef]
- Odena, A.; Olah, C.; Shlens, J. Conditional image synthesis with auxiliary classifier gans. In Proceedings of the 34th International Conference on Machine Learning (ICML), Sydney, Australia, 6–11 August 2017; pp. 2642–2651. [Google Scholar]
- Son, J.; Park, S.J.; Jung, K.-H. Retinal vessel segmentation in fundoscopic images with generative adversarial networks. arXiv 2017, arXiv:1706.09318. [Google Scholar]
- Pan, Z.; Yu, W.; Yi, X.; Khan, A.; Yuan, F.; Zheng, Y. Recent progress on generative adversarial networks (GANs): A survey. IEEE Access 2019, 7, 36322–36333. [Google Scholar] [CrossRef]
- Mirza, M.; Osindero, S. Conditional generative adversarial nets. arXiv 2014, arXiv:1411.1784. [Google Scholar]
- Shams, S.; Platania, R.; Zhang, J.; Kim, J.; Lee, K.; Park, S.-J. Deep generative breast cancer screening and diagnosis. In Proceedings of the 21st International Conference on Medical Image Computing and Computer-Assisted Intervention, Granada, Spain, 16–20 September 2018; pp. 859–867. [Google Scholar]
- Singh, V.K.; Romani, S.; Rashwan, H.A.; Akram, F.; Pandey, N.; Sarker, M.M.K.; Abdulwahab, S.; Torrents-Barrena, J.; Saleh, A.; Arquez, M. Conditional generative adversarial and convolutional networks for X-ray breast mass segmentation and shape classification. In Proceedings of the 21st International Conference on Medical Image Computing and Computer-Assisted Intervention, Granada, Spain, 16–20 September 2018; pp. 833–840. [Google Scholar]
- Wu, E.; Wu, K.; Cox, D.; Lotter, W. Conditional infilling GANs for data augmentation in mammogram classification. In Image Analysis for Moving Organ, Breast, and Thoracic Images; Springer: Berlin/Heidelberg, Germany, 2018; pp. 98–106. [Google Scholar]
- Guan, S.; Loew, M. Breast cancer detection using synthetic mammograms from generative adversarial networks in convolutional neural networks. J. Med. Imaging 2019, 6, 031411. [Google Scholar] [CrossRef] [PubMed]
- Karthik, S.; Perumal, R.S.; Mouli, P.C. Breast cancer classification using deep neural networks. In Knowledge Computing and Its Applications; Springer: Berlin/Heidelberg, Germany, 2018; pp. 227–241. [Google Scholar]
- Hadad, O.; Bakalo, R.; Ben-Ari, R.; Hashoul, S.; Amit, G. Classification of breast lesions using cross-modal deep learning. In Proceedings of the IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017), Melbourne, Australia, 18–21 April 2017; pp. 109–112. [Google Scholar]
