From Detection to Motion-Based Classification: A Two-Stage Approach for T. cruzi Identification in Video Sequences
Abstract
1. Introduction
2. Related Work
3. Materials and Methods
3.1. Database
3.2. From Video Processing to Motion-Based Classification
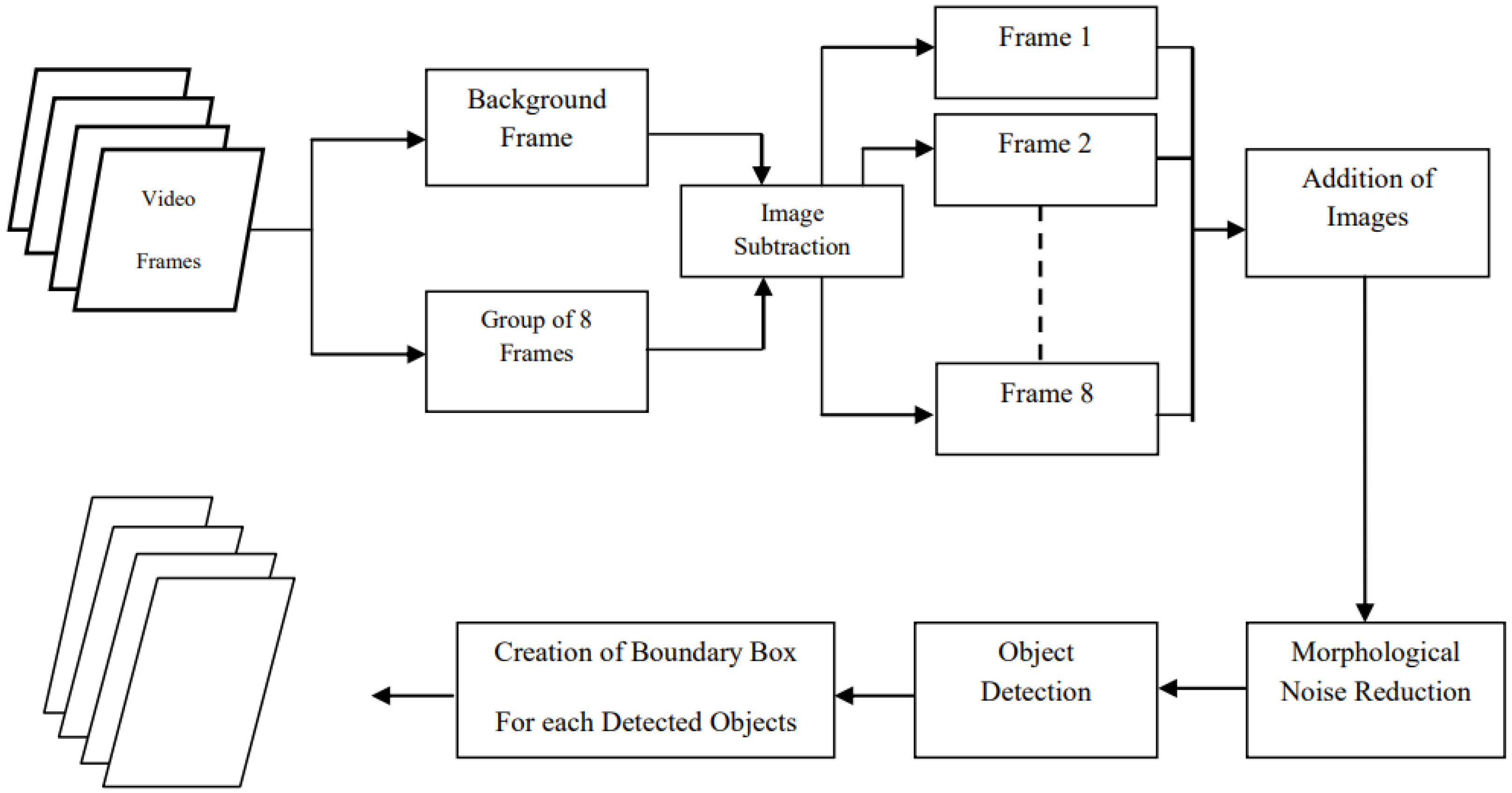
3.2.1. Adaptive Motion Detection Algorithm and Preprocessing
Frame Differencing Technique
3.2.2. Algorithm Implementation
Frame Differencing Algorithm
| Algorithm 1 Motion Detection using Frame Differencing |
| Require: Video sequence Ensure: Motion saliency maps 1: for each frame pair do 2: Convert frames to grayscale 3: Apply Gaussian smoothing ( kernel) 4: Calculate absolute difference: 5: Apply threshold: 6: Morphological filtering (opening + closing, ellipse) 7: Generate saliency map: 8: end for |
