U-Net Convolutional Neural Network for Mapping Natural Vegetation and Forest Types from Landsat Imagery in Southeastern Australia
Abstract
1. Introduction
- Test if a U-Net CNN model using Landsat data can map natural vegetation and forest types and help to understand their change over time in SE Australia;
- Quantify the effectiveness of the method and its temporal stability;
- Compare CNN results with vegetation maps generated using RF.
2. Material and Methods
2.1. Input Data
2.2. Label Data
2.3. Modelling Methodology
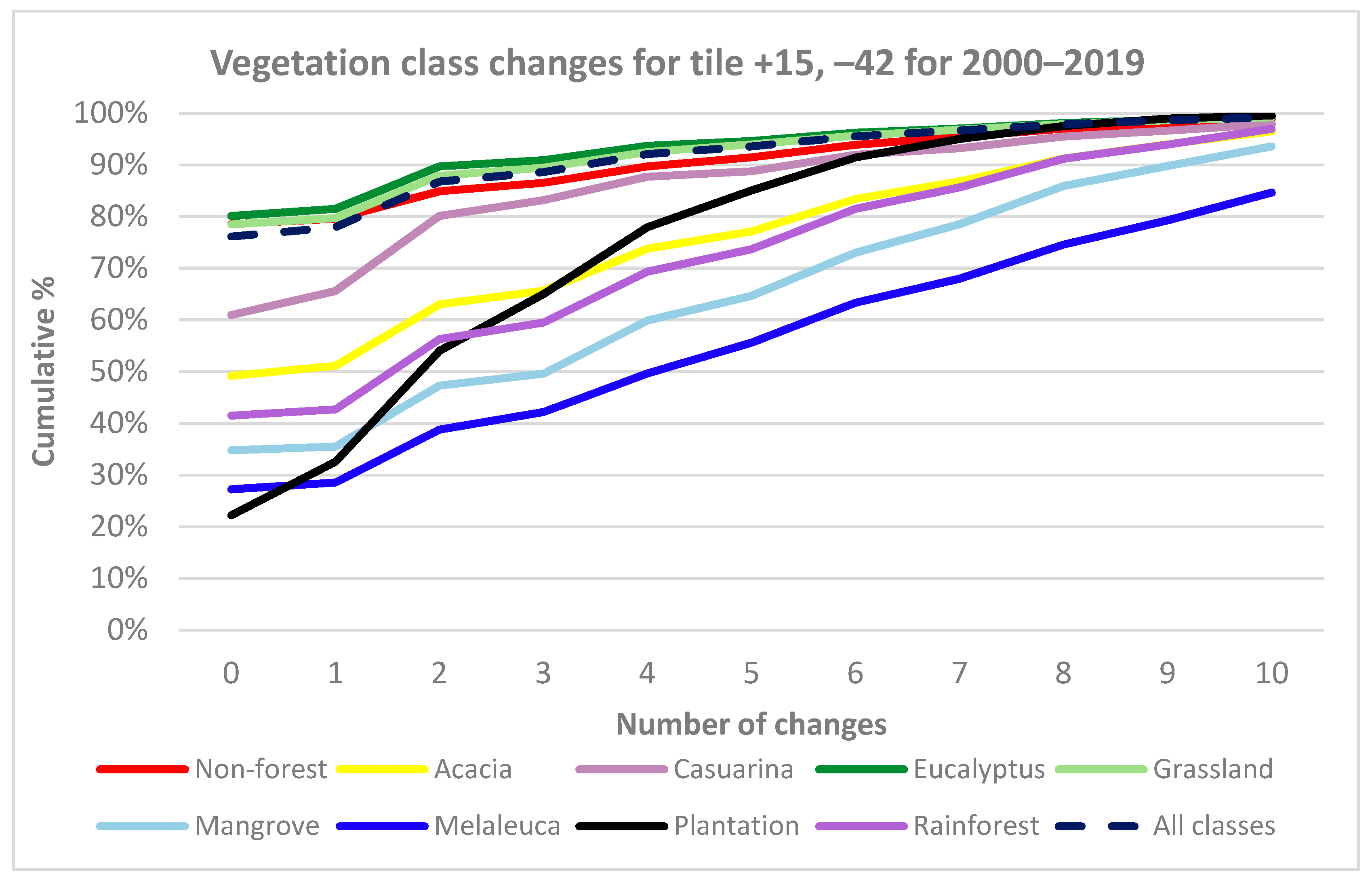
- Year-on-year changes for all years (2000–2001, 2001–2002, …, 2018–2019) were compared using confusion matrices to determine the similarity of model results for each of the 19 annual transitions.
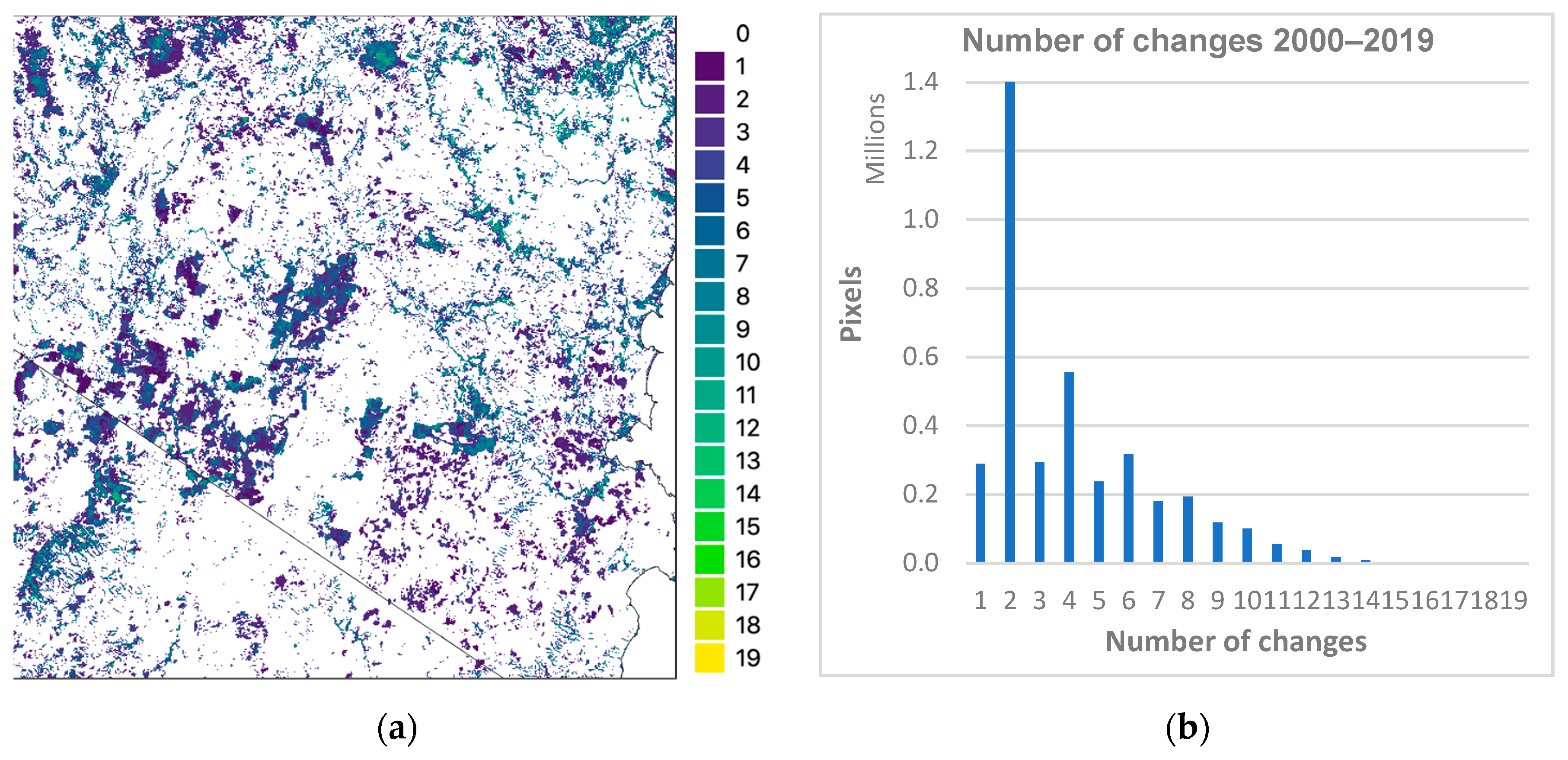
- The total number of CNN class changes for all 16 M pixels in the test tile for the 20 years was determined. These results were further subdivided by vegetation class, using the 2018 label data for the tile as the reference to examine the temporal stability of each predicted model class.