- Wu, E.; Wu, K.; Lotter, W. Synthesizing lesions using contextual GANs improves breast cancer classification on mammograms. arXiv 2020, arXiv:2006.00086. [Google Scholar]
- Sutton, S.R.; Barto, G.A. Reinforcement Learning: An Introduction. In A Bradford Book; The MIT Press: Cambridge, MA, USA, 2015. [Google Scholar]
- Kato, I.; Koganezawa, K.; Takanishi, A. Automatic breast cancer palpation robot: WAPRO-4. Adv. Robot. 1988, 3, 251–261. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Suzuki, M.; Kato, A.; Konishi, K.; Hashizume, M.; Fujie, M.G. A robotic palpation-based needle insertion method for diagnostic biopsy and treatment of breast cancer. In Proceedings of the IEEE/RSJ International Conference on Intelligent Robots and Systems, St Louis, MO, USA, 11–15 October 2009; pp. 5534–5539. [Google Scholar]
- Larson, B.T.; Tsekos, N.V.; Erdman, A.G. A robotic device for minimally invasive breast interventions with real-time MRI guidance. In Proceedings of the Third IEEE Symposium on Bioinformatics and Bioengineering, Bethesda, MD, USA, 10–12 March 2003; pp. 190–197. [Google Scholar]
- Maicas, G.; Carneiro, G.; Bradley, A.P.; Nascimento, J.C.; Reid, I. Deep reinforcement learning for active breast lesion detection from DCE-MRI. In Proceedings of the 20th International Conference on Medical Image Computing and Computer-Assisted Intervention, Quebec City, QC, Canada, 10–14 September 2017; pp. 665–673. [Google Scholar]
- Tsekos, N.V.; Shudy, J.; Yacoub, E.; Tsekos, P.V.; Koutlas, I.G. Development of a robotic device for MRI-guided interventions in the breast. In Proceedings of the 2nd Annual IEEE International Symposium on Bioinformatics and Bioengineering (BIBE 2001), Bethesda, MD, USA, 4–6 November 2001; pp. 201–208. [Google Scholar]
- Mallapragada, V.; Sarkar, N.; Podder, T.K. Toward a robot-assisted breast intervention system. IEEE/ASME Trans. Mechatron. 2010, 16, 1011–1020. [Google Scholar] [CrossRef]
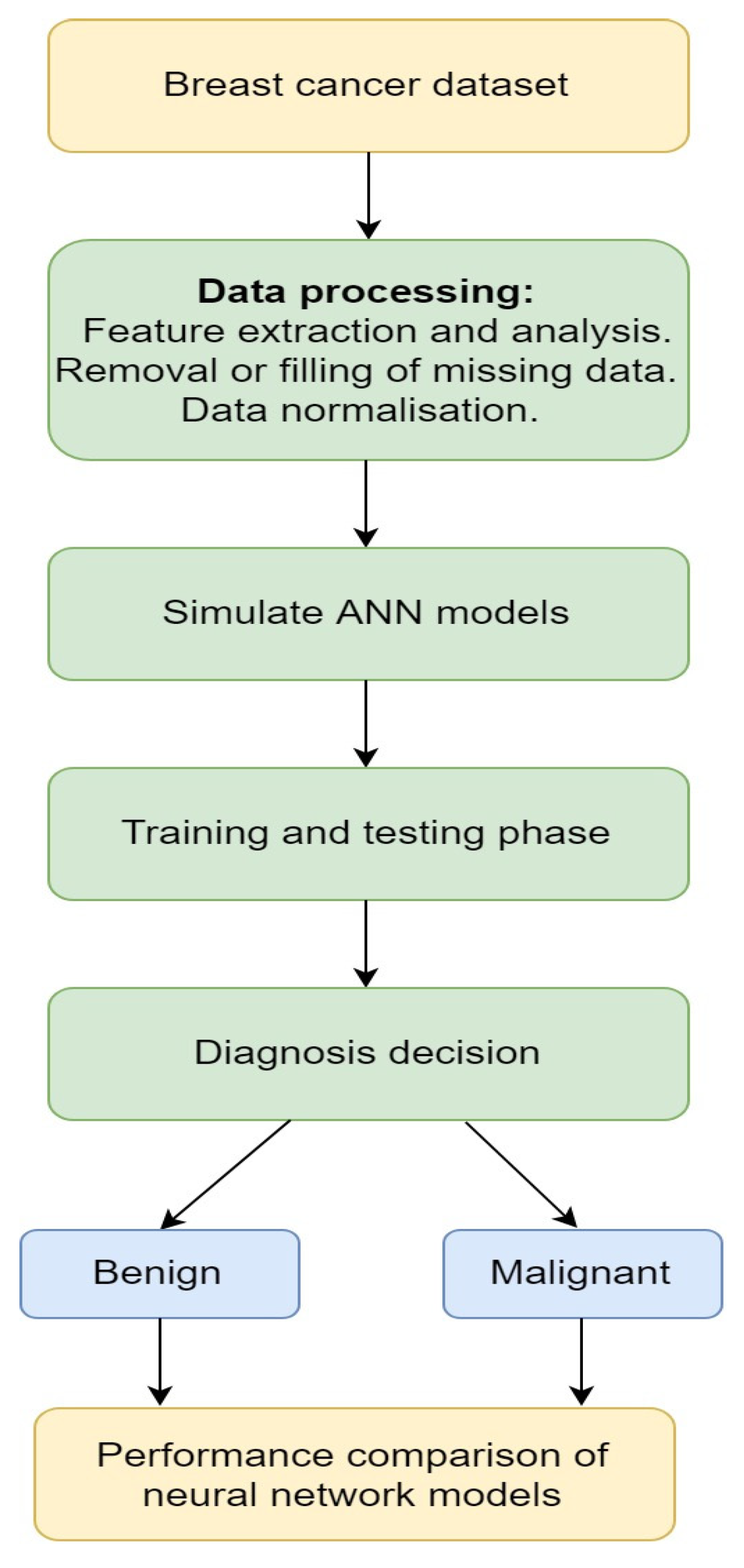
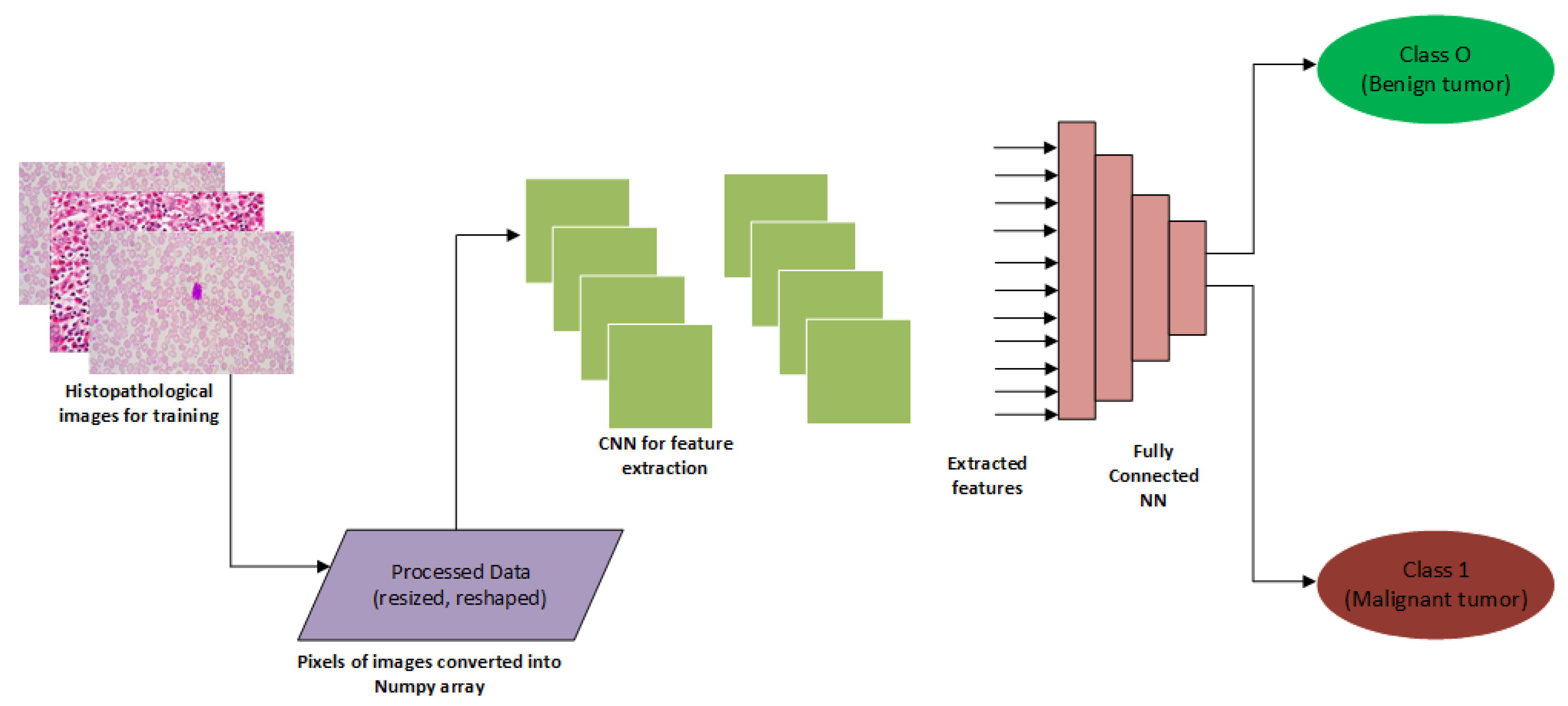

| Reference | Computation Technique | Scope | Evaluation Results | Datasets |
|---|---|---|---|---|
| Acharya et al. [96] | Texture features + SVM | Breast cancer detection using thermal imaging | Accuracy = 88.10%, specificity = 90.48%, sensitivity = 85.71% | 25 normal and 25 cancerous collected from Singapore General Hospital, Singapore |
