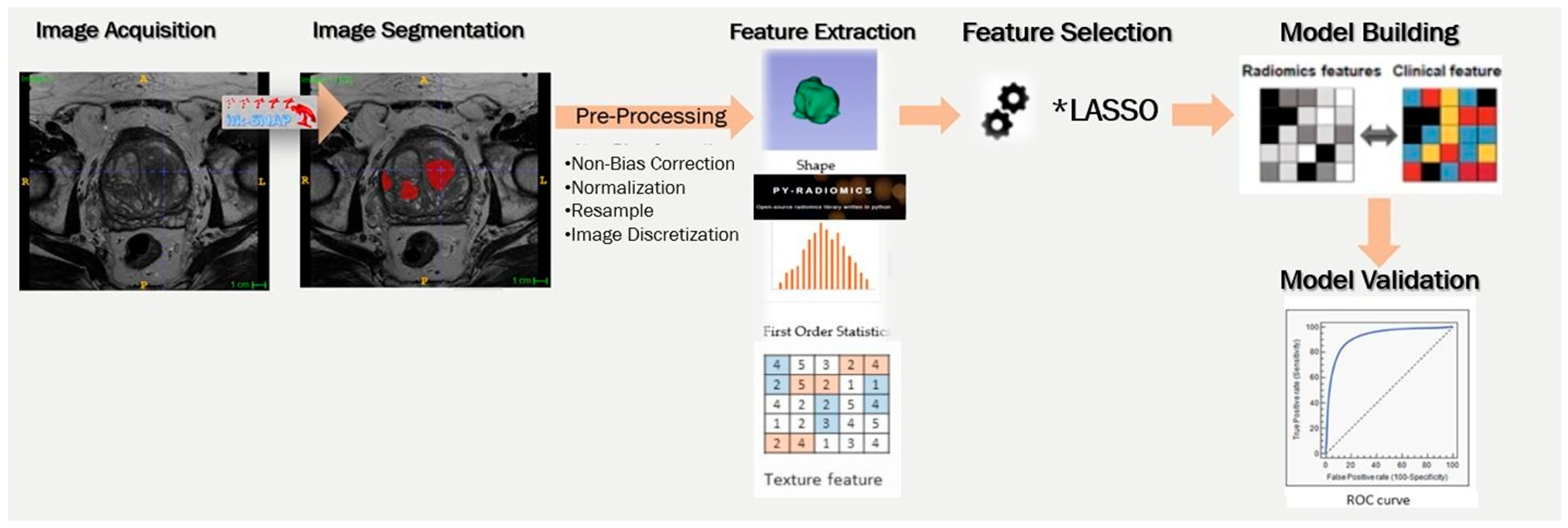
1. Introduction
Prostate cancer (PCa) is the second-most common cancer in men worldwide and the fifth leading cause of cancer-related deaths among men [
1]. The early detection and grading of PCa play a crucial role in patient management, therapy planning, and long-term survival evaluation. Serum Prostate-Specific Antigen (PSA) and Digital Rectal Examination (DRE) are the most widely used PCa screenings in clinical practice, following the European Association of Urology (EAU)—European Society for Radiotherapy and Oncology (ESTRO)—International Society of Geriatric Oncology (SIOG) Guidelines [
2]. The traditional PSA cutoff of 4 ng/mL imposes histopathological verification through biopsy [
3,
4]. However, the study by Marriel et al. indicates that PSA has a low specificity of 20%, disputing the usefulness of PSA in the accurate diagnosis of clinically significant prostate cancer (cs-PCa), since they recorded many cases with low PSA value and cs-PCa and, contrarily, high PSA value in benign pathologies like prostate hypertrophy [
5].
In the studies of Gershmann et al., only approximately 18% of men with elevated PSA were diagnosed with cancer. The remaining 82% of men underwent biopsies without actually having prostate cancer and were exposed to potential complications such as bleeding, infection, and urinary retention [
6]. Thus, there is a need to develop an algorithm that, by taking clinical, demographic, and imaging information, will more accurately define the cases that truly need a biopsy [
7].