- All plots where only one species of interest (Table 3) was observed (4620 plots);
- Plots with ≥50% crown cover (3456 plots);
- Plots of ≥400 m2 area − generally as 20 m × 20 m (5439 plots);
- Plots with one species of interest and ≥50% crown cover (1667 plots);
- Plots with one species of interest and ≥400 m2 (3785 plots);
- Plots with ≥50% crown cover and ≥400 m2 (3043 plots);
- Plots with one species of interest, ≥50% crown cover and ≥400 m2 (1436 plots).
3. Results
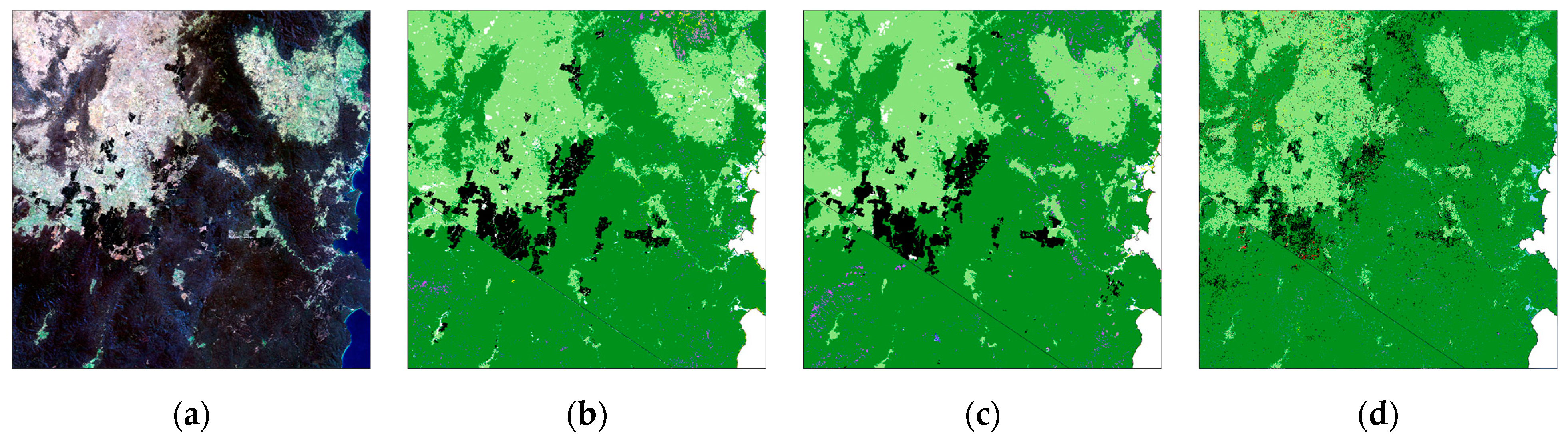
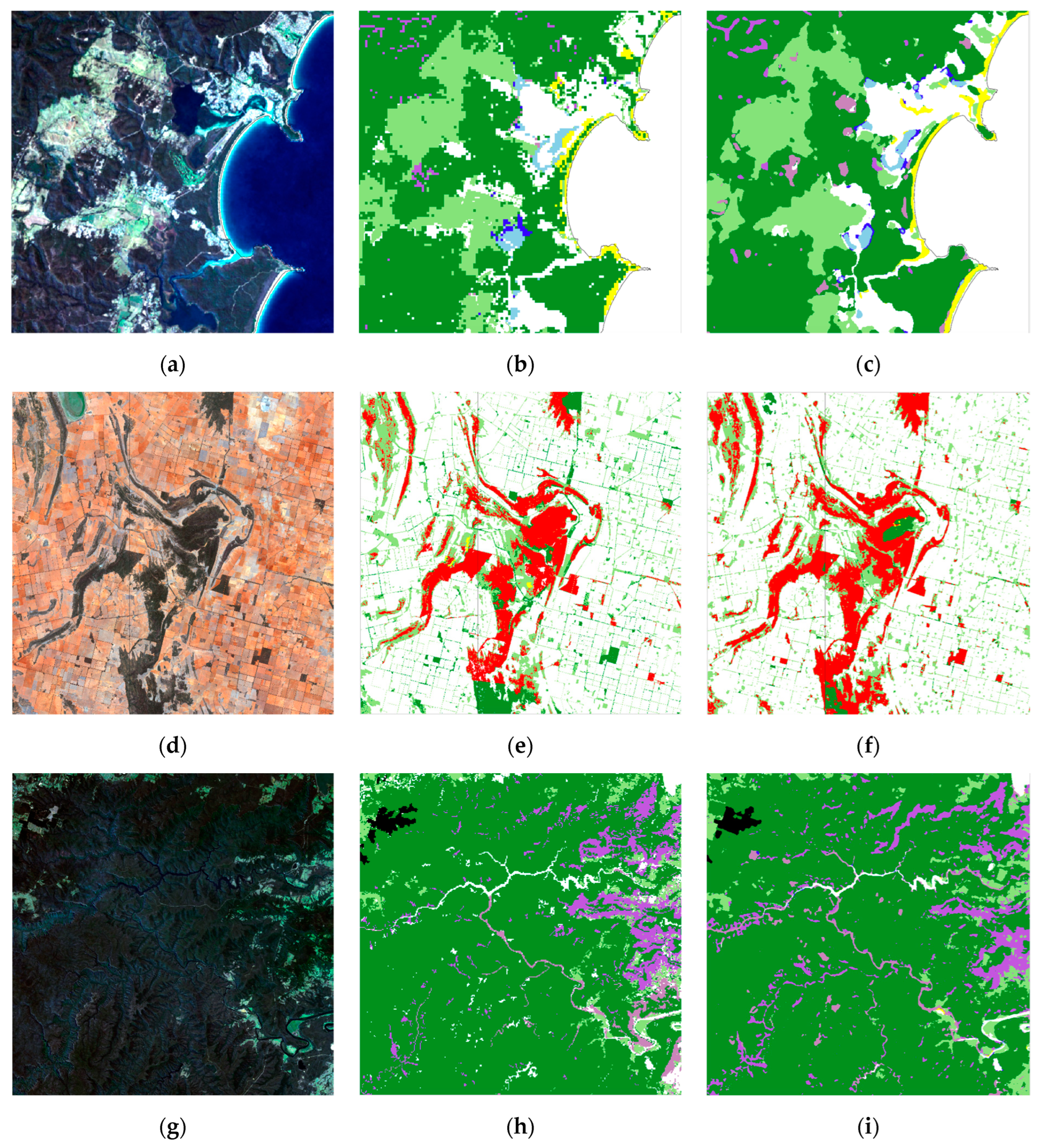
3.1. Temporal Stability of CNN
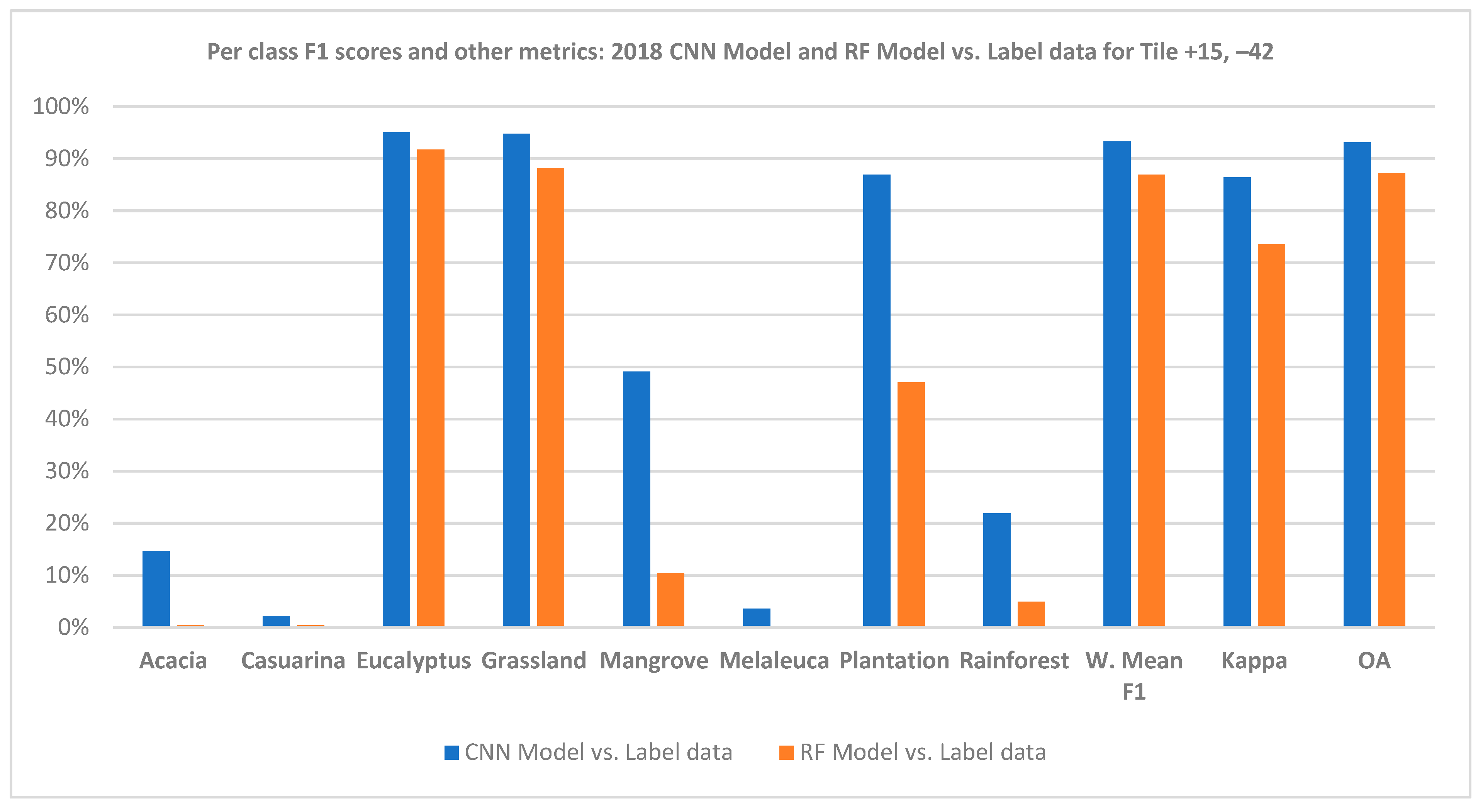
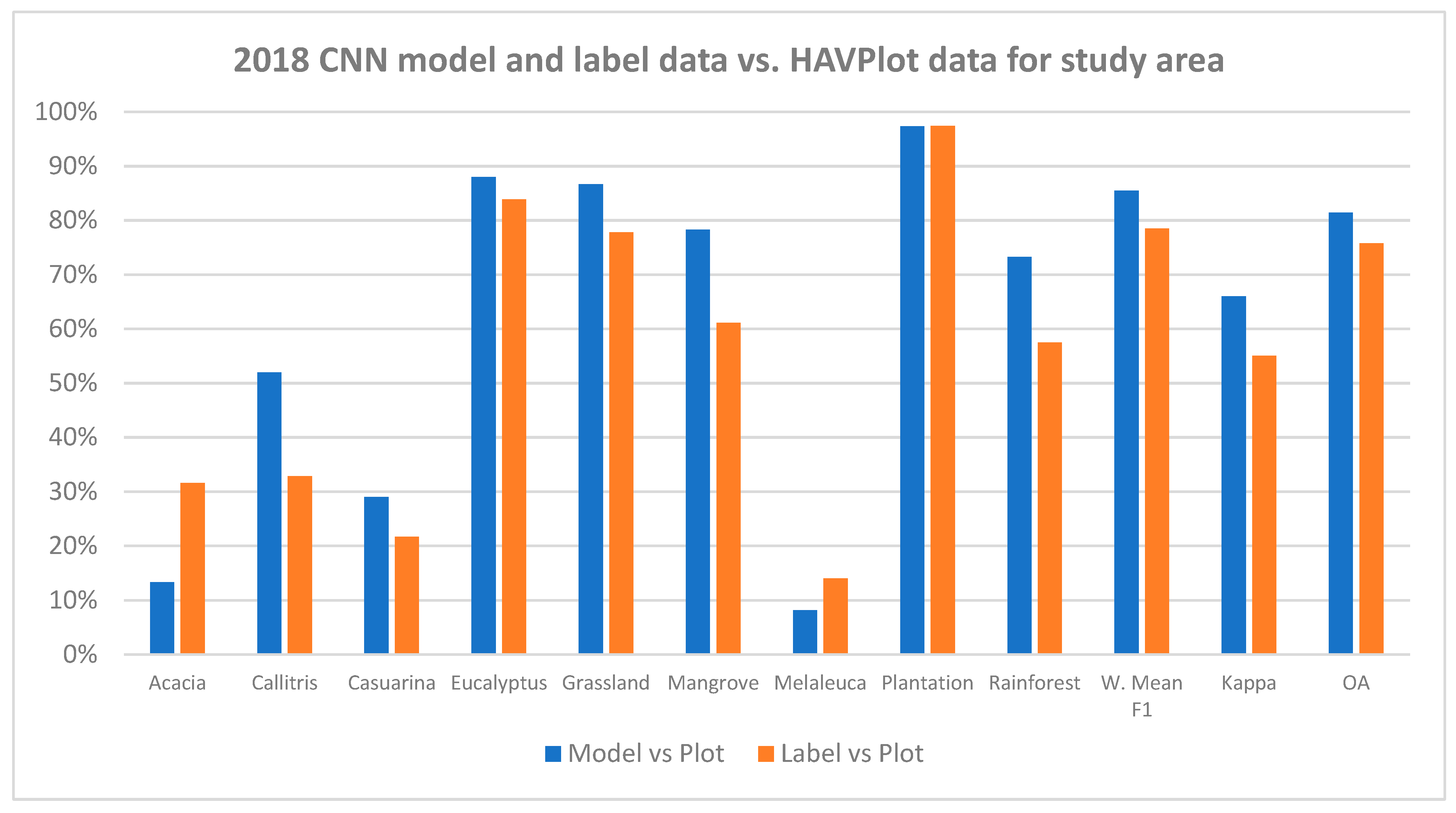
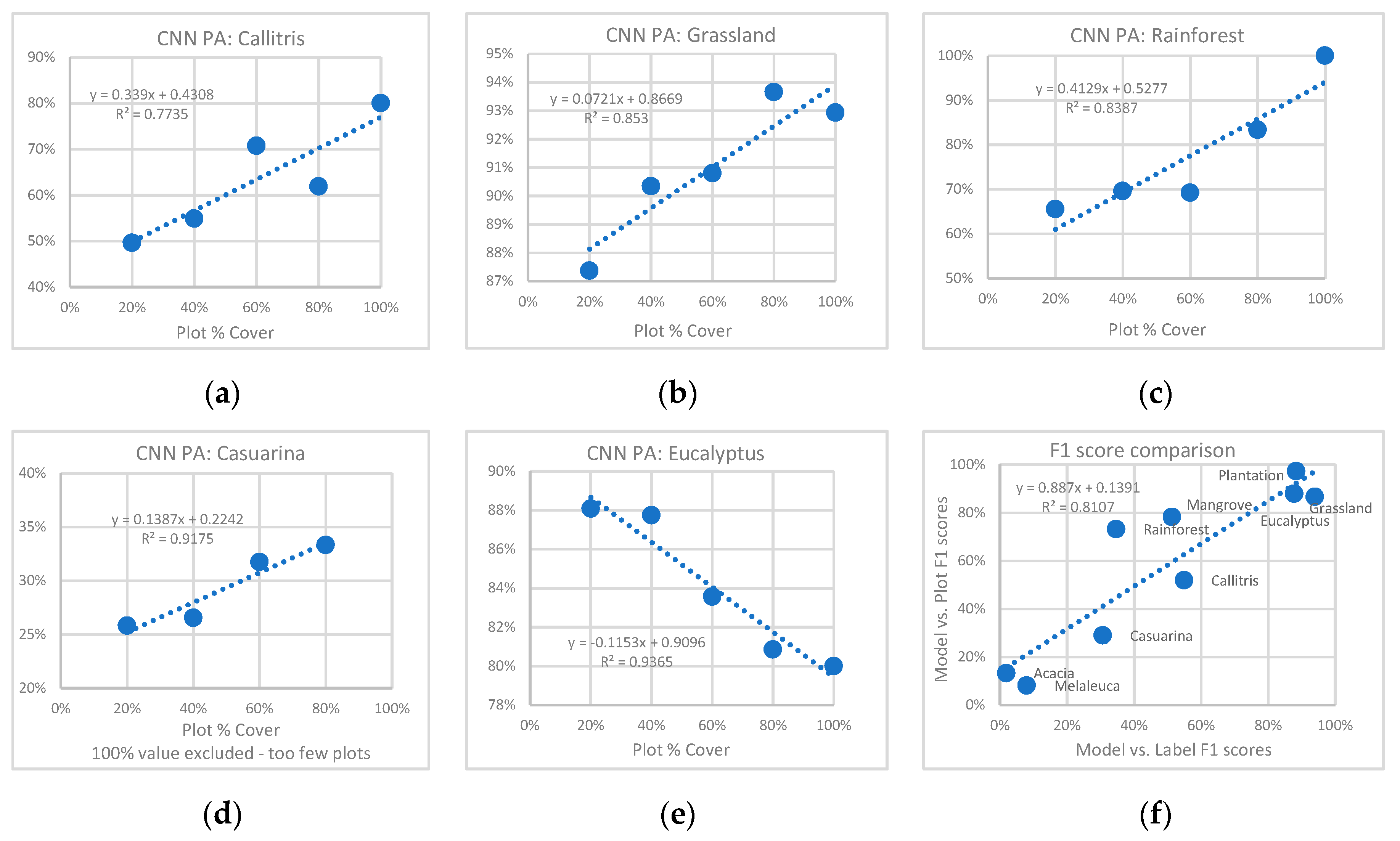
3.2. Comparison of CNN Results to HAVPlot Vegetation Survey Data
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| All Plots in Study Area | Plots with One Species of Interest (SOI) | Plots >= 50% Cover | Plots >= 400 m2 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Class Name | Ref. | No. Plots | Match | % | No. Plots | Match | % | No. Plots | Match | % | No. Plots | Match | % |
| Acacia | Label | 78 | 24 | 30.8% | 65 | 20 | 30.8% | 30 | 9 | 30.0% | 63 | 23 | 36.5% |
| Model | 78 | 5 | 6.4% | 65 | 3 | 4.6% | 30 | 3 | 10.0% | 63 | 5 | 7.9% | |
| Callitris | Label | 536 | 136 | 25.4% | 391 | 113 | 28.9% | 244 | 56 | 23.0% | 389 | 78 | 20.1% |
| Model | 536 | 237 | 44.2% | 391 | 194 | 49.6% | 244 | 99 | 40.6% | 389 | 147 | 37.8% | |
| Casuarina | Label | 344 | 60 | 17.4% | 244 | 47 | 19.3% | 209 | 37 | 17.7% | 315 | 60 | 19.0% |
| Model | 344 | 75 | 21.8% | 244 | 63 | 25.8% | 209 | 47 | 22.5% | 315 | 75 | 23.8% | |
| Eucalyptus | Label | 4097 | 3521 | 85.9% | 2601 | 2200 | 84.6% | 2161 | 1871 | 86.6% | 3546 | 3030 | 85.4% |
| Model | 4097 | 3658 | 89.3% | 2601 | 2291 | 88.1% | 2161 | 1953 | 90.4% | 3546 | 3150 | 88.8% | |
| Grassland | Label | 768 | 615 | 80.1% | 768 | 615 | 80.1% | 317 | 270 | 85.2% | 515 | 445 | 86.4% |
| Model | 768 | 671 | 87.4% | 768 | 671 | 87.4% | 317 | 283 | 89.3% | 515 | 457 | 88.7% | |
| Mangrove | Label | 33 | 13 | 39.4% | 32 | 13 | 40.6% | 29 | 11 | 37.9% | 30 | 13 | 43.3% |
| Model | 33 | 20 | 60.6% | 32 | 19 | 59.4% | 29 | 18 | 62.1% | 30 | 19 | 63.3% | |
| Melaleuca | Label | 76 | 5 | 6.6% | 56 | 3 | 5.4% | 42 | 4 | 9.5% | 68 | 4 | 5.9% |
| Model | 76 | 2 | 2.6% | 56 | 1 | 1.8% | 42 | 2 | 4.8% | 68 | 2 | 2.9% | |
| Plantation | Label | 190 | 188 | 98.9% | 190 | 188 | 98.9% | 190 | 188 | 98.9% | 190 | 188 | 98.9% |
| Model | 190 | 184 | 96.8% | 190 | 184 | 96.8% | 190 | 184 | 96.8% | 190 | 184 | 96.8% | |