Frame Enhancement
| Algorithm 2 Adaptive Frame Enhancement |
| Require: RGB frame I Ensure: Enhanced frame 1: Convert I from BGR to LAB colour space 2: Split into L, A, B channels 3: Apply CLAHE to L-channel: 4: 5: Merge enhanced with original A, B channels 6: Convert back to BGR colour space 7: return |
Parasite Detection and Localization
| Algorithm 3 Parasite Detection Pipeline |
| Require: Saliency map , Enhanced frame I Ensure: Refined parasite locations 1: Threshold saliency map (threshold = 30) 2: Apply morphological operations ( kernel) 3: Find contours using Suzuki-Abe algorithm 4: Filter contours by area ( pixels) 5:for each valid contour do 6: Calculate moments , , 7: Centroid: 8: end for 9: Apply DBSCAN (, min_samples = 2) to centroids 10: Calculate cluster centroids as final detections 11: return refined parasite locations |
Training Data Generation
| Algorithm 4 Training Sample Generation. |
| Require: Enhanced frame , Parasite locations Ensure: Positive samples , Negative samples 1: for each parasite location in P do 2: Extract crop with 50-pixel padding 3: Save as positive training sample 4:end for 5:while and attempts do 6: Generate random location 7: if to all parasites px then 8: Extract crop at 9: Add to 10: end if 11: end while 12: return , |
Dataset Splitting and Augmentation
3.2.3. Deep Learning Models and Training
Model Selection
3.3. Video Processing for Object Detection
3.3.1. Dataset Preparation