| Maglogiannis et al. [97] | SVM | Diagnosis and prognosis | Accuracy = 96.91%, specificity = 97.67%, Sensitivity = 97.84% | Wisconsin prognostic breast cancer (WPBC) |
| Huang et al. [98] | SVM | Classifying benign and malignant | Accuracy = 94.4%, specificity = 94.4%, Sensitivity = 94.3% | 250 images of benign breast tumors from 215 patients and carcinomas from 35 patients. |
| Wang et al. [7] | SVM | Reduce the diagnosis variance and increase the diagnostic accuracy of breast cancer | Variance = 97.89%, increase in accuracy by 33.34% | Wisconsin Breast Cancer, Wisconsin Diagnostic Breast Cancer, and the U.S. National Cancer Institute’s Surveillance, Epidemiology, and End Results (SEER) program |
| Abbass [101] | EANN | Diagnosis | Average accuracy = 0.981 ± 0.005 | Wisconsin |
| Bhardwaj et al. [108] | Genetically optimized neural network | Classification | Accuracy of 98.24%, 99.63% and 100% for 50–50, 60–40, 70–30 training–testing partition, respectively | WBCD |
| Tourassi et al. [102] | CSNN | Diagnosis | CSNN ROC area index = 0.84 ± 0.02 | 500 private images |
| Çakır et al. [106] | Weka | Treatment methods | Accuracy = 92% | 462 patients data |
| Karabatak [109] | Weighted Naïve Bayesian | Detection | Sensitivity = 99.11%, specificity = 98.25%, accuracy = 98.54% | WBCD |
| Şahan et al. [107] | Fuzzy + KNN | Diagnosis | Accuracy = 99.14% | WBCD |
| Bagui et al. [110] | Rank nearest neighbor | Diagnosis | Accuracy = 98.1% | WBCD |
| Chen et al. [111] | Rough set_SVM | Distinguishing benign breast tumour from malignant one | Accuracy = 99.41%, Sensitivity = 100%, specificity = 100% | WBCD |
| Polat et al. [112] | Least square SVM | Classification | Accuracy = 94.87%, Sensitivity = 96.42%, specificity = 95.86% | WBCD |
| Reference | Deep Learning Technique | Scope | Evaluation Results | Datasets |
|---|---|---|---|---|
| Tan et al. [126] | CNN | Classification | Accuracy = 82% | mini-Mammographic Image Analysis Society (mini-MIAS) |
| Amit et al. [122] | CNN | Classification | Accuracy = 83%, Area under the curve = 0.91 | ED (MRI) |
| Byra et al. [127] | CNN | Classification | Accuracy = 83%, Area under the curve = 0.912 | ED (US, Nakagami) |
| Gao et al. [128] | CNN | Classification | Accuracy = 90% Area under the curve = 0.92 | INbreast |
| Wang et al. [129] | CNN | Classification | Accuracy = 76.5% | Private |
| Tan et al. [126] | CNN | Classification | Accuracy = 95%, Area under the curve = 0.97 | BreakHis |