| Rainforest | Label | 345 | 144 | 41.7% | 273 | 119 | 43.6% | 234 | 98 | 41.9% | 323 | 141 | 43.7% |
| Model | 345 | 224 | 64.9% | 273 | 179 | 65.6% | 234 | 156 | 66.7% | 323 | 215 | 66.6% | |
| Total Plots | 6467 | 4620 | 3456 | 5439 | |||||||||
| Label match | 4706 | 72.8% | 3318 | 71.8% | 2544 | 73.6% | 3982 | 73.2% | |||||
| Model match | 5076 | 78.5% | 3605 | 78.0% | 2745 | 79.4% | 4254 | 78.2% | |||||
| OneSOI + >= 50% Cover | OneSOI + >= 400 m2 | Plots >= 50% Cover + >= 400 m2 | Plots OneSOI + >= 50% Cover + >= 400 m2 | ||||||||||
| Class Name | Ref. | No. Plots | Match | % | No. Plots | Match | % | No. Plots | Match | % | No. Plots | Match | % |
| Acacia | Label | 18 | 6 | 33.3% | 51 | 20 | 39.2% | 27 | 8 | 29.6% | 16 | 6 | 37.5% |
| Model | 18 | 1 | 5.6% | 51 | 3 | 5.9% | 27 | 3 | 11.1% | 16 | 1 | 6.3% | |
| Callitris | Label | 113 | 33 | 29.2% | 264 | 62 | 23.5% | 207 | 38 | 18.4% | 91 | 22 | 24.2% |
| Model | 113 | 65 | 57.5% | 264 | 119 | 45.1% | 207 | 73 | 35.3% | 91 | 50 | 54.9% | |
| Casuarina | Label | 111 | 24 | 21.6% | 224 | 47 | 21.0% | 192 | 37 | 19.3% | 103 | 24 | 23.3% |
| Model | 111 | 35 | 31.5% | 224 | 63 | 28.1% | 192 | 47 | 24.5% | 103 | 35 | 34.0% | |
| Eucalyptus | Label | 706 | 587 | 83.1% | 2202 | 1851 | 84.1% | 1933 | 1665 | 86.1% | 624 | 518 | 83.0% |
| Model | 706 | 624 | 88.4% | 2202 | 1928 | 87.6% | 1933 | 1737 | 89.9% | 624 | 548 | 87.8% | |
| Grassland | Label | 317 | 270 | 85.2% | 515 | 445 | 86.4% | 211 | 184 | 87.2% | 211 | 184 | 87.2% |
| Model | 317 | 283 | 89.3% | 515 | 457 | 88.7% | 211 | 186 | 88.2% | 211 | 186 | 88.2% | |
| Mangrove | Label | 28 | 11 | 39.3% | 30 | 13 | 43.3% | 28 | 11 | 39.3% | 28 | 11 | 39.3% |
| Model | 28 | 17 | 60.7% | 30 | 19 | 63.3% | 28 | 17 | 60.7% | 28 | 17 | 60.7% | |
| Melaleuca | Label | 22 | 2 | 9.1% | 49 | 2 | 4.1% | 37 | 4 | 10.8% | 18 | 2 | 11.1% |
| Model | 22 | 1 | 4.5% | 49 | 1 | 2.0% | 37 | 2 | 5.4% | 18 | 1 | 5.6% | |
| Plantation | Label | 190 | 188 | 98.9% | 190 | 188 | 98.9% | 190 | 188 | 98.9% | 190 | 188 | 98.9% |
| Model | 190 | 184 | 96.8% | 190 | 184 | 96.8% | 190 | 184 | 96.8% | 190 | 184 | 96.8% | |
| Rainforest | Label | 162 | 73 | 45.1% | 260 | 119 | 45.8% | 218 | 95 | 43.6% | 155 | 73 | 47.1% |
| Model | 162 | 111 | 68.5% | 260 | 176 | 67.7% | 218 | 149 | 68.3% | 155 | 110 | 71.0% | |
| Total Plots | 1667 | 3785 | 3043 | 1436 | |||||||||
| Label match | 1194 | 71.6% | 2747 | 72.6% | 2230 | 73.3% | 1028 | 71.6% | |||||
| Model match | 1321 | 79.2% | 2950 | 77.9% | 2398 | 78.8% | 1132 | 78.8% | |||||