Manual Annotation and Ground Truth Generation
Data Splitting Strategy
Model Training Using YOLO Architectures
Performance Evaluation
Model Selection
Model Architecture Variants
Inference on Unlabelled Videos
4. Experimental Results
4.1. Implementation Details
4.2. Evaluation Metrics
4.3. Motion-Based Parasite Detection and Sample Classification
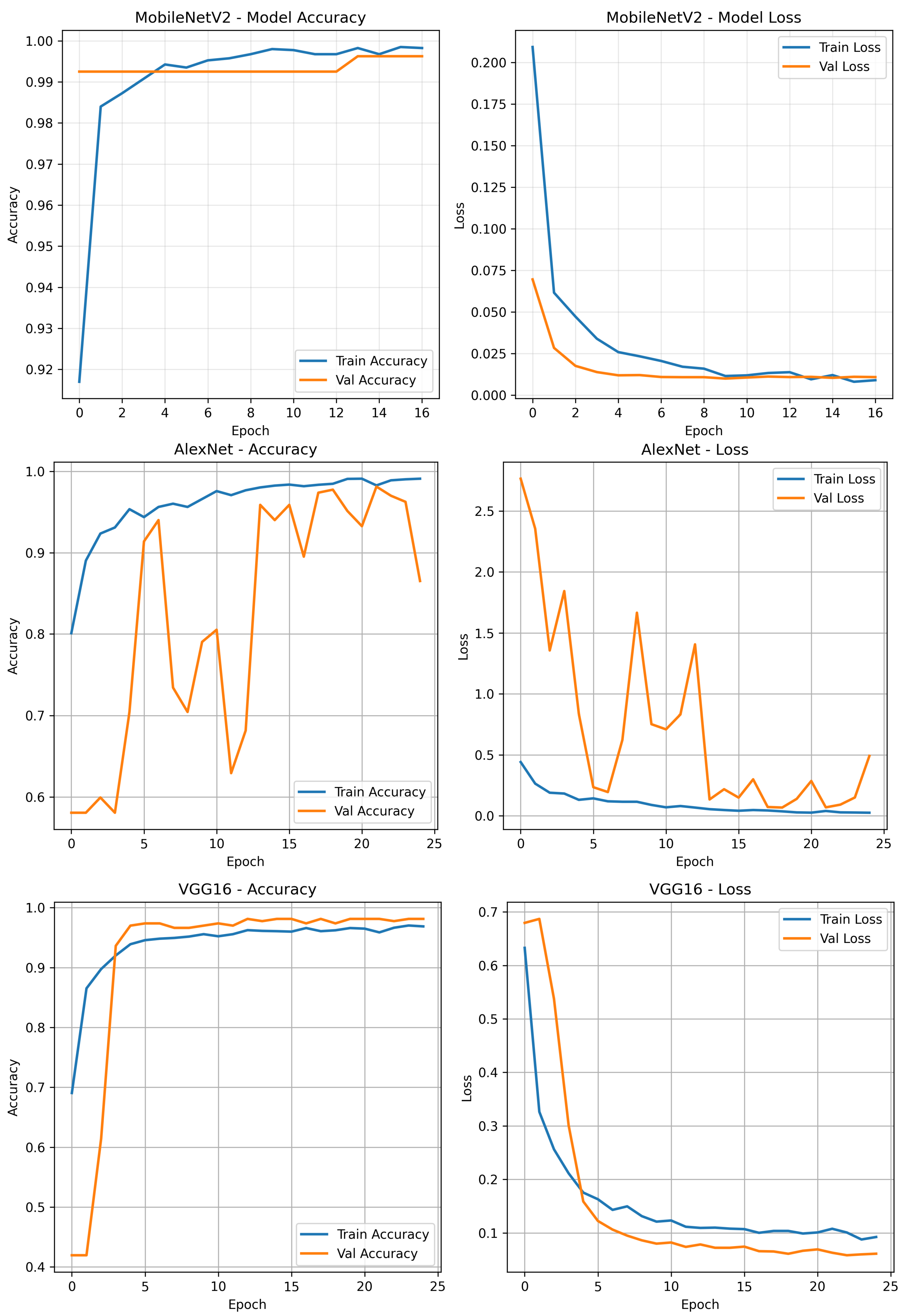
Deep Learning Classification Results
4.4. Video-Based Parasite Localization Using YOLO Detection Models
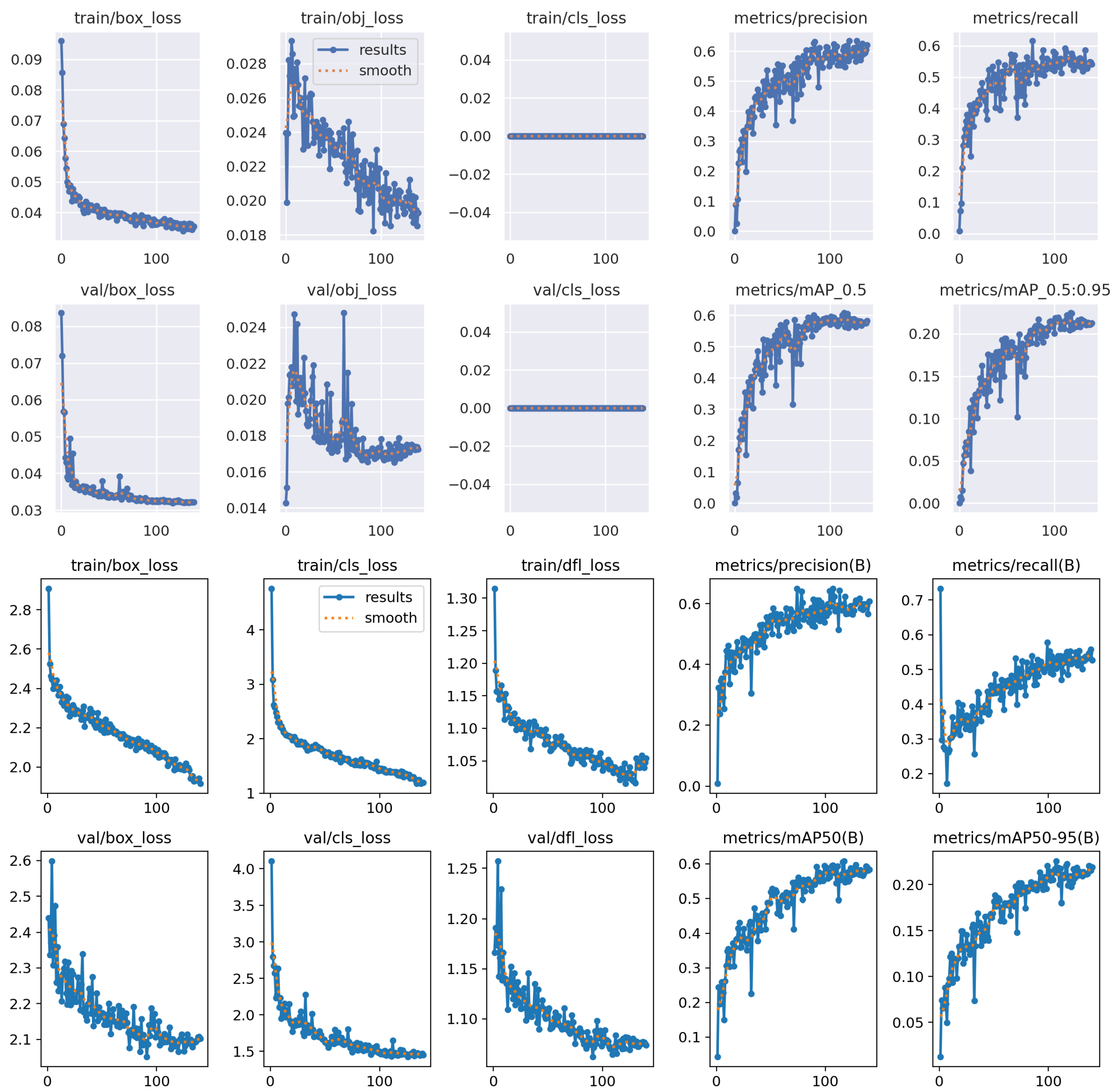
4.4.1. Training Performance for Video-Based Parasite Detection
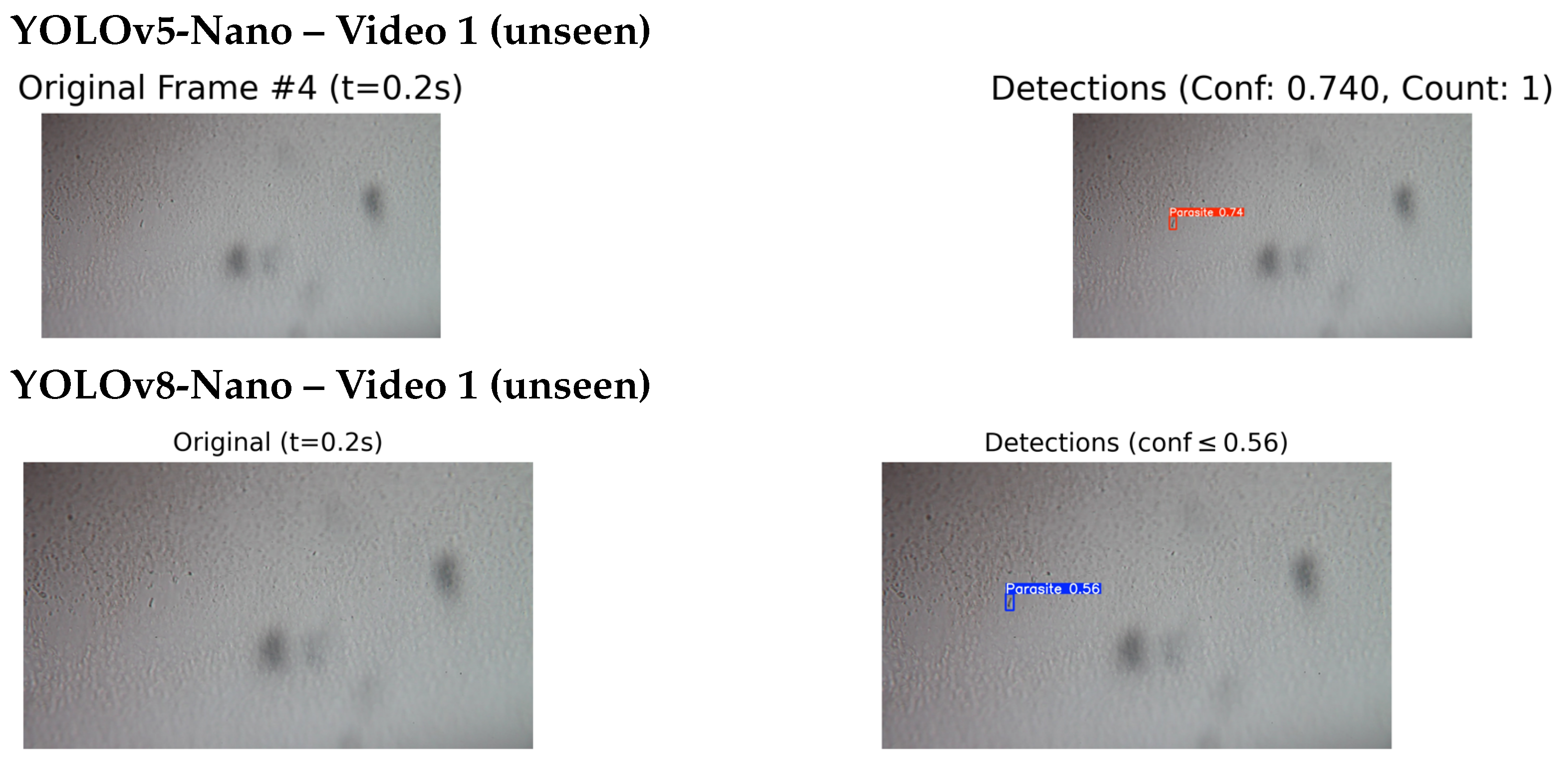
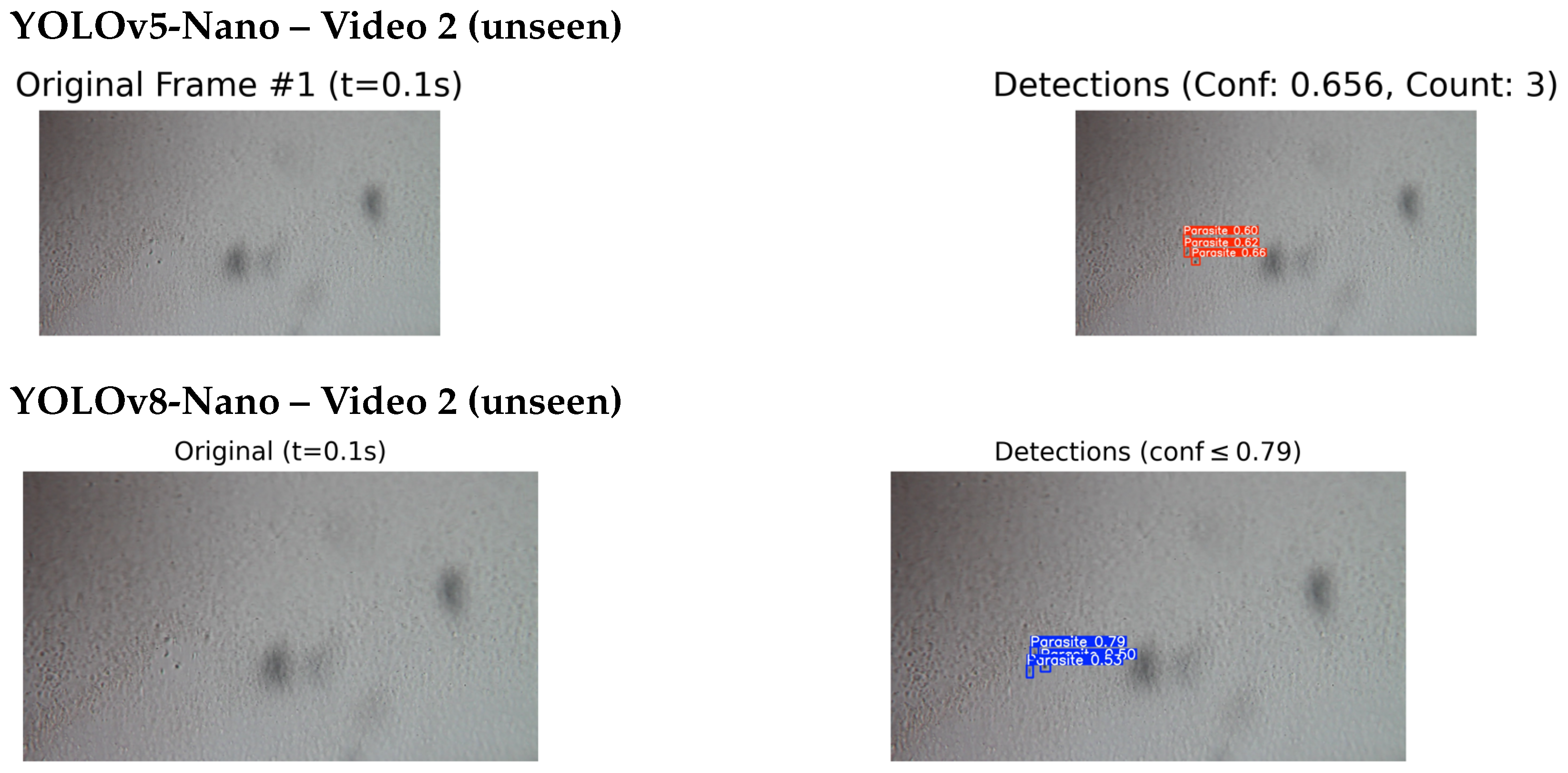
4.4.2. Qualitative Testing on Unseen and External Videos
5. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AUC | Area under ROC Curve |
| CIR | Laboratory of Zoonotic Diseases |
| CNN | Convolutional Neural Network |
| DL | Deep Learning |
| FP | False Positive |
| FN | False Negative |
| IACUC | The Institutional Animal Care and Use Committee of the University |
| IoU | Intersection over Union |
| JImaging | Journal of Imaging |
| mAP | Mean Average Precision |
| ROC | Receiver Operating Characteristic |
| ROI | Region of Interest |
| TP | True Positive |
| T. cruzi | T. cruzi |
| TN | True Negative |
References
- World Health Organization. Chagas Disease (American Trypanosomiasis). 2022. Available online: https://www.who.int/news-room/fact-sheets/detail/chagas-disease-(american-trypanosomiasis) (accessed on 10 December 2024).
