A 2.5D Self-Training Strategy for Carotid Artery Segmentation in T1-Weighted Brain Magnetic Resonance Images
Abstract
1. Introduction
2. Related Work
3. Materials and Methods
3.1. Datasets
3.2. Preprocessing and Data Augmentation
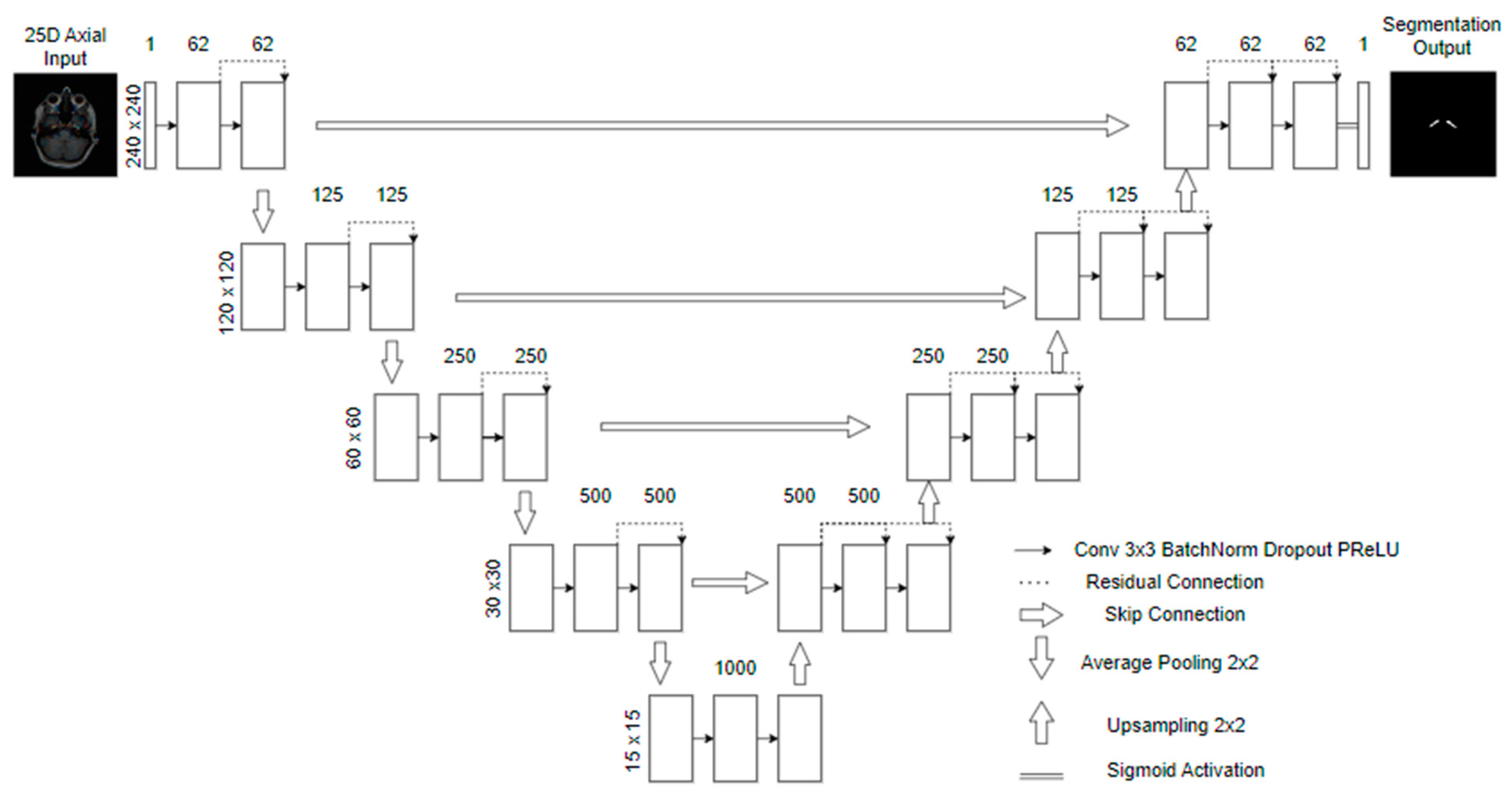
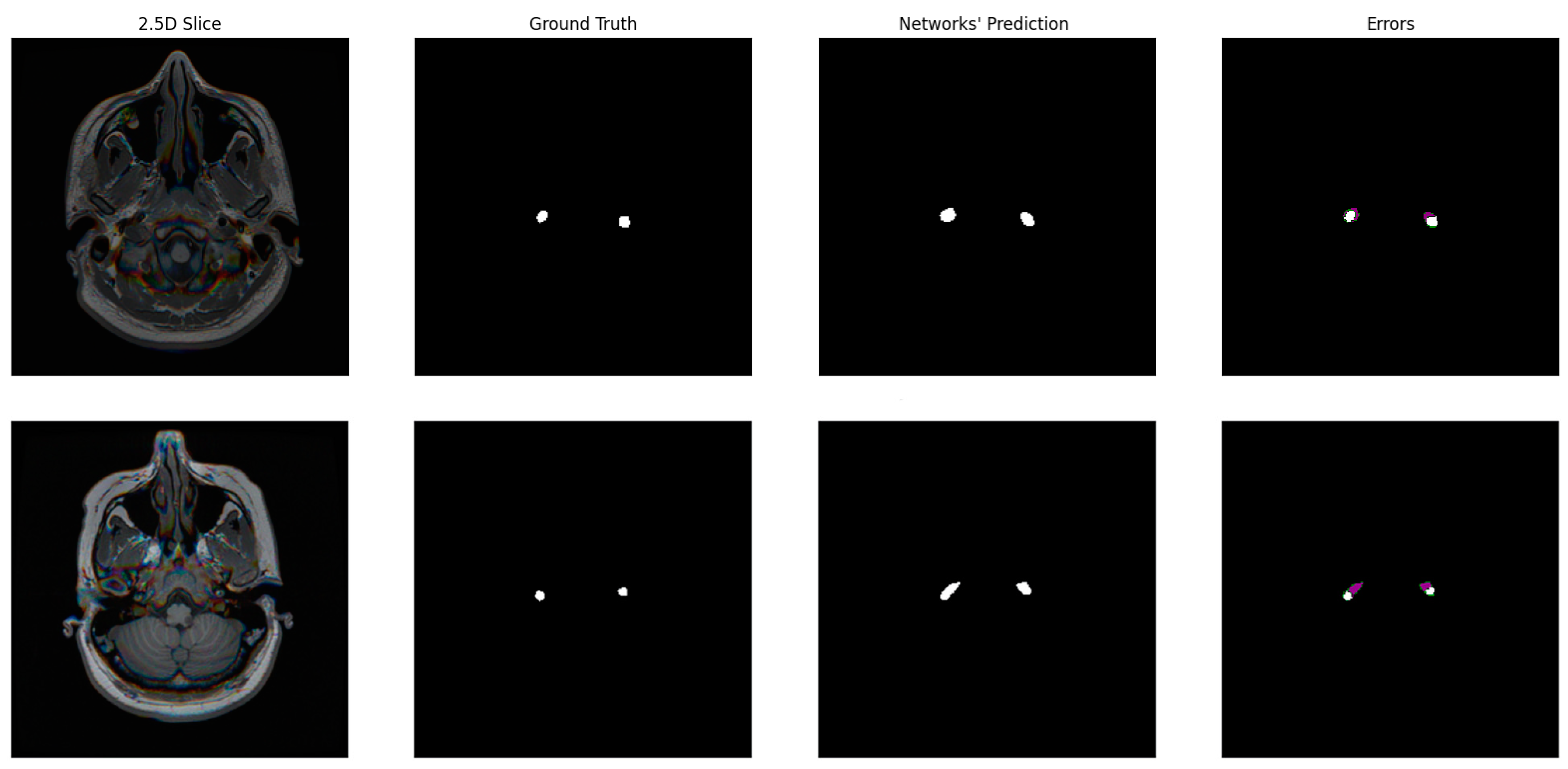
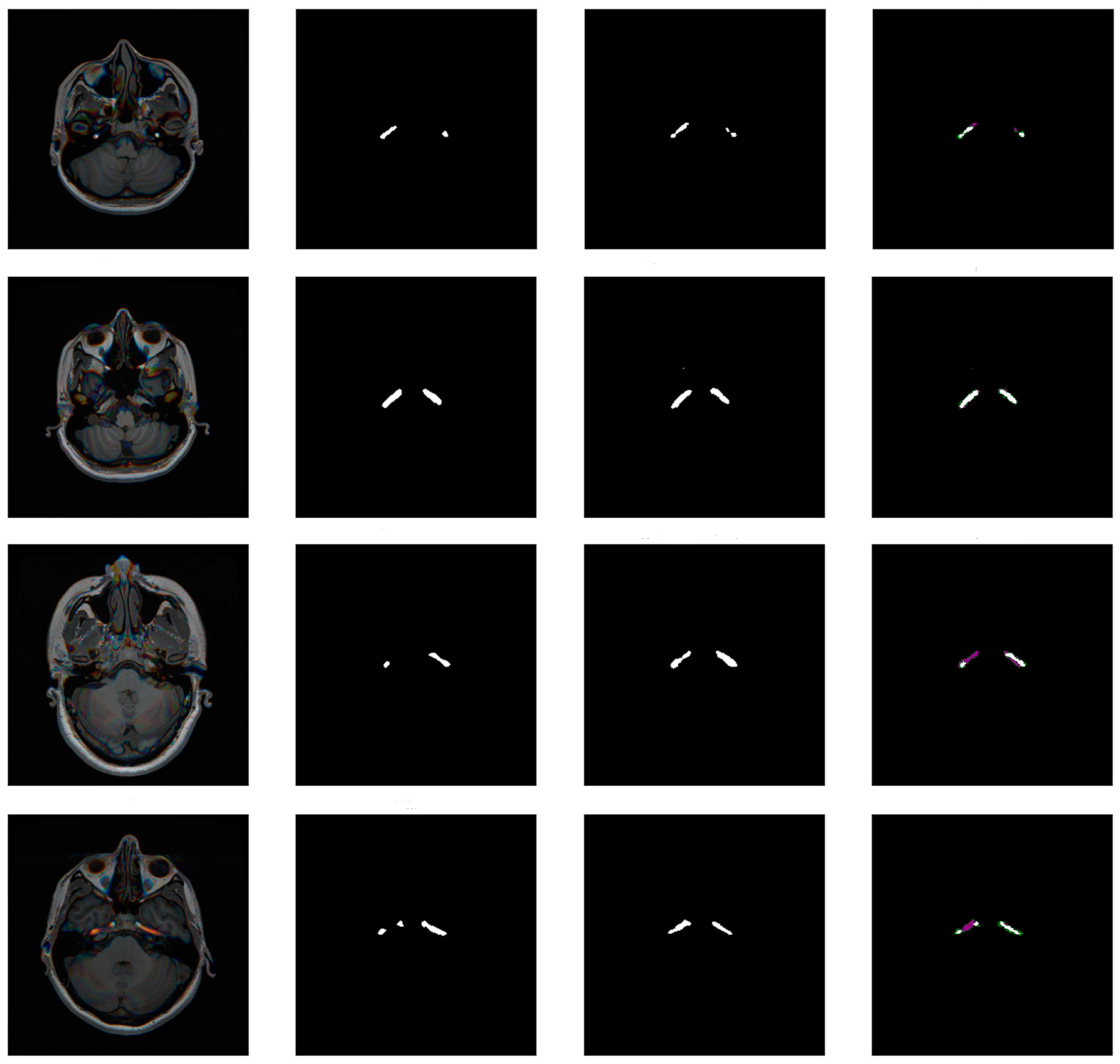
3.3. Model
3.4. Mask Update Scheme
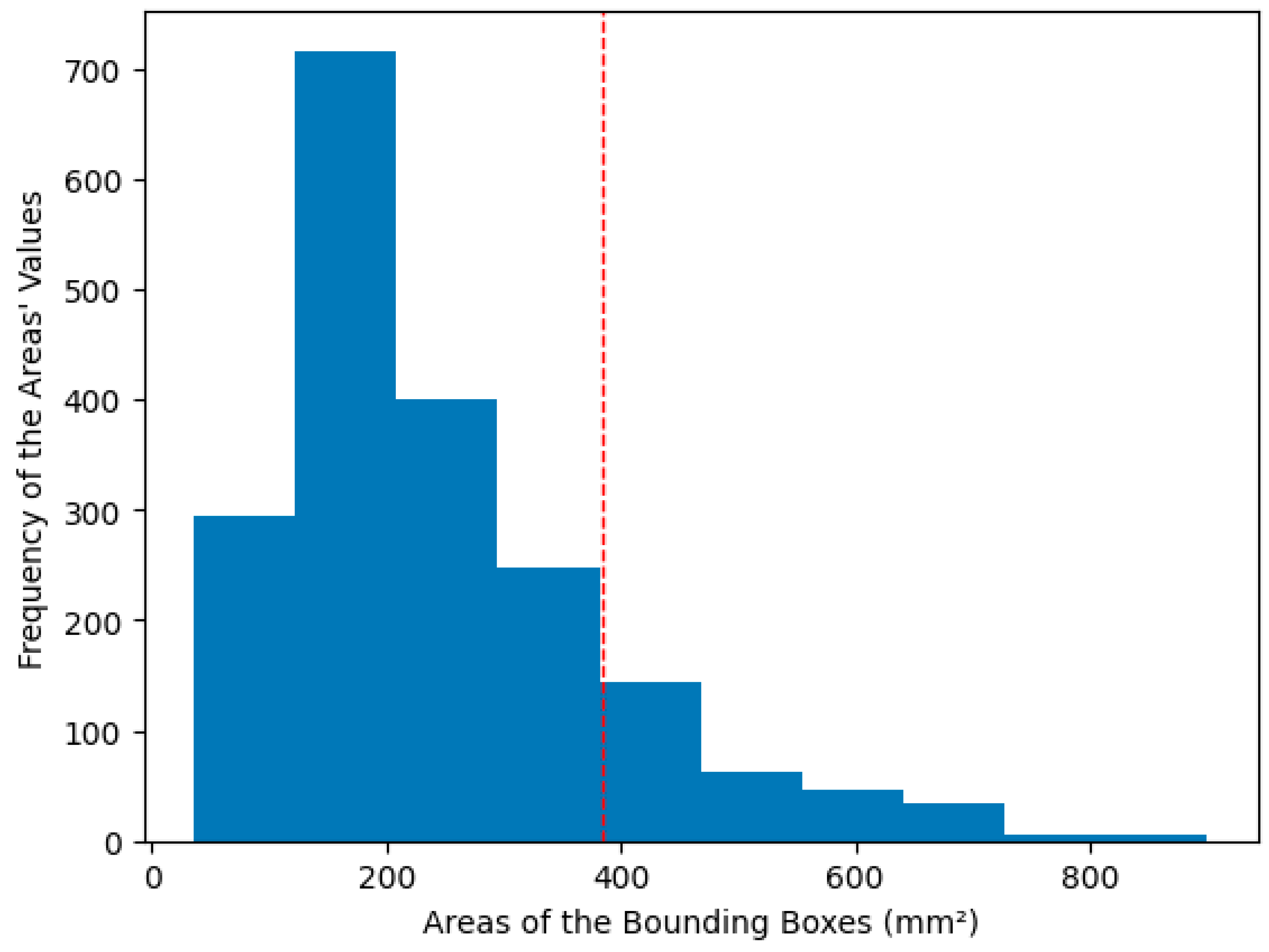
- We first train five models with the bounding boxes as the target for the semantic segmentation using 5-fold cross-validation (Round 0). We stratify the fold so that each fold has the same dataset separation. The model is trained for a maximum of 100 epochs. The training stops if the network does not improve the validation’s mean Intersection over Union (IoU) [34] in 10 epochs. We also only save the best weights in the validation.
- We used the trained models in an ensemble (average of the five cross-validation models’ predictions) to perform the segmentation in all the training dataset images, including those used to train them. After, we post-process the predictions using an erosion morphological operation, with a disk of radius one as the structuring element. This operation was performed only during the first four training rounds to eliminate a bit more of the false positives that naturally occur because of the initial bounding boxes.