| Band 1—Blue (B) | Band 2—Green (G) | Band 3—Red (R) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Class Name | Min | Max | Mean | Std. Dev. | Min | Max | Mean | Std. Dev. | Min | Max | Mean | Std. Dev. |
| Acacia | 0 | 0.386 | 0.0523613 | 0.0305669 | 0 | 0.4553 | 0.076023 | 0.043626 | 0 | 0.5183 | 0.0851198 | 0.0487858 |
| Callitris | 0.014 | 0.3308 | 0.0342461 | 0.0056443 | 0.0218 | 0.4181 | 0.053862 | 0.008623 | 0.023 | 0.4603 | 0.0764481 | 0.0186796 |
| Casuarina | 0 | 0.2179 | 0.0281768 | 0.0092408 | 0 | 0.2542 | 0.0385615 | 0.0127618 | 0 | 0.2807 | 0.0411939 | 0.0166458 |
| Eucalyptus | 0 | 0.3911 | 0.0255216 | 0.0095069 | 0 | 0.446 | 0.0381438 | 0.011564 | 0 | 0.4807 | 0.0428946 | 0.016918 |
| Grassland | 0 | 0.5751 | 0.0596124 | 0.0142985 | 0 | 0.6337 | 0.0877194 | 0.0186126 | 0 | 0.6665 | 0.114662 | 0.0306597 |
| Mangrove | 0 | 0.1643 | 0.1023459 | 0.0542211 | 0 | 0.237 | 0.1394232 | 0.0765441 | 0 | 0.2726 | 0.1493367 | 0.086076 |
| Melaleuca | 0 | 0.1627 | 0.0318912 | 0.0152693 | 0 | 0.203 | 0.0403129 | 0.0203437 | 0 | 0.2472 | 0.0390473 | 0.023417 |
| Plantation | 0.0009 | 0.2357 | 0.0197081 | 0.0143993 | 0.0054 | 0.3203 | 0.034739 | 0.0191936 | 0.0045 | 0.3959 | 0.0349825 | 0.0281191 |
| Rainforest | 0 | 0.1295 | 0.0202462 | 0.0066634 | 0 | 0.1764 | 0.0314772 | 0.0095101 | 0 | 0.2205 | 0.0260377 | 0.009938 |
| All classes | 0 | 0.5751 | 0.0502893 | 0.0207012 | 0 | 0.6337 | 0.0742551 | 0.0303376 | 0 | 0.6892 | 0.0989609 | 0.0482741 |
| Band 4—NIR | Band 5—SWIR1 | Band 6—SWIR2 | ||||||||||
| Min | Max | Mean | Std. Dev. | Min | Max | Mean | Std. Dev. | Min | Max | Mean | Std. Dev. | |
| Acacia | 0 | 0.6203 | 0.2073108 | 0.063295 | 0 | 0.7281 | 0.15896 | 0.0732649 | 0 | 0.5691 | 0.1045825 | 0.0581059 |
| Callitris | 0.0204 | 0.533 | 0.194315 | 0.0206494 | 0.0193 | 0.4865 | 0.2204281 | 0.0396873 | 0.013 | 0.3946 | 0.152838 | 0.0371982 |
| Casuarina | 0 | 0.4742 | 0.1758893 | 0.0439502 | 0 | 0.5422 | 0.1354448 | 0.0475161 | 0 | 0.4237 | 0.0802761 | 0.0365492 |
| Eucalyptus | 0 | 0.5561 | 0.1999782 | 0.0328359 | 0 | 0.6103 | 0.1366978 | 0.0447655 | 0 | 0.4604 | 0.0762808 | 0.0351649 |
| Grassland | 0 | 0.6797 | 0.2543644 | 0.0382521 | 0 | 0.7416 | 0.3229756 | 0.0615734 | 0 | 0.6705 | 0.2293733 | 0.0612242 |
| Mangrove | 0 | 0.4128 | 0.1243975 | 0.0510203 | 0 | 0.3683 | 0.0319198 | 0.0423791 | 0 | 0.3092 | 0.0193094 | 0.0257986 |
| Melaleuca | 0 | 0.4582 | 0.1512467 | 0.0762302 | 0 | 0.3717 | 0.0932456 | 0.0544463 | 0 | 0.3212 | 0.0546035 | 0.0388007 |
| Plantation | 0.0088 | 0.5034 | 0.230016 | 0.0380534 | 0.0049 | 0.573 | 0.1209631 | 0.0714456 | 0.0035 | 0.4487 | 0.0673211 | 0.055262 |