- Coura, J.R.; Viñas, P.A. Chagas disease: A new worldwide challenge. Nature 2010, 465, S6–S7. [Google Scholar] [CrossRef]
- Rassi, A.; Marin-Neto, J.A. Chagas disease. Lancet 2010, 375, 1388–1402. [Google Scholar] [CrossRef]
- Jagu, E.; Pomel, S.; Pethe, S.; Loiseau, P.M.; Labruère, R. Polyamine-based analogs and conjugates as antikinetoplastid agents. Eur. J. Med. Chem. 2017, 139, 982–1015. [Google Scholar] [CrossRef]
- Tyler, K.; Engman, D. The life cycle of Trypanosoma cruzi revisited. Int. J. Parasitol. 2001, 31, 472–481. [Google Scholar] [CrossRef]
- Souza, W.D. Basic cell biology of Trypanosoma cruzi. Curr. Pharm. Des. 2002, 8, 269–285. [Google Scholar] [CrossRef]
- Andrade, L.O.; Andrews, N.W. The Trypanosoma cruzi–host-cell interplay: Location, invasion, retention. Nat. Rev. Microbiol. 2005, 3, 819–823. [Google Scholar] [CrossRef] [PubMed]
- de Fuentes-Vicente, J.A.; Gutiérrez-Cabrera, A.E.; Flores-Villegas, A.L.; Lowenberger, C.; Benelli, G.; Salazar-Schettino, P.M.; Cordoba-Aguilar, A. What makes an effective Chagas disease vector? Factors underlying Trypanosoma cruzi-triatomine interactions. Acta Trop. 2018, 183, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Rassi, A., Jr.; Rassi, A.; Little, W.C. Chagas’ heart disease. Clin. Cardiol. 2000, 23, 883–889. [Google Scholar] [CrossRef] [PubMed]
- Moncayo, Á.; Silveira, A.C. Current trends and future prospects for control of Chagas disease. In American Trypanosomiasis; Elsevier: Amsterdam, The Netherlands, 2010; pp. 55–82. [Google Scholar]
- Rassi, A.; de Rezende, J.M. American trypanosomiasis (Chagas disease). Infect. Dis. Clin. 2012, 26, 275–291. [Google Scholar] [CrossRef]
- Gomes, Y.M.; Lorena, V.; Luquetti, A.O. Diagnosis of Chagas disease: What has been achieved? What remains to be done with regard to diagnosis and follow up studies? Mem. Do Inst. Oswaldo Cruz 2009, 104, 115–121. [Google Scholar] [CrossRef]
- Bern, C.; Messenger, L.A.; Whitman, J.D.; Maguire, J.H. Chagas disease in the United States: A public health approach. Clin. Microbiol. Rev. 2019, 33, 10–1128. [Google Scholar] [CrossRef]
- Zingales, B.; Miles, M.A.; Campbell, D.A.; Tibayrenc, M.; Macedo, A.M.; Teixeira, M.M.; Schijman, A.G.; Llewellyn, M.S.; Lages-Silva, E.; Machado, C.R.; et al. The revised Trypanosoma cruzi subspecific nomenclature: Rationale, epidemiological relevance and research applications. Infect. Genet. Evol. 2012, 12, 240–253. [Google Scholar] [CrossRef]
- Pereira, A.C.; Pyrrho, A.S.; Vanzan, D.; Mazza, L.O.; Gomes, J.G.R.C. Deep Convolutional Neural Network applied to Chagas Disease Parasitemia Assessment. In Proceedings of the Anais do 14. Congresso Brasileiro de Inteligência Computacional, Belém, Brazil, 3–6 November 2019. [Google Scholar]
- Soberanis-Mukul, R.; Uc-Cetina, V.; Brito-Loeza, C.; Ruiz-Piña, H. An automatic algorithm for the detection of Trypanosoma cruzi parasites in blood sample images. Comput. Methods Programs Biomed. 2013, 112, 633–639. [Google Scholar] [CrossRef]
- Uc-Cetina, V.; Brito-Loeza, C.; Ruiz-Piña, H. Chagas parasite detection in blood images using AdaBoost. Comput. Math. Methods Med. 2015, 2015, 139681. [Google Scholar] [CrossRef]
- Rosado, L.; Correia da Costa, J.M.; Elias, D.; Cardoso, J.S. A review of automatic malaria parasites detection and segmentation in microscopic images. Anti-Infect. Agents 2016, 14, 11–22. [Google Scholar] [CrossRef]
- Morais, M.C.C.; Silva, D.; Milagre, M.M.; de Oliveira, M.T.; Pereira, T.; Silva, J.S.; Costa, L.d.F.; Minoprio, P.; Junior, R.M.C.; Gazzinelli, R.; et al. Automatic detection of the parasite Trypanosoma cruzi in blood smears using a machine learning approach applied to mobile phone images. PeerJ 2022, 10, e13470. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Mucherino, A.; Papajorgji, P.J.; Pardalos, P.M. k-Nearest Neighbor Classification. In Data Mining in Agriculture; Springer: New York, NY, USA, 2009; pp. 83–106. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Martins, G.L.; Ferreira, D.S.; Ramalho, G.L. Collateral motion saliency-based model for Trypanosoma cruzi detection in dye-free blood microscopy. Comput. Biol. Med. 2021, 132, 104220. [Google Scholar] [CrossRef] [PubMed]
- Thapa, G.; Sharma, K.; Ghose, M.K. Moving object detection and segmentation using frame differencing and summing technique. Int. J. Comput. Appl. 2014, 102, 20–25. [Google Scholar] [CrossRef]
- Husein, A.; Halim, D.; Leo, R. Motion detect application with frame difference method on a surveillance camera. J. Phys. Conf. Ser. 2019, 1230, 012017. [Google Scholar] [CrossRef]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.C. Mobilenetv2: Inverted residuals and linear bottlenecks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 4510–4520. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012, 25, 84–90. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Kakkar, B.; Goyal, M.; Johri, P.; Kumar, Y. Artificial intelligence-based approaches for detection and classification of different classes of malaria parasites using microscopic images: A systematic review. Arch. Comput. Methods Eng. 2023, 30, 4781–4800. [Google Scholar] [CrossRef]
- Ultralytics. YOLOv8 vs YOLOv5: A Detailed Comparison. Available online: https://docs.ultralytics.com/compare/yolov8-vs-yolov5/ (accessed on 15 August 2025).