Multiparametric Magnetic Resonance Imaging (mpMRI) can be considered a sophisticated diagnostic approach for the detection, differentiation, and risk classification of PCa since it provides imaging biomarkers from conventional and advanced imaging techniques, such as high-resolution T2-weighted (T2W), diffusion-weighted (DWI) and dynamic contrast-enhanced sequences (DCE) [
8,
9]. Mp-MRI diagnostic accuracy in PCa further increased the area under the receiver operating characteristic curve (AUC = 0.893) value when expert radiologists followed the Prostate Imaging Reporting and the Data System Version 2 (PI-RADS v2), which is considered the most promising approach for PCa screening with high diagnostic accuracy AUC = 0.893 to PCa differentiation [
10].
The lack of expert prostate imaging radiologists and the interobserver variability in the interpretation of mp-MRI, the large spectrum of acquisition parameters, and the heterogeneity of PCa tumors are factors that significantly reduce the sensitivity and specificity of the imaging method [
11,
12,
13].
Consequently, there is a need for objective indices to mitigate radiologists’ faults. Radiomic analysis and machine learning (ML) methods offer an objective approach for evaluating MRI data by extracting imaging features usually not easily detectable by the radiologist’s eye [
14,
15]. Radiomic analysis allows the mining of quantitative characteristics like texture, size, and shape from clinical images, like MRI, useful to diagnose and differentiate PCa [
14]. ML is adept at analyzing vast, complex datasets without prior biomedical hypotheses, uncovering insights that may be clinically relevant. As a result, ML, particularly in the area of classification, is being integrated into radiomic research to refine prostate cancer evaluations and reduce subjectivity [
16]. Although ML and radiomics combined are promising diagnosis tools in prostate cancer, they face limitations related to the high susceptibility to variations in acquisition parameters, the sample size, the statistical methodological approach, and the heterogeneous datasets mixing peripheral zone (PZ) with transition zone (TZ) tumors [
17].
The main purpose of this study was to evaluate the diagnostic performance of different ML approaches to detect and assess PCa aggressiveness using standardized MRI protocol across many centers. In particular, we investigated the diagnostic performance of ML to differentiate the different PCa grades by (a) applying seven ML models and (b) changing the input variables.
4. Discussion
The past decade, the research community has stratified post-processing methods such as radiomic analysis combined with ML models to diagnose clinically significant prostate cancer. In this study, we validated the noteworthy contribution of radiomics, trying to reveal the impact of different methodological approaches in the final model’s efficiency. Specifically, we investigated the effect of the different inputs in ML models and the effect of the applied ML methods in different clinical queries.
Our results have shown that the models’ efficiency is highly dependent on the input variables, as expected. In most of the examined scenarios, the T2W and DWI-weighted-derived radiomics either as independent or combined inputs shown limited clinical usability. The models’ efficiency was significantly improved by introducing PSA clinical index independently of lesion location or the degree of malignancy.
The positive effect of introducing a clinical variable on the model performance is in line with the existing literature. Marvaso et al. created four different models. Model 1, including only clinical variables (PSA, pre-operative GS, ISUP, Tumor Nodule Metastasis (TNM) stage and age), achieved AUC = 0.68. Model 2 combined the aforementioned clinical and radiological features (ADC, PI-RADS, lesion volume) and showed a significant improvement, with an AUC of 0.79. Model 3, which integrated prior clinical data with radiomic features, achieved an AUC of 0.71. Finally, Model 4, which combined all features, achieved the highest AUC of 0.81, indicating that the most accurate predictions of PCa pathology were obtained when all variables were incorporated [
34].
Similar results were observed in the study by Dominguez et al., where an LR classifier was used to distinguish clinical insignificant (ciPCa) and csPCa, and its performance was improved notably with the inclusion of both radiological (T2W- and Apparent Diffusion Coefficient (ADC)-derived radiomics, prostate volume) and PSA clinical feature (CL) [
35]. Specifically, the individual variables, CL, T2W, ADC showed AUC 0.76, 0.85, and 0.81, respectively, and their combination 0.91 [
35].
Controversially, there are studies in which the integration of PSA with radiologically derived quantitative metrics did not contribute to further increasing and in some cases decreasing the model’s rendering. In the study conducted by Gong et al., the addition of PSA to the T2W-DWI yielded restricted improvement in model performance. Specifically, the clinical model achieved AUCs of 0.723, while the T2W-DWI model reached 0.788, and the combined T2W-DWI-clinical model slightly improved to 0.780 [
36].