- Using each post-processed mask, we calculate the IoU for the bounding boxes: if it is above 50%, we use the prediction of the post-processed mask as a new mask. If not, we return to the initial bounding box as a mask. We calculate the IoU for each carotid, separating the images into two parts, evaluating the image for each artery separately, and concatenating the results.
- Finally, we multiply the resulting mask by the bounding boxes, erasing pixels outside them.
4. Results
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhu, K.; Xiong, N.N.; Lu, M. A Survey of Weakly-supervised Semantic Segmentation. In Proceedings of the 2023 IEEE 9th International Conference on Big Data Security on Cloud, IEEE International Conference on High Performance and Smart Computing, and IEEE International Conference on Intelligent Data and Security, BigDataSecurity-HPSC-IDS, New York, NY, USA, 6–8 May 2023. [Google Scholar] [CrossRef]
- Chan, L.; Hosseini, M.S.; Plataniotis, K.N. A Comprehensive Analysis of Weakly-Supervised Semantic Segmentation in Different Image Domains. Int. J. Comput. Vis. 2021, 129, 361–384. [Google Scholar] [CrossRef]
- Kumar, A.; Jiang, H.; Imran, M.; Valdes, C.; Leon, G.; Kang, D.; Nataraj, P.; Zhou, Y.; Weiss, M.D.; Shao, W. A Flexible 2.5D Medical Image Segmentation Approach with In-Slice and Cross-Slice Attention. arXiv 2024, arXiv:2405.00130. [Google Scholar] [CrossRef]
- Carmo, D.; Rittner, L.; Lotufo, R. Open-source tool for Airway Segmentation in Computed Tomography using 2.5D Modified EfficientDet: Contribution to the ATM22 Challenge. arXiv 2022, arXiv:2209.15094. [Google Scholar] [CrossRef]
- Avesta, A.; Hossain, S.; Lin, M.; de Aboian, M.; Krumholz, H.M.; Aneja, S. Comparing 3D, 2.5D, and 2D Approaches to Brain Image Auto-Segmentation. Bioengineering 2023, 10, 181. [Google Scholar] [CrossRef] [PubMed]
- Ou, Y.; Yuan, Y.; Huang, X.; Wong, K.; Volpi, J.; Wang, J.Z.; Wong, S.T.C. LambdaUNet: 2.5D Stroke Lesion Segmentation of Diffusion-Weighted MR Images. In Proceedings of the 24th International Conference on Medical Image Computing and Computer Assisted Intervention–MICCAI 2021, Strasbourg, France, 27 September–1 October 2021; Proceedings, Part I 24. Springer International Publishing: Cham, Switzerland, 2021. [Google Scholar] [CrossRef]
- Huang, Q.; Tian, H.; Jia, L.; Li, Z.; Zhou, Z. A review of deep learning segmentation methods for carotid artery ultrasound images. Neurocomputing 2023, 545, 126298. [Google Scholar] [CrossRef]
- Wang, Y.; Yao, Y. Application of Artificial Intelligence Methods in Carotid Artery Segmentation: A Review. IEEE Access 2023, 11, 13846–13858. [Google Scholar] [CrossRef]
- Sari, H.; Erlandsson, K.; Law, I.; Larsson, H.B.; Ourselin, S.; Arridge, S.; Atkinson, D.; Hutton, B.F. Estimation of an image derived input function with MR-defined carotid arteries in FDG-PET human studies using a novel partial volume correction method. J. Cereb. Blood Flow Metab. 2017, 37, 1398–1409. [Google Scholar] [CrossRef]
- Galovic, M.; Erlandsson, K.; Fryer, T.D.; Hong, Y.T.; Manavaki, R.; Sari, H.; Chetcuti, S.; Thomas, B.A.; Fisher, M.; Sephton, S.; et al. Validation of a combined image derived input function and venous sampling approach for the quantification of [18F]GE-179 PET binding in the brain. NeuroImage 2021, 237, 118194. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Yang, X.; Li, Y.; Jiang, G.; Jia, S.; Gong, Z.; Mao, Y.; Zhang, S.; Teng, Y.; Zhu, J.; et al. Deep Learning-Based Automated Detection of Arterial Vessel Wall and Plaque on Magnetic Resonance Vessel Wall Images. Front. Neurosci. 2022, 16, 888814. [Google Scholar] [CrossRef]
- Chen, Y.-F.; Chen, Z.-J.; Lin, Y.-Y.; Lin, Z.-Q.; Chen, C.-N.; Yang, M.-L.; Zhang, J.-Y.; Li, Y.-Z.; Wang, Y.; Huang, Y.-H. Stroke risk study based on deep learning-based magnetic resonance imaging carotid plaque automatic segmentation algorithm. Front. Cardiovasc. Med. 2023, 10, 1101765. [Google Scholar] [CrossRef]
- Shapey, J.; Wang, G.; Dorent, R.; Dimitriadis, A.; Li, W.; Paddick, I.; Kitchen, N.; Bisdas, S.; Saeed, S.R.; Ourselin, S.; et al. An artificial intelligence framework for automatic segmentation and volumetry of vestibular schwannomas from contrast-enhanced T1-weighted and high-resolution T2-weighted MRI. J. Neurosurg. 2021, 134, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Shapey, J.; Li, W.; Dorent, R.; Dimitriadis, A.; Bisdas, S.; Paddick, I.; Bradford, R.; Zhang, S.; Ourselin, S.; et al. Automatic Segmentation of Vestibular Schwannoma from T2-Weighted MRI by Deep Spatial Attention with Hardness-Weighted Loss. In Proceedings of the 22nd International Conference on Medical Image Computing and Computer Assisted Intervention—MICCAI 2019, Shenzhen, China, 13–17 October 2019; Shen, D., Liu, T., Peters, T.M., Staib, L.H., Essert, C., Zhou, S., Yap, P.-T., Khan, A., Eds.; Springer International Publishing: Cham, Switzerland, 2019; Volume 11765, pp. 264–272. [Google Scholar] [CrossRef]