| Rainforest | 0 | 0.5718 | 0.2449978 | 0.0540823 | 0 | 0.3887 | 0.1034989 | 0.0329448 | 0 | 0.3114 | 0.0468167 | 0.020886 |
| All classes | 0 | 0.7573 | 0.2270545 | 0.0583411 | 0 | 0.9071 | 0.2666144 | 0.1124712 | 0 | 0.9025 | 0.192971 | 0.099816 |
| G/R | (R + G + B)/3 | G/(R + G + B) | R/(R + G + B) | B/(R + G + B) | NDVI | NDWI | ||||||
| Acacia | 0.8931295 | 0.1785965 | 0.3560728 | 0.3986799 | 0.2452473 | 0.42 | −0.46 | |||||
| Callitris | 0.704556 | 0.1417254 | 0.3273165 | 0.4645713 | 0.2081121 | 0.44 | −0.57 | |||||
| Casuarina | 0.9360971 | 0.0891477 | 0.3572752 | 0.3816647 | 0.26106 | 0.62 | −0.64 | |||||
| Eucalyptus | 0.8892447 | 0.0895455 | 0.3579559 | 0.4025392 | 0.2395049 | 0.65 | −0.68 | |||||
| Grassland | 0.7650259 | 0.2222522 | 0.3348148 | 0.4376515 | 0.2275337 | 0.38 | −0.49 | |||||
| Mangrove | 0.9336167 | 0.3228752 | 0.3564847 | 0.3818319 | 0.2616834 | −0.09 | 0.06 | |||||
| Melaleuca | 1.0324124 | 0.0899907 | 0.3623588 | 0.3509826 | 0.2866585 | 0.59 | −0.58 | |||||
| Plantation | 0.9930386 | 0.0762908 | 0.3884504 | 0.3911735 | 0.2203761 | 0.74 | −0.74 | |||||
| Rainforest | 1.2089066 | 0.0642636 | 0.4047935 | 0.3348427 | 0.2603638 | 0.81 | −0.77 | |||||
| All classes | 0.750348 | 0.1899791 | 0.3322298 | 0.4427675 | 0.2250027 | 0.39 | −0.51 | |||||
References
- DCCEEW (Department of Climate Change, Energy, the Environment and Water). NVIS Data Products. 2020. Available online: https://www.dcceew.gov.au/environment/land/native-vegetation/national-vegetation-information-system/data-products (accessed on 10 May 2024).
- Scarth, P.; Armston, J.; Lucas, R.; Bunting, P. A Structural Classification of Australian Vegetation Using ICESat/GLAS, ALOS PALSAR, and Landsat Sensor Data. Remote Sens. 2019, 11, 147. [Google Scholar] [CrossRef]
- ABARES. Forests of Australia (2018). 2019. Available online: https://www.agriculture.gov.au/abares/forestsaustralia/forest-data-maps-and-tools/spatial-data/forest-cover#forests-of-australia-2018 (accessed on 10 May 2024).
- Flood, N.; Watson, F.; Collett, L. Using a U-Net Convolutional Neural Network to Map Woody Vegetation Extent from High Resolution Satellite Imagery across Queensland, Australia. Int. J. Appl. Earth Obs. Geoinf. 2019, 82, 101897. [Google Scholar] [CrossRef]
- Bhandari, S.; Phinn, S.; Gill, T. Preparing Landsat Image Time Series (LITS) for Monitoring Changes in Vegetation Phenology in Queensland, Australia. Remote Sens. 2012, 4, 1856–1886. [Google Scholar] [CrossRef]