- Martin, D.L.; Goodhew, B.; Czaicki, N.; Foster, K.; Rajbhandary, S.; Hunter, S.; Brubaker, S.A. Trypanosoma cruzi survival following cold storage: Possible implications for tissue banking. PLoS ONE 2014, 9, e95398. [Google Scholar] [CrossRef]


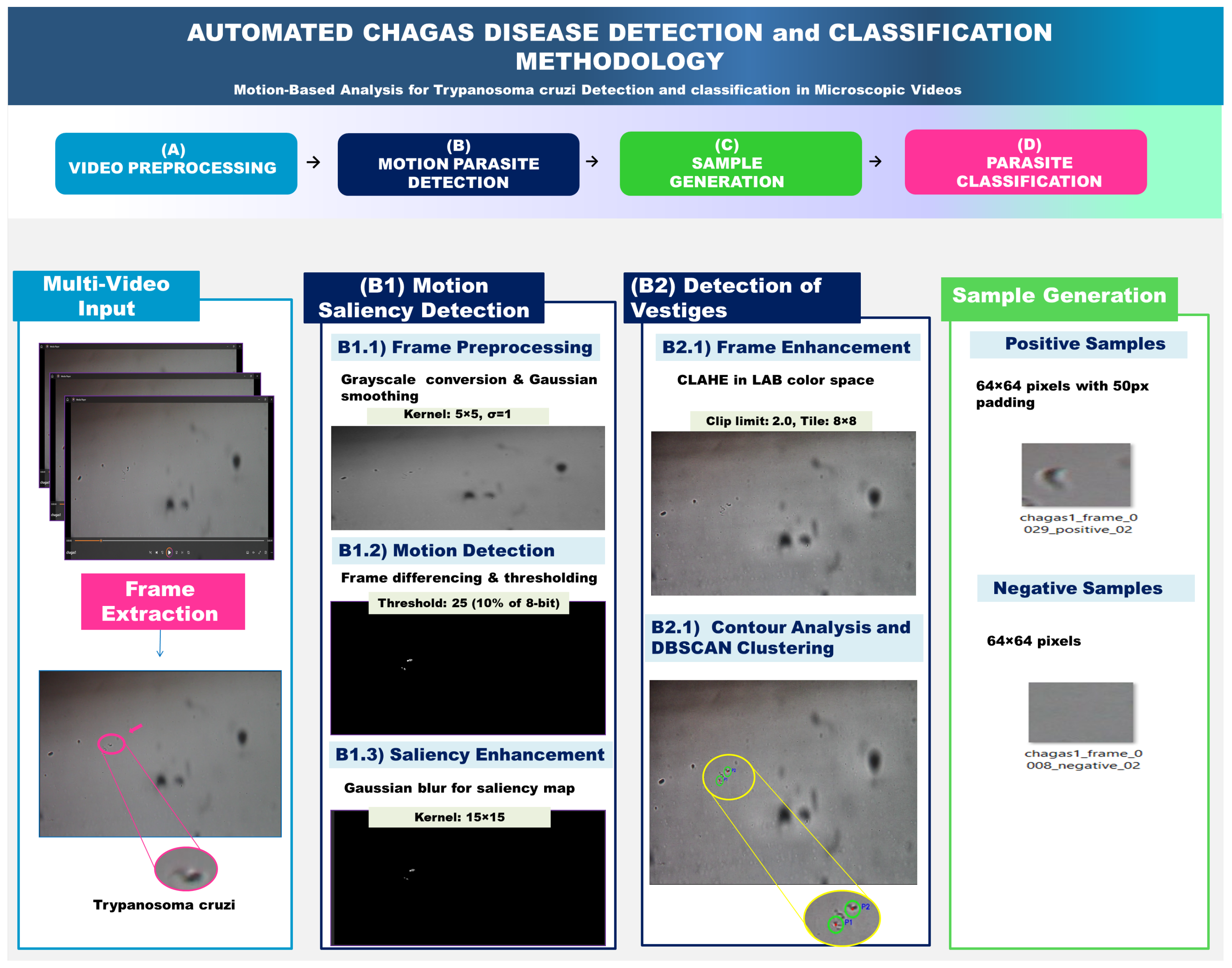






| Parameter | Value | Description |
|---|---|---|
| Motion Threshold (T) | 25 | ∼10% of 8-bit intensity range |
| Gaussian Kernel | Noise reduction filter |
| Technique | Parameters | Purpose |
|---|---|---|
| CLAHE | clip_limit = 2.0, tile_size = | Contrast enhancement |
| Colour Space | LAB (L-channel only) | Preserve chromaticity |
| Component | Parameter | Value | Purpose |
|---|---|---|---|
| Contour Analysis | Min Area | 50 pixels | Filter Noise |
| Morphological | Kernel Size | ellipse | Shape Refinement |
| DBSCAN | Epsilon () | 30 pixels | Clustering Radius |
| DBSCAN | Min Samples | 2 points | Min Cluster Size |
| Sample Generation | Negative Distance | 80 pixels | Avoid Parasite Regions |
| Total Time | Videos | Avg. Time/Video |
|---|---|---|
| 75.3 s | 23 | 3.27 s |
| Sample Type | Count per Frame | Size | Constraints |
|---|---|---|---|
| Positive | Variable | + 50 px padding | Around detected parasites |
| Negative | 3 | >80 px from any parasite |
| Model | Parameters (M) | FLOPs (G) |
|---|---|---|
| MobileNetV2 | 3.4 | 0.30 |
| AlexNet | 61.0 | 0.72 |
| VGG16 | 138.0 | 15.3 |
| Model | Size (Pixels) | mAPval 50–95 | Speed CPU ONNX (ms) | Speed T4 TensorRT (ms) | Params (M) | FLOPs (B) |
|---|---|---|---|---|---|---|
| YOLOv8n | 640 | 37.3 | 80.4 | 1.47 | 3.2 | 8.7 |
| YOLOv8s | 640 | 44.9 | 128.4 | 2.66 | 11.2 | 28.6 |
| YOLOv8m | 640 | 50.2 | 234.7 | 5.86 | 25.9 | 78.9 |
| YOLOv8l | 640 | 52.9 | 375.2 | 9.06 | 43.7 | 165.2 |
| YOLOv8x | 640 | 53.9 | 479.1 | 14.37 | 68.2 | 257.8 |
| YOLOv5n | 640 | 28.0 | 73.6 | 1.12 | 2.6 | 7.7 |
| YOLOv5s | 640 | 37.4 | 120.7 | 1.92 | 9.1 | 24.0 |
| YOLOv5m | 640 | 45.4 | 233.9 | 4.03 | 25.1 | 64.2 |
| YOLOv5l | 640 | 49.0 | 408.4 | 6.61 | 53.2 | 135.0 |
| YOLOv5x | 640 | 50.7 | 763.2 | 11.89 | 97.2 | 246.4 |