- Elsheikh, S.; Urbach, H.; Reisert, M. Intracranial Vessel Segmentation in 3D High-Resolution T1 Black-Blood MRI. Am. J. Neuroradiol. 2022, 43, 1719–1721. [Google Scholar] [CrossRef] [PubMed]
- Quon, J.L.; Chen, L.C.; Kim, L.; Grant, G.A.; Edwards, M.S.B.; Cheshier, S.H.; Yeom, K.W. Deep Learning for Automated Delineation of Pediatric Cerebral Arteries on Pre-operative Brain Magnetic Resonance Imaging. Front. Surg. 2020, 7, 517375. [Google Scholar] [CrossRef] [PubMed]
- Shi, F.; Yang, Q.; Guo, X.; Qureshi, T.A.; Tian, Z.; Miao, H.; Dey, D.; Li, D.; Fan, Z. Intracranial Vessel Wall Segmentation Using Convolutional Neural Networks. IEEE Trans. Biomed. Eng. 2019, 66, 2840–2847. [Google Scholar] [CrossRef] [PubMed]
- Samber, D.D.; Ramachandran, S.; Sahota, A.; Naidu, S.; Pruzan, A.; Fayad, Z.A.; Mani, V. Segmentation of carotid arterial walls using neural networks. World J. Radiol. 2020, 12, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Hong, Y. Scribble2D5: Weakly-Supervised Volumetric Image Segmentation via Scribble Annotations. In Proceedings of the 25th International Conference on Medical Image Computing and Computer Assisted Intervention—MICCAI 2022, Singapore, 18–22 September 2022; Wang, L., Dou, Q., Fletcher, P.T., Speidel, S., Li, S., Eds.; Springer Nature: Cham, Switzerland, 2022; pp. 234–243. [Google Scholar] [CrossRef]
- Zareba, M.R.; Fafrowicz, M.; Marek, T.; Beldzik, E.; Oginska, H.; Beres, A.; Faba, P.; Janik, J.; Lewandowska, K.; Ostrogorska, M.; et al. Neuroimaging of chronotype, sleep quality and daytime sleepiness: Structural T1-weighted magnetic resonance brain imaging data from 136 young adults. Data Brief 2022, 41, 107956. [Google Scholar] [CrossRef]
- Van Schuerbeek, P.; Baeken, C.; de Mey, J. The Heterogeneity in Retrieved Relations between the Personality Trait “Harm Avoidance” and Gray Matter Volumes Due to Variations in the VBM and ROI Labeling Processing Settings. PLoS ONE 2016, 11, e0153865. [Google Scholar] [CrossRef]
- Koenders, L.; Cousijn, J.; Vingerhoets, W.A.M.; van den Brink, W.; Wiers, R.W.; Meijer, C.J.; Machielsen, M.W.J.; Veltman, D.J.; Goudriaan, A.E.; de Haan, L. Grey matter changes associated with heavy cannabis use: A longitudinal sMRI study. PLoS ONE 2016, 11, e0152482. [Google Scholar] [CrossRef] [PubMed]
- LaMontagne, P.J.; Benzinger, T.L.; Morris, J.C.; Keefe, S.; Hornbeck, R.; Xiong, C.; Grant, E.; Hassenstab, J.; Moulder, K.; Vlassenko, A.G.; et al. OASIS-3: Longitudinal Neuroimaging, Clinical, and Cognitive Dataset for Normal Aging and Alzheimer Disease. medRxiv 2019. [Google Scholar] [CrossRef]
- Bouthillier, A.; van Loveren, H.R.; Keller, J.T. Segments of the internal carotid artery: A new classification. Neurosurgery 1996, 38, 425–433. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the 18th International Conference, Munich, Germany, 5–9 October 2015; Volume 9351, pp. 234–241. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Delving Deep into Rectifiers: Surpassing Human-Level Performance on ImageNet Classification. In Proceedings of the International Conference on Computer Vision, Las Condes, Chile, 11–18 December 2015; Available online: http://arxiv.org/abs/1502.01852 (accessed on 20 May 2024).
- Ioffe, S.; Szegedy, C. Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. In Proceedings of the 32nd International Conference on International Conference on Machine Learning, Lille, France, 6 July–11 July 2015; Volume 37, pp. 448–456. [Google Scholar]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Salakhutdinov, R. Dropout: A Simple Way to Prevent Neural Networks from Overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Zhang, Z.; Liu, Q.; Wang, Y. Road Extraction by Deep Residual U-Net. IEEE Geosci. Remote Sens. Lett. 2018, 15, 749–753. [Google Scholar] [CrossRef]
- Diakogiannis, F.I.; Waldner, F.; Caccetta, P.; Wu, C. ResUNet-a: A deep learning framework for semantic segmentation of remotely sensed data. ISPRS J. Photogramm. Remote Sens. 2020, 162, 94–114. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J.L. Adam: A Method for Stochastic Optimization. In Proceedings of the 3rd International Conference on Learning Representations, ICLR 2015—Conference Track Proceedings, San Diego, CA, USA, 7–9 May 2015; Available online: https://arxiv.org/abs/1412.6980v9 (accessed on 17 May 2024).