- Kalinaki, K.; Malik, O.A.; Lai, D.T.C.; Sukri, R.S.; Wahab, R.B.H.A. Spatial-Temporal Mapping of Forest Vegetation Cover Changes along Highways in Brunei Using Deep Learning Techniques and Sentinel-2 Images. Ecol. Inform. 2023, 77, 102193. [Google Scholar] [CrossRef]
- Busby, J.R. A Biogeoclimatic Analysis of Nothofagus Cunninghamii (Hook.) Oerst. in Southeastern Australia. Aust. J. Ecol. 1986, 11, 1–7. [Google Scholar] [CrossRef]
- Booth, T.H.; Nix, H.A.; Busby, J.R.; Hutchinson, M.F. bioclim: The First Species Distribution Modelling Package, Its Early Applications and Relevance to Most Current MaxEnt Studies. Divers. Distrib. 2014, 20, 1–9. [Google Scholar] [CrossRef]
- Stockwell, D. The GARP Modelling System: Problems and Solutions to Automated Spatial Prediction. Int. J. Geogr. Inf. Sci. 1999, 13, 143–158. [Google Scholar] [CrossRef]
- Boston, A.N.; Stockwell, D.R.B. Interactive Species Distribution Reporting, Mapping and Modelling Using the World Wide Web. Comput. Netw. ISDN Syst. 1995, 28, 231–238. [Google Scholar] [CrossRef]
- Kattenborn, T.; Eichel, J.; Wiser, S.; Burrows, L.; Fassnacht, F.E.; Schmidtlein, S. Convolutional Neural Networks Accurately Predict Cover Fractions of Plant Species and Communities in Unmanned Aerial Vehicle Imagery. Remote Sens. Ecol. 2020, 6, 472–486. [Google Scholar] [CrossRef]
- Bhatt, P.; Maclean, A.L. Comparison of High-Resolution NAIP and Unmanned Aerial Vehicle (UAV) Imagery for Natural Vegetation Communities Classification Using Machine Learning Approaches. GIScience Remote Sens. 2023, 60, 2177448. [Google Scholar] [CrossRef]
- Xie, Y.; Sha, Z.; Yu, M. Remote Sensing Imagery in Vegetation Mapping: A Review. J. Plant Ecol. 2008, 1, 9–23. [Google Scholar] [CrossRef]
- Defries, R.S.; Townshend, J.R.G. Global Land Cover Characterization from Satellite Data: From Research to Operational Implementation? GCTE/LUCC RESEARCH REVIEW. Glob. Ecol. Biogeogr. 1999, 8, 367–379. [Google Scholar] [CrossRef]
- Hansen, M.C.; Stehman, S.V.; Potapov, P.V. Quantification of Global Gross Forest Cover Loss. Proc. Natl. Acad. Sci. USA 2010, 107, 8650–8655. [Google Scholar] [CrossRef]
- Caffaratti, G.D.; Marchetta, M.G.; Euillades, L.D.; Euillades, P.A.; Forradellas, R.Q. Improving Forest Detection with Machine Learning in Remote Sensing Data. Remote Sens. Appl. Soc. Environ. 2021, 24, 100654. [Google Scholar] [CrossRef]
- Lymburner, L.; Tan, P.; Mueller, N.; Thackway, R.; Lewis, A.; Thankappan, M.; Randall, L.; Islam, A.; Senarath, U. The National Dynamic Land Cover Dataset. Geoscience Australia; ACT: Symonston, Australia, 2011; pp. 3297–3300. Available online: https://ecat.ga.gov.au/geonetwork/srv/eng/catalog.search#/metadata/71071 (accessed on 10 May 2024).