However, Lu et al. compared multiple models for PCa prediction in a validation cohort, where the TZ-PSA density model yielded a relatively low AUC of 0.592. In contrast, radiomic models with the ADC-based radscore reaching 0.779, the T2W-based radscore 0.808, the fusion radscore 0.844, and the radiomic nomogram incorporating TZ volume achieving the highest AUC of 0.872. This discrepancy may be attributed to differences in dataset composition (57.4% of their cases located in ΤΖ) [
37].
Moreover, there are numerous studies including only radiological metrics in their models and achieving palatable efficiency. A recent review of Antonil et al. presented 14 studies that introduced only radiological features in computational models to discriminate cs-PCa and ciPCa. In line with our results, efficiency was improved when they were introduced to more than one source of feature (in most cases T2w and DWI). AUC ranged from 0.68 to 0.81 when DWI or T2w imaging data were introduced as individual inputs, while their combination achieved AUC 0.73 to 0.98 [
38].
Our AUC values were observed to be lower than those reported by some studies in the literature. We assume that this is because most studies used data from a single MR system and applied higher, more sensitive to lesion detection b-values than ours. For example, Jin et al. used b = 2000 s/mm
2, Jing et al. b = 1500 s/mm
2, and Hamm et al. b = 1400 s/mm
2—all acquired on 3T scanners. A notable exception is Castillo et al., who used data from both 1.5T and 3T systems with b-values ranging from 600 to 1000, reporting an AUC of 0.72, similar to our results [
39,
40,
41,
42].
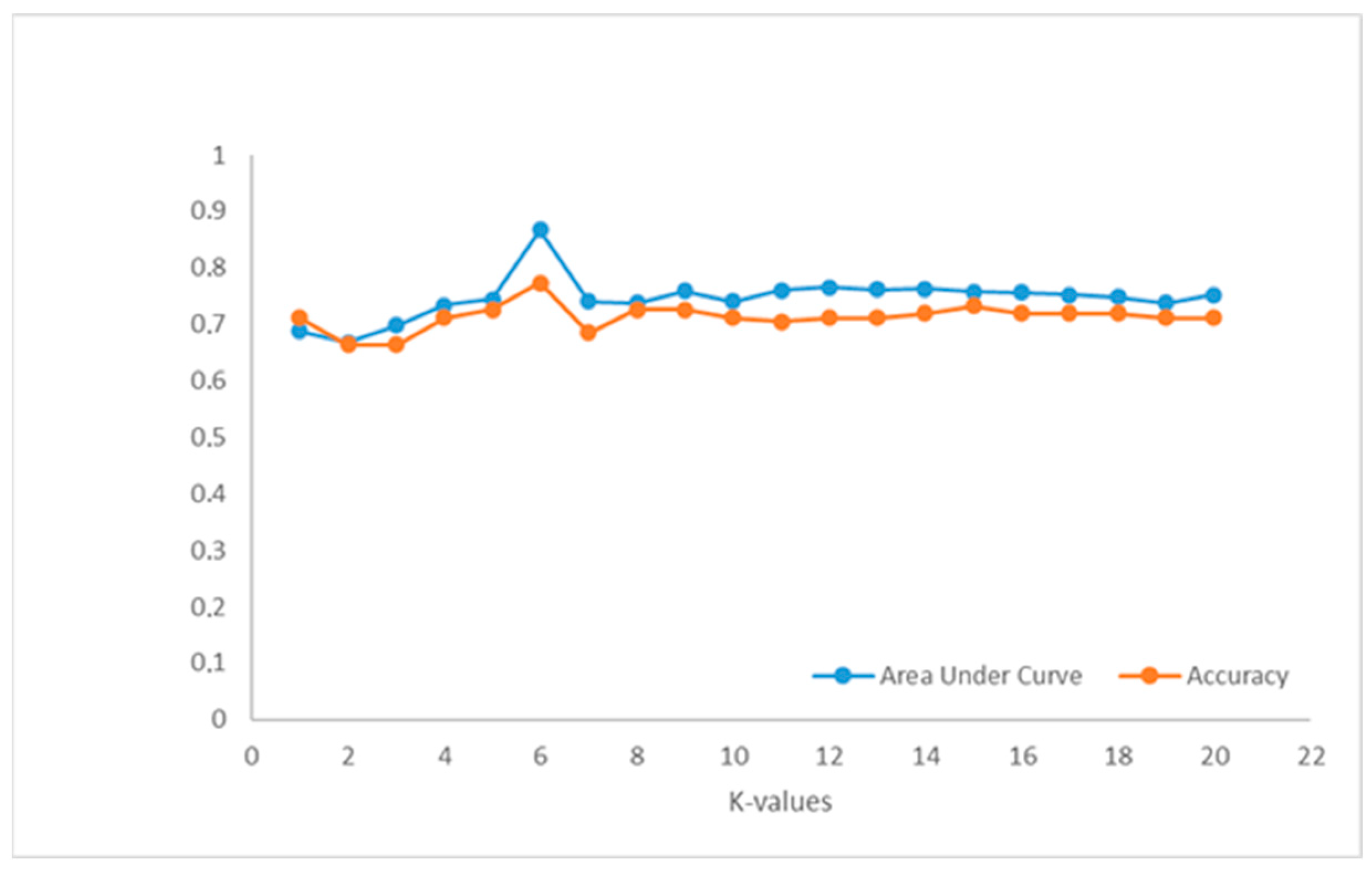