| Model | Train Accuracy (%) | Train Loss | Val Accuracy (%) | Val Loss |
|---|---|---|---|---|
| MobileNetV2 | 99.86 | 0.0113 | 99.25 | 0.0100 |
| AlexNet | 98.42 | 0.0412 | 97.75 | 0.0686 |
| VGG16 | 96.41 | 0.1039 | 97.75 | 0.0581 |
| Model | Accuracy (%) | Precision (%) | Recall (%) | F1-Score (%) | Loss | AUC |
|---|---|---|---|---|---|---|
| MobileNetV2 | 99.63 | 100.00 | 99.12 | 99.56 | 0.0099 | 1.00 |
| AlexNet | 97.40 | 96.49 | 97.35 | 96.92 | 0.0794 | 0.9969 |
| VGG16 | 98.14 | 97.37 | 98.23 | 97.80 | 0.0730 | 0.9986 |
| Model | Inference Time (ms/Image) |
|---|---|
| MobileNetV2 | 13.6 |
| AlexNet | 3.6 |
| VGG16 | 7.8 |
| Model | Precision (P) | Recall (R) | mAP@0.5 | mAP@0.5:0.95 |
|---|---|---|---|---|
| YOLOv5-Nano | 0.581 | 0.575 | 0.604 | 0.224 |
| YOLOv5-Small | 0.631 | 0.525 | 0.590 | 0.215 |
| YOLOv5-Medium | 0.549 | 0.559 | 0.595 | 0.219 |
| YOLOv8-Nano | 0.651 | 0.504 | 0.595 | 0.226 |
| YOLOv8-Small | 0.573 | 0.544 | 0.592 | 0.214 |
| YOLOv8-Medium | 0.560 | 0.515 | 0.553 | 0.208 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chenni, K.; Brito-Loeza, C.; Karabağ, C.; Rada, L. From Detection to Motion-Based Classification: A Two-Stage Approach for T. cruzi Identification in Video Sequences. J. Imaging 2025, 11, 315. https://doi.org/10.3390/jimaging11090315
Chenni K, Brito-Loeza C, Karabağ C, Rada L. From Detection to Motion-Based Classification: A Two-Stage Approach for T. cruzi Identification in Video Sequences. Journal of Imaging. 2025; 11(9):315. https://doi.org/10.3390/jimaging11090315
Chicago/Turabian StyleChenni, Kenza, Carlos Brito-Loeza, Cefa Karabağ, and Lavdie Rada. 2025. "From Detection to Motion-Based Classification: A Two-Stage Approach for T. cruzi Identification in Video Sequences" Journal of Imaging 11, no. 9: 315. https://doi.org/10.3390/jimaging11090315
APA StyleChenni, K., Brito-Loeza, C., Karabağ, C., & Rada, L. (2025). From Detection to Motion-Based Classification: A Two-Stage Approach for T. cruzi Identification in Video Sequences. Journal of Imaging, 11(9), 315. https://doi.org/10.3390/jimaging11090315