- Kuhnell, C.A.; Goulevitch, B.M.; Danaher, T.J.; Harris, D.P. Mapping Woody Vegetation Cover over the State of Queensland Using Landsat TM Imagery. In Proceedings of the 9th Australasian Remote Sensing and Photogrammetry Conference, Sydney, Australia, 20–24 July 1998; pp. 20–24. [Google Scholar]
- Gill, T.; Johansen, K.; Phinn, S.; Trevithick, R.; Scarth, P.; Armston, J. A Method for Mapping Australian Woody Vegetation Cover by Linking Continental-Scale Field Data and Long-Term Landsat Time Series. Int. J. Remote Sens. 2017, 38, 679–705. [Google Scholar] [CrossRef]
- Calderón-Loor, M.; Hadjikakou, M.; Bryan, B.A. High-Resolution Wall-to-Wall Land-Cover Mapping and Land Change Assessment for Australia from 1985 to 2015. Remote Sens. Environ. 2021, 252, 112148. [Google Scholar] [CrossRef]
- Lucas, R.; Bunting, P.; Paterson, M.; Chisholm, L. Classification of Australian Forest Communities Using Aerial Photography, CASI and HyMap Data. Remote Sens. Environ. 2008, 112, 2088–2103. [Google Scholar] [CrossRef]
- Goodwin, N.; Turner, R.; Merton, R. Classifying Eucalyptus Forests with High Spatial and Spectral Resolution Imagery: An Investigation of Individual Species and Vegetation Communities. Aust. J. Bot. 2005, 53, 337. [Google Scholar] [CrossRef]
- Shang, X.; Chisholm, L.A. Classification of Australian Native Forest Species Using Hyperspectral Remote Sensing and Machine-Learning Classification Algorithms. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 2481–2489. [Google Scholar] [CrossRef]
- Fassnacht, F.E.; Latifi, H.; Stereńczak, K.; Modzelewska, A.; Lefsky, M.; Waser, L.T.; Straub, C.; Ghosh, A. Review of Studies on Tree Species Classification from Remotely Sensed Data. Remote Sens. Environ. 2016, 186, 64–87. [Google Scholar] [CrossRef]
- Johansen, K.; Phinn, S. Mapping Structural Parameters and Species Composition of Riparian Vegetation Using IKONOS and Landsat ETM+ Data in Australian Tropical Savannahs. Photogramm. Eng. Remote Sens. 2006, 72, 71–80. [Google Scholar] [CrossRef]
- De Bem, P.; De Carvalho Junior, O.; Fontes Guimarães, R.; Trancoso Gomes, R. Change Detection of Deforestation in the Brazilian Amazon Using Landsat Data and Convolutional Neural Networks. Remote Sens. 2020, 12, 901. [Google Scholar] [CrossRef]
- Guirado, E.; Alcaraz-Segura, D.; Cabello, J.; Puertas-Ruíz, S.; Herrera, F.; Tabik, S. Tree Cover Estimation in Global Drylands from Space Using Deep Learning. Remote Sens. 2020, 12, 343. [Google Scholar] [CrossRef]
- Zhou, X.; Zhou, W.; Li, F.; Shao, Z.; Fu, X. Vegetation Type Classification Based on 3D Convolutional Neural Network Model: A Case Study of Baishuijiang National Nature Reserve. Forests 2022, 13, 906. [Google Scholar] [CrossRef]
- Kussul, N.; Lavreniuk, M.; Skakun, S.; Shelestov, A. Deep Learning Classification of Land Cover and Crop Types Using Remote Sensing Data. IEEE Geosci. Remote Sens. Lett. 2017, 14, 778–782. [Google Scholar] [CrossRef]
- Bragagnolo, L.; Da Silva, R.V.; Grzybowski, J.M.V. Amazon Forest Cover Change Mapping Based on Semantic Segmentation by U-Nets. Ecol. Inform. 2021, 62, 101279. [Google Scholar] [CrossRef]
- Belgiu, M.; Drăguţ, L. Random Forest in Remote Sensing: A Review of Applications and Future Directions. ISPRS J. Photogramm. Remote Sens. 2016, 114, 24–31. [Google Scholar] [CrossRef]
- Talukdar, S.; Singha, P.; Mahato, S.; Shahfahad; Pal, S.; Liou, Y.-A.; Rahman, A. Land-Use Land-Cover Classification by Machine Learning Classifiers for Satellite Observations—A Review. Remote Sens. 2020, 12, 1135. [Google Scholar] [CrossRef]
- Stoian, A.; Poulain, V.; Inglada, J.; Poughon, V.; Derksen, D. Land Cover Maps Production with High Resolution Satellite Image Time Series and Convolutional Neural Networks: Adaptations and Limits for Operational Systems. Remote Sens. 2019, 11, 1986. [Google Scholar] [CrossRef]
- Syrris, V.; Hasenohr, P.; Delipetrev, B.; Kotsev, A.; Kempeneers, P.; Soille, P. Evaluation of the Potential of Convolutional Neural Networks and Random Forests for Multi-Class Segmentation of Sentinel-2 Imagery. Remote Sens. 2019, 11, 907. [Google Scholar] [CrossRef]
- Ma, L.; Liu, Y.; Zhang, X.; Ye, Y.; Yin, G.; Johnson, B.A. Deep Learning in Remote Sensing Applications: A Meta-Analysis and Review. ISPRS J. Photogramm. Remote Sens. 2019, 152, 166–177. [Google Scholar] [CrossRef]
- Boston, T.; Van Dijk, A.; Larraondo, P.R.; Thackway, R. Comparing CNNs and Random Forests for Landsat Image Segmentation Trained on a Large Proxy Land Cover Dataset. Remote Sens. 2022, 14, 3396. [Google Scholar] [CrossRef]
- Boston, T.; Van Dijk, A.; Thackway, R. Convolutional Neural Network Shows Greater Spatial and Temporal Stability in Multi-Annual Land Cover Mapping Than Pixel-Based Methods. Remote Sens. 2023, 15, 2132. [Google Scholar] [CrossRef]
- Moore, C.E.; Brown, T.; Keenan, T.F.; Duursma, R.A.; Van Dijk, A.I.J.M.; Beringer, J.; Culvenor, D.; Evans, B.; Huete, A.; Hutley, L.B.; et al. Reviews and Syntheses: Australian Vegetation Phenology: New Insights from Satellite Remote Sensing and Digital Repeat Photography. Biogeosciences 2016, 13, 5085–5102. [Google Scholar] [CrossRef]
- Geoscience Australia. Digital Earth Australia—Public Data—Surface Reflectance 25m Geomedian v2.1.0. 2019. Available online: https://data.dea.ga.gov.au/?prefix=geomedian-australia/v2.1.0/ (accessed on 10 May 2024).