- Dice, L.R. Measures of the Amount of Ecologic Association Between Species. Ecology 1945, 26, 297–302. [Google Scholar] [CrossRef]
- Sudre, C.H.; Li, W.; Vercauteren, T.; Ourselin, S.; Jorge Cardoso, M. Generalised dice overlap as a deep learning loss function for highly unbalanced segmentations. arXiv 2017, arXiv:1707.03237. [Google Scholar] [CrossRef]
- Müller, D.; Soto-Rey, I.; Kramer, F. Towards a guideline for evaluation metrics in medical image segmentation. BMC Res. Notes 2022, 15, 210. [Google Scholar] [CrossRef] [PubMed]
- Ni, Z.-L.; Bian, G.-B.; Zhou, X.-H.; Hou, Z.-G.; Xie, X.-L.; Wang, C.; Zhou, Y.-J.; Li, R.-Q.; Li, Z. RAUNet: Residual Attention U-Net for Semantic Segmentation of Cataract Surgical Instruments. In Proceedings of the International Conference on Neural Information Processing, Sydney, NSW, Australia, 12–15 December 2019. [Google Scholar] [CrossRef]
- Kundu, S.; Karale, V.; Ghorai, G.; Sarkar, G.; Ghosh, S.; Dhara, A.K. Nested U-Net for Segmentation of Red Lesions in Retinal Fundus Images and Sub-image Classification for Removal of False Positives. J. Digit. Imaging 2022, 35, 1111–1119. [Google Scholar] [CrossRef]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16 × 16 words: Transformers for image recognition at scale. In Proceedings of the ICLR 2021—9th International Conference on Learning Representations, Virtual, 3–7 May 2021. [Google Scholar]