The selection of ML algorithm in PCa classification depends on data characteristics like data dimensionality, feature correlations, and computational resources. Performance evaluation through cross-validation and performance metrics is crucial to determine the most suitable algorithm [
43]. The comparison of seven ML algorithms in this survey provides greater reliability for our model.
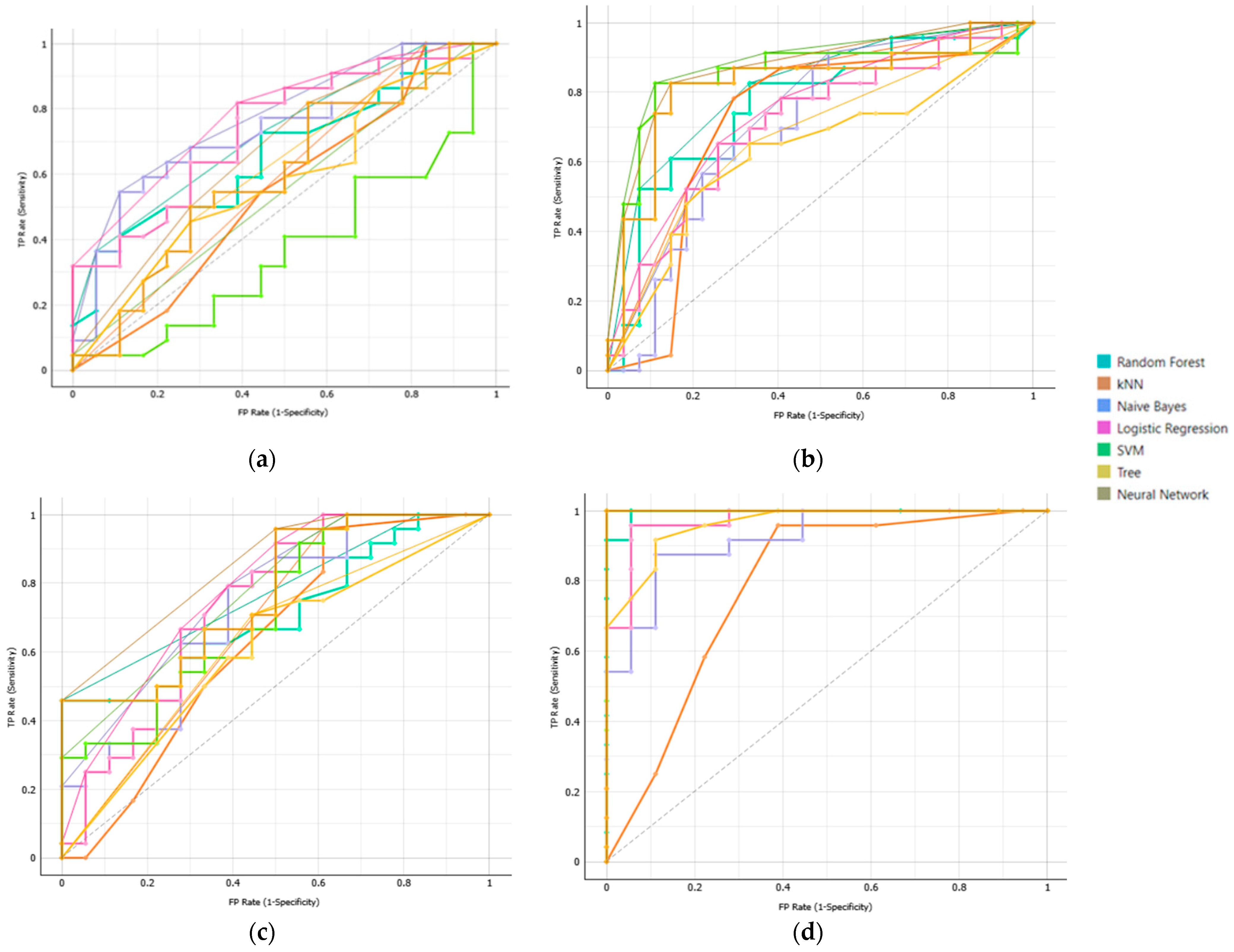
Classification performance of our ML models in the prediction of csPCa was shown to be improved, incorporating T2W, DWI and PSA. Among models, NN achieved the highest performance (AUC = 0.992), followed by SVM (AUC = 0.957), DT (AUC = 0.953), and RF (AUC = 0.946). The efficiency of these models was consistent across different clinical questions posed, highlighting their robustness and generalizability compared to the traditional ML models LR, kNN, and NB, whose performance varied across different tasks. Specifically, LR and kNN showed moderate performance AUC = 0.884 and 0.868, respectively, whereas NB had the lowest performance (AUC = 0.830).
The models’ generalization performance approved relatively consistently across the various evaluation strategies employed, including cross-validation and a held-out validation set. NN exhibited the highest performance during model development, achieving an AUC of 0.992 in cross-validation. Its performance on the held-out set remained robust (AUC = 0.936), indicating strong generalization during initial validation. Similarly, SVM presented a perfect AUC of 1.000 on the held-out set and high performance of AUC = 0.957 in cross-validation. RF and DT delivered strong results during cross-validation (AUCs of 0.946 and 0.953, respectively). While their performance saw a drop on the held-out set (AUCs of 0.814 and 0.929), they still exhibited solid generalization. In contrast, LR, kNN, and NB presented moderate performance during model development (AUC_cross_validation, kNN:0.868; NB: 0.830 and LR: 0.884). However, they maintained consistent performance across datasets (AUC_held_out, kNN: 0.764; NB: 0.700; LR: 0.764) These findings indicate the high predictive abilities of deep learning models compared with traditional ML models, which have less capture ability to detect complex feature interactions.
According to Nematollahi et al. and other related studies, the performance of various supervised ML models using mpMRI or bpMRI data for prostate cancer (PCa) diagnosis varies considerably [
31]. Across the published studies different methodological approaches were observed, as regards to the input variables, data sample, and pre- and post-processing analysis steps. Therefore, logistic regression (LR) consistently demonstrates strong performance, with reported AUCs ranging from 0.82 to 0.97 [
31]. SVM also perform well, with AUCs between 0.727 and 0.89 for mpMRI and up to 0.85 for bpMRI [
38,
44,
45]. kNN achieves AUCs of 0.82–0.88 (mpMRI) and up to 0.84 (bpMRI), while RF shows AUCs ranging from 0.76 to 0.94 [
38,
46,
47,
48]. NB, although still effective, presents the lowest AUCs overall (0.80–0.83 in mpMRI and 0.695–0.80 in bpMRI) [
37,
48,
49]. NN, DT, and LR models using bpMRI yield AUCs ranging from 0.71 to 0.936, depending on the study and configuration [
38,
43,
48,
50,
51,
52,
53,
54]. The literature review and the results of our study indicate the need to optimize the analysis process, regarding input variables and model choice and the need to standardize the pre-processing analysis steps.