| Litjens et al. [130] | CNN | Classification | Area under the curve = 0.99 | Private |
| Araújo et al. [132] | CNN | Classification | Accuracy = 77.8% (four classes), Accuracy = 83.3% (two classes) | BICBH |
| Ragab et al. [135] | CNN with SVM | Feature extraction | Accuracy = 73%, Area under the curve = 0.94 | Digital Database for Screening Mammography (DDSM), CBIS-DDSM |
| Acharya et al. [136] | CNN with K-means | Feature extraction | Accuracy = 97% | Private |
| Karthik et al. [147] | DNN | Classification | Accuracy = 98% | WBC |
| Yu et al. [134] | DNN + CNN | Classification | Accuracy = 81%, Area under the curve = 0.88 | BCDR |
| Sun et al. [131] | CNN | Classification | Accuracy = 82.43%, Area under the curve = 0.8818 | ED(Mg) |
| Hadad et al. [148] | CNN | Classification | Accuracy = 94%, Area under the curve = 0.98 | ED(Mg, MRI) |
| Nahid et al. [123] | CNN | Classification | Accuracy = 91% | BreakHis |
| Shams et al. [143] | GANs | Classification | Area under the curve = 0.88, Area under the curve = 0.925 | DDSM, INbreast |
| Singh et al. [144] | GANs + CNN | Classification | Accuracy = 72% | DDSM and Private |
| Wu et al. [149] | GANs | Classification | Accuracy = 89% | DDSM |
| Guan et al. [146] | GANs | Classification | Accuracy = 79.8% | DDSM |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aruleba, K.; Obaido, G.; Ogbuokiri, B.; Fadaka, A.O.; Klein, A.; Adekiya, T.A.; Aruleba, R.T. Applications of Computational Methods in Biomedical Breast Cancer Imaging Diagnostics: A Review. J. Imaging 2020, 6, 105. https://doi.org/10.3390/jimaging6100105
Aruleba K, Obaido G, Ogbuokiri B, Fadaka AO, Klein A, Adekiya TA, Aruleba RT. Applications of Computational Methods in Biomedical Breast Cancer Imaging Diagnostics: A Review. Journal of Imaging. 2020; 6(10):105. https://doi.org/10.3390/jimaging6100105
Chicago/Turabian StyleAruleba, Kehinde, George Obaido, Blessing Ogbuokiri, Adewale Oluwaseun Fadaka, Ashwil Klein, Tayo Alex Adekiya, and Raphael Taiwo Aruleba. 2020. "Applications of Computational Methods in Biomedical Breast Cancer Imaging Diagnostics: A Review" Journal of Imaging 6, no. 10: 105. https://doi.org/10.3390/jimaging6100105
APA StyleAruleba, K., Obaido, G., Ogbuokiri, B., Fadaka, A. O., Klein, A., Adekiya, T. A., & Aruleba, R. T. (2020). Applications of Computational Methods in Biomedical Breast Cancer Imaging Diagnostics: A Review. Journal of Imaging, 6(10), 105. https://doi.org/10.3390/jimaging6100105