- Huang, Z.; Wang, H.; Deng, Z.; Ye, J.; Su, Y.; Sun, H.; He, J.; Gu, Y.; Gu, L.; Zhang, S.; et al. STU-Net: Scalable and Transferable Medical Image Segmentation Models Empowered by Large-Scale Supervised Pre-training. arXiv 2023, arXiv:2304.06716. [Google Scholar] [CrossRef]
- Qin, Z.; Chen, Y.; Zhu, G.; Zhou, E.; Zhou, Y.; Zhou, Y.; Zhu, C. Enhanced Pseudo-Label Generation with Self-supervised Training for Weakly-supervised Semantic Segmentation. IEEE Trans. Circuits Syst. Video Technol. 2024. early access. [Google Scholar] [CrossRef]
- Feng, J.; Li, C.; Wang, J. CAM-TMIL: A Weakly-Supervised Segmentation Framework for Histopathology based on CAMs and MIL. J. Phys. Conf. Ser. 2023, 2547, 012014. [Google Scholar] [CrossRef]
- Cheng, H.; Gu, C.; Wu, K. Weakly-Supervised Semantic Segmentation via Self-training. J. Phys. Conf. Ser. 2020, 1487, 012001. [Google Scholar] [CrossRef]
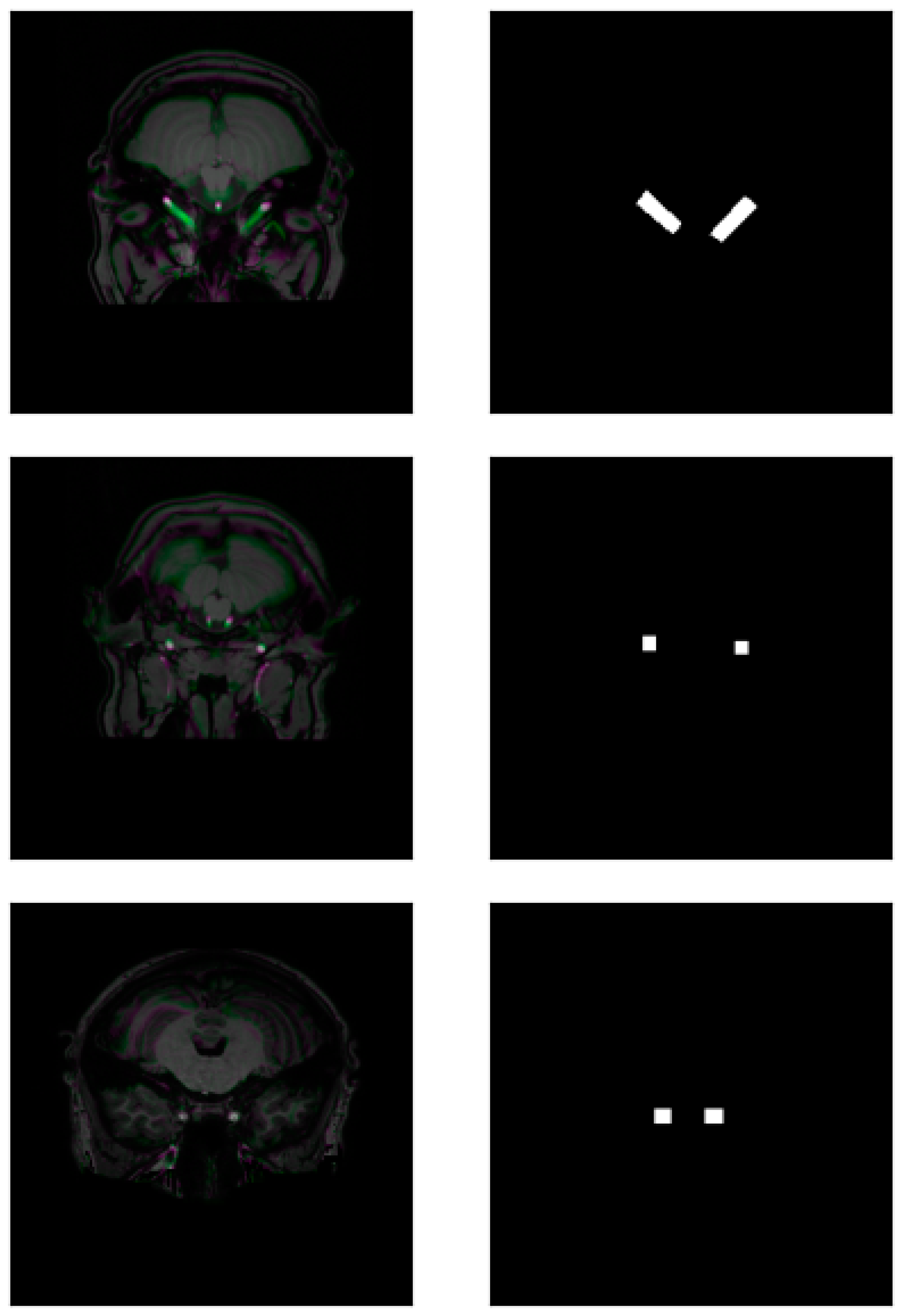
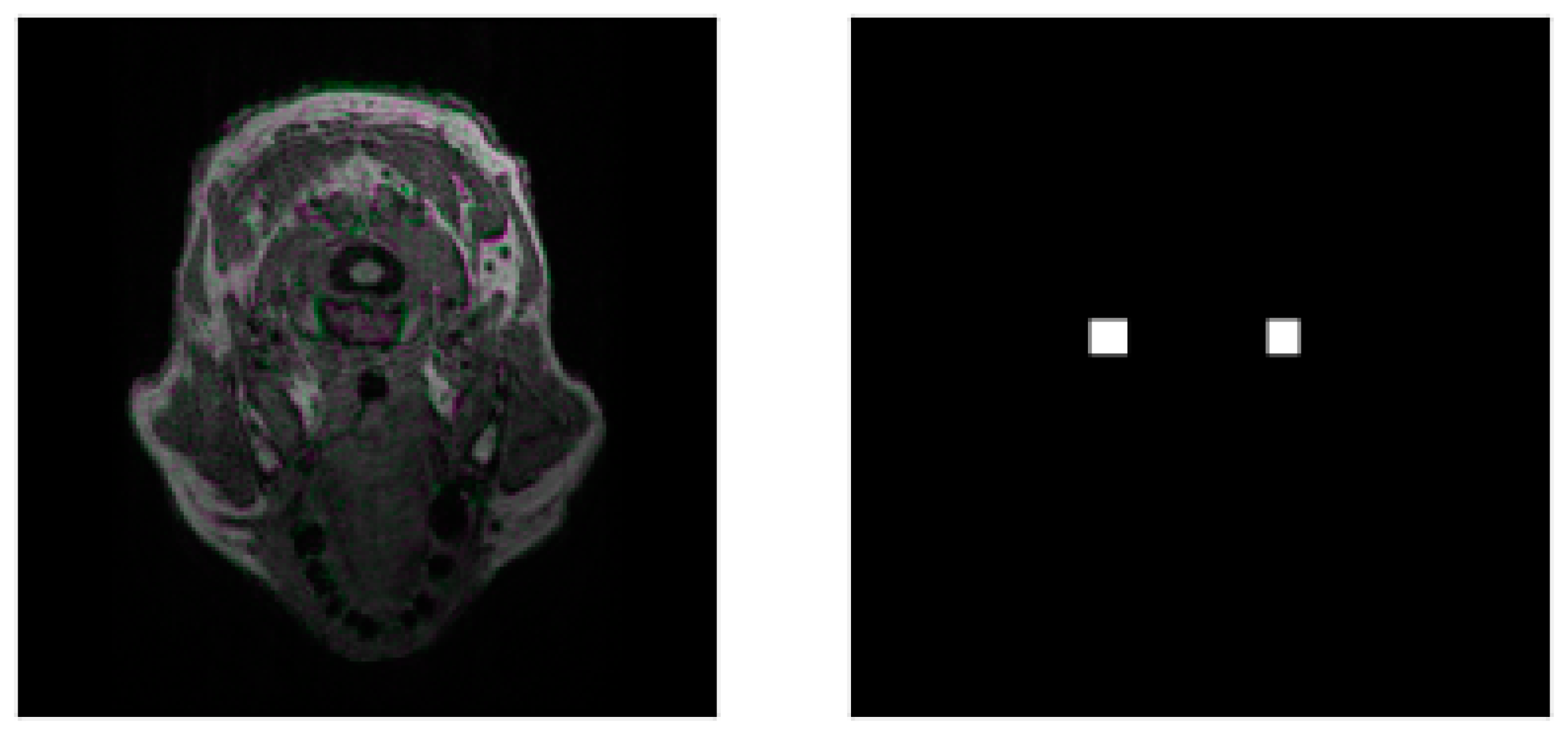


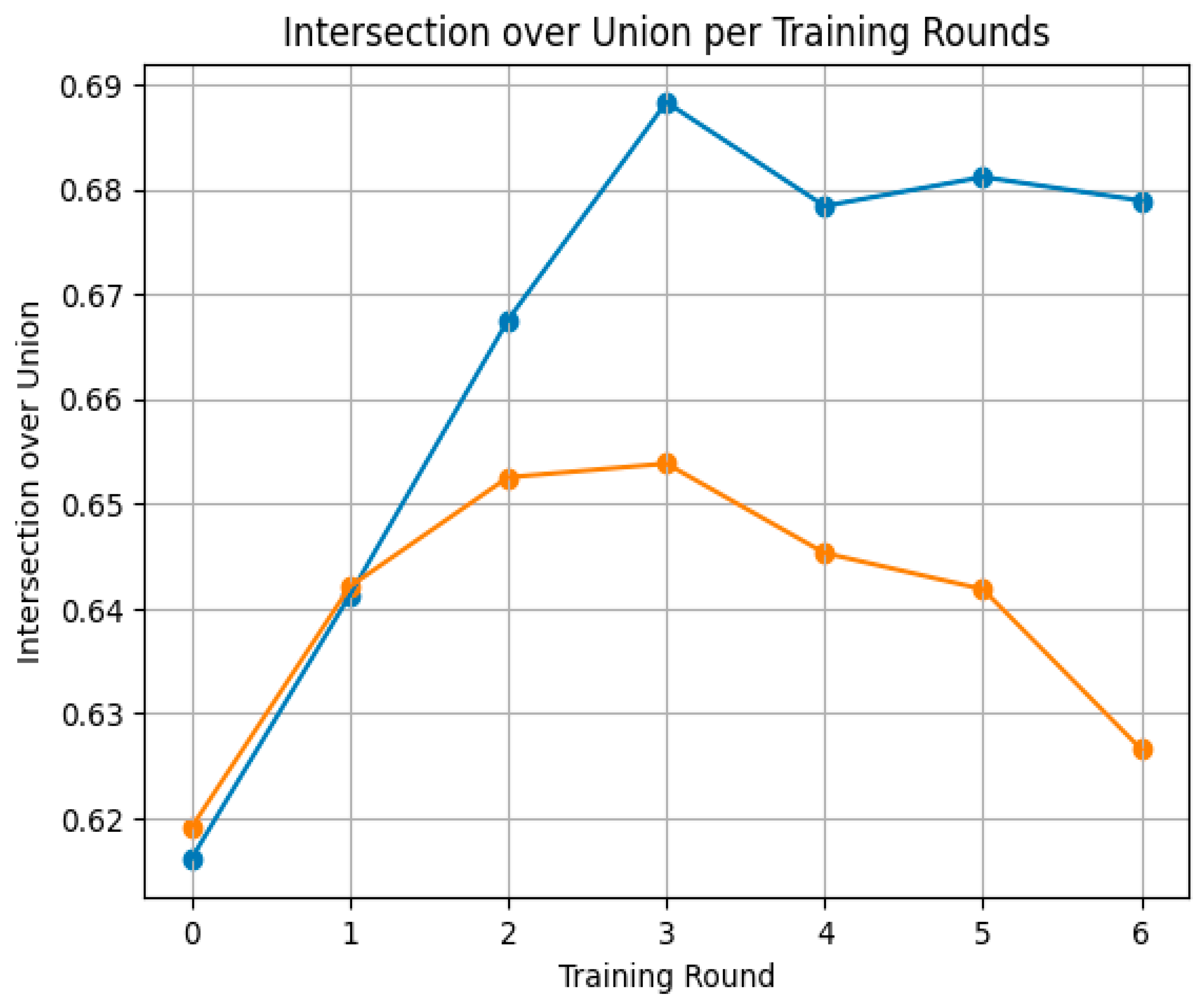

| Round of Training | IoU | DSC |
|---|---|---|
| Round 0 | 0.616 ± 0.066 | 0.365 ± 0.169 |
| Round 1 | 0.641 ± 0.073 | 0.426 ± 0.174 |
| Round 2 | 0.668 ± 0.085 | 0.480 ± 0.197 |
| Round 3 | 0.688 ± 0.085 | 0.526 ± 0.194 |