Differentiating ciPCa from csPCa represents an initial critical step in PCa management. However, within the csPCa spectrum, accurate grading—especially the distinction between ISUP grades 1, 2, and ≥3—is essential because it significantly influences treatment strategies. [
55] ISUP grade 1 (GS 6, 3 + 3) is often suitable for active surveillance, while ISUP grade 2 (GS 7, 3 + 4) may necessitate treatment for its limited aggressiveness, and ISUP grade ≥ 3 denotes more aggressive disease that warrants immediate intervention [
56]. Accurate risk stratification is therefore essential to prevent both overtreatment of low-risk cases and under-management of potentially aggressive disease [
57].
A significant disparity exists in PCa research concerned more with detection methods than with the grading and management of low-grade tumors. Twilt et al. observed that only a minority of studies employ ML for ISUP grade prediction using radiomic features. Algorithms’ efficiency to detect high-grade lesions ISUP ≥ 4 is usually high, while that to distinguish intermediate from low-grade lesions is not consistent across studies [
58]. Indicatively, Abraham et al., applying a Convolution Neural Network (CNN) to T2W-, DWI-, and ADC-derived metrics, reported low AUC values, especially in low-grade lesions [AUC: 0.626 (GS 6~ISUP = 1), 0.535 (GS 3 + 4~ISUP = 2), 0.379 (GS 4 + 3~ISUP = 3), 0.761 (GS 8~ISUP = 4), and 0.847 (GS ≥ 9~ISUP = 5)] [
59]. Low efficiencies were also reported by McGarry et al., who combined four MRI contrasts (T2W, ADC 0–1000, ADC 50–2000, and DCE) to generate Gleason probability maps, achieving low AUC (0.56) for distinguishing GS 4–5 from GS 3, but higher performance (AUC = 0.79) for benign vs. malignant classification [
60]. Improved performance was reported by Chaddad et al. in two different studies, where they used two different methodological approaches to lesion grading. At first, they used Joint Intensity Matrix and Gray-Level Co-Occurrence Matrix features from The Cancer Imaging Archive (TCIA) dataset and reported lower than their expectation AUC values of 78.4% (GS ≤ 6), 82.35% (GS 3 + 4), and 64.76% (GS ≥ 4 + 3), which they attribute to omission of key clinical and morphological features [
61]. Later, they applied an RF classifier with zone-based features achieving better AUC value in low-grade lesions and high-grade lesions (GS 6 AUC = 0.83 and GS ≥ 4 + 3 AUC = 0.77, respectively), while AUC was importantly decreased in intermediate lesions of GS 3 + 4 (AUC = 0.73). Similar performance was reported by Nketiah et al., who used logistic regression on texture features from T2W, ADC, and DCE, [AUCs of 0.83 Angular Second Moment (ASM) for GS 3 + 4 vs. 4 + 3] [
62]. Higher AUC values were achieved by Jensen et al., who used a kNN model in which they were introduced T2WI- and DWI-derived features, highlighting the effect of lesion location, since AUC values were 0.96 in PZ and 0.83 in TZ to identify ISUP 1 or 2, 0.98 in PZ and 0.94 in TZ for ISUP 3, and 0.91 in PZ and 0.87 in TZ for ISUP ≥ 4 [
63]. Also, high performance was published by Fehr et al., who employed a Recursive Feature Selection–Support Vector Machine (RFS-SVM) with Synthetic Minority Oversampling Technique (SMOTE), achieving AUCs of 0.93 (GS 6 vs. ≥7) and 0.92 (GS 3 + 4 vs. 4 + 3), including both TZ and PZ lesions [
64].
Our results are comparable to those reported in the literature when only radiology-derived features are used, but significantly higher when PSA values are included. Therefore, all these findings highlight considerable variability in ML-based ISUP grading. Standardized radiomic workflows, larger multicenter datasets, and prospective validation are critical to improving model reliability and clinical integration.
This study has several strengths, which mainly concern the methodology used. We tried to deploy a high-performance model, including the optimal combination of input parameters and discovering the most effective algorithm. Also, models’ generality was improved, including imaging data from four different MRI systems, of which acquisition protocols were not standardized and tested, applying both cross-validation and held-out tests. However, our study has several limitations. First, the held-out test was only performed in the model that differentiated benign from malignant prostate lesions regardless of their location. Second, the relatively small sample size may affect the robustness of our findings. Third, the study lacks an assessment of the impact of conventional radiological parameters such as prostate volume and does not incorporate other clinical variables or patient history data. Finally, features related to lesion perfusion were not extracted, as the imaging protocol did not include DCE sequences.