| Round 4 | 0.678 ± 0.082 | 0.504 ± 0.198 |
| Round 5 | 0.681 ± 0.080 | 0.512 ± 0.191 |
| Round 6 | 0.679 ± 0.081 | 0.506 ± 0.193 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Araújo, A.S.; Pinho, M.S.; Marques da Silva, A.M.; Fiorentini, L.F.; Becker, J. A 2.5D Self-Training Strategy for Carotid Artery Segmentation in T1-Weighted Brain Magnetic Resonance Images. J. Imaging 2024, 10, 161. https://doi.org/10.3390/jimaging10070161
de Araújo AS, Pinho MS, Marques da Silva AM, Fiorentini LF, Becker J. A 2.5D Self-Training Strategy for Carotid Artery Segmentation in T1-Weighted Brain Magnetic Resonance Images. Journal of Imaging. 2024; 10(7):161. https://doi.org/10.3390/jimaging10070161
Chicago/Turabian Stylede Araújo, Adriel Silva, Márcio Sarroglia Pinho, Ana Maria Marques da Silva, Luis Felipe Fiorentini, and Jefferson Becker. 2024. "A 2.5D Self-Training Strategy for Carotid Artery Segmentation in T1-Weighted Brain Magnetic Resonance Images" Journal of Imaging 10, no. 7: 161. https://doi.org/10.3390/jimaging10070161
APA Stylede Araújo, A. S., Pinho, M. S., Marques da Silva, A. M., Fiorentini, L. F., & Becker, J. (2024). A 2.5D Self-Training Strategy for Carotid Artery Segmentation in T1-Weighted Brain Magnetic Resonance Images. Journal of Imaging, 10(7), 161. https://doi.org/10.3390/jimaging10070161