- Griffiths, P.; van der Linden, S.; Kuemmerle, T.; Hostert, P. A Pixel-Based Landsat Compositing Algorithm for Large Area Land Cover Mapping. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2013, 6, 2088–2101. [Google Scholar] [CrossRef]
- White, J.C.; Wulder, M.A.; Hobart, G.W.; Luther, J.E.; Hermosilla, T.; Griffiths, P.; Coops, N.C.; Hall, R.J.; Hostert, P.; Dyk, A.; et al. Pixel-Based Image Compositing for Large-Area Dense Time Series Applications and Science. Can. J. Remote Sens. 2014, 40, 192–212. [Google Scholar] [CrossRef]
- Roberts, D.; Mueller, N.; Mcintyre, A. High-Dimensional Pixel Composites From Earth Observation Time Series. IEEE Trans. Geosci. Remote Sens. 2017, 55, 6254–6264. [Google Scholar] [CrossRef]
- Van der Walt, S.; Colbert, S.C.; Varoquaux, G. The NumPy Array: A Structure for Efficient Numerical Computation. Comput. Sci. Eng. 2011, 13, 22–30. [Google Scholar] [CrossRef]
- ABARES. Catchment Scale Land Use of Australia—Update December 2018. 2019. Available online: https://www.agriculture.gov.au/abares/aclump/land-use/catchment-scale-land-use-of-australia-update-december-2018 (accessed on 10 May 2024).
- ABARES. Australian Forest Profiles. 2019. Available online: https://www.agriculture.gov.au/abares/forestsaustralia/australias-forests/profiles (accessed on 10 May 2024).
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar] [CrossRef]
- Yakubovskiy, P. Segmentation Models. GitHub Repository. 2019. Available online: https://github.com/qubvel/segmentation_models (accessed on 10 May 2024).
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. Available online: https://openaccess.thecvf.com/content_cvpr_2016/html/He_Deep_Residual_Learning_CVPR_2016_paper.html (accessed on 10 May 2024).
- Tan, C.; Sun, F.; Kong, T.; Zhang, W.; Yang, C.; Liu, C. A Survey on Deep Transfer Learning. In Artificial Neural Networks and Machine Learning—ICANN 2018. ICANN 2018; Kůrková, V., Manolopoulos, Y., Hammer, B., Iliadis, L., Maglogiannis, I., Eds.; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2018; Volume 11141. [Google Scholar] [CrossRef]
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; et al. ImageNet Large Scale Visual Recognition Challenge. Int. J. Comput. Vis. 2015, 115, 211–252. [Google Scholar] [CrossRef]
- Kingma, D.; Ba, J. Adam: A Method for Stochastic Optimization. In Proceedings of the 3rd International Conference for Learning Representations, ICLR 2015, San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- Zhu, W.; Huang, Y.; Zeng, L.; Chen, X.; Liu, Y.; Qian, Z.; Du, N.; Fan, W.; Xie, X. AnatomyNet: Deep Learning for Fast and Fully Automated Whole-Volume Segmentation of Head and Neck Anatomy. arXiv 2018, arXiv:1808.05238. [Google Scholar] [CrossRef]
- Buslaev, A.; Iglovikov, V.I.; Khvedchenya, E.; Parinov, A.; Druzhinin, M.; Kalinin, A.A. Albumentations: Fast and Flexible Image Augmentations. Information 2020, 11, 125. [Google Scholar] [CrossRef]
- Chollet, F. Keras: The Python Deep Learning Library. 2015. Available online: https://keras.io/ (accessed on 10 May 2024).
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Hossin, M.; Sulaiman, M.N. A Review on Evaluation Metrics for Data Classification Evaluations. IJDKP 2015, 5, 1–11. [Google Scholar] [CrossRef]
- Cohen, J.A. Coefficient of Agreement for Nominal Scales. Educ. Psychol. Meas. 1960, 20, 37–46. [Google Scholar] [CrossRef]
- Mokany, K.; McCarthy, J.K.; Falster, D.S.; Gallagher, R.V.; Harwood, T.D.; Kooyman, R.; Westoby, M. Patterns and Drivers of Plant Diversity across Australia. Ecography 2022, 2022, e06426. [Google Scholar] [CrossRef]
- Fisher, A.; Scarth, P.; Armston, J.; Danaher, T. Relating Foliage and Crown Projective Cover in Australian Tree Stands. Agric. For. Meteorol. 2018, 259, 39–47. [Google Scholar] [CrossRef]
- NFI, 1998. Australia’s State of the Forests Report 1998. Bureau of Rural Sciences, Canberra. Available online: https://www.agriculture.gov.au/sites/default/files/documents/Australia%27s_State_of_the_Forests_Report_1998_v1.0.0.pdf (accessed on 10 May 2024).
- Yapp, G.A.; Thackway, R. Responding to Change—Criteria and Indicators for Managing the Transformation of Vegetated Landscapes to Maintain or Restore Ecosystem Diversity. In Biodiversity in Ecosystems-Linking Structure and Function; IntechOpen: London, UK, 2015; pp. 111–141. [Google Scholar] [CrossRef]
- Bureau of Meteorology. Record-Breaking La Niña Events: An Analysis of the La Niña: A Life Cycle and the Impacts and Significance of the 2010–2011 and 2011–2012 La Niña Events in Australia. 2012. Available online: http://www.bom.gov.au/climate/enso/history/La-Nina-2010-12.pdf (accessed on 10 May 2024).
- Defries, R.S.; Townshend, J.R.G. NDVI-Derived Land Cover Classifications at a Global Scale. Int. J. Remote Sens. 1994, 15, 3567–3586. [Google Scholar] [CrossRef]
- McFeeters, S.K. The Use of the Normalized Difference Water Index (NDWI) in the Delineation of Open Water Features. Int. J. Remote Sens. 1996, 17, 1425–1432. [Google Scholar] [CrossRef]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image Is Worth 16 × 16 Words: Transformers for Image Recognition at Scale. arXiv 2021, arXiv:2010.1192. [Google Scholar]
- Ma, J.; Li, F.; Wang, B. U-Mamba: Enhancing Long-Range Dependency for Biomedical Image Segmentation. arXiv 2024, arXiv:2401.04722. [Google Scholar]
- Zhu, L.; Liao, B.; Zhang, Q.; Wang, X.; Liu, W.; Wang, X. Vision Mamba: Efficient Visual Representation Learning with Bidirectional State Space Model. arXiv 2024, arXiv:2401.09417. [Google Scholar]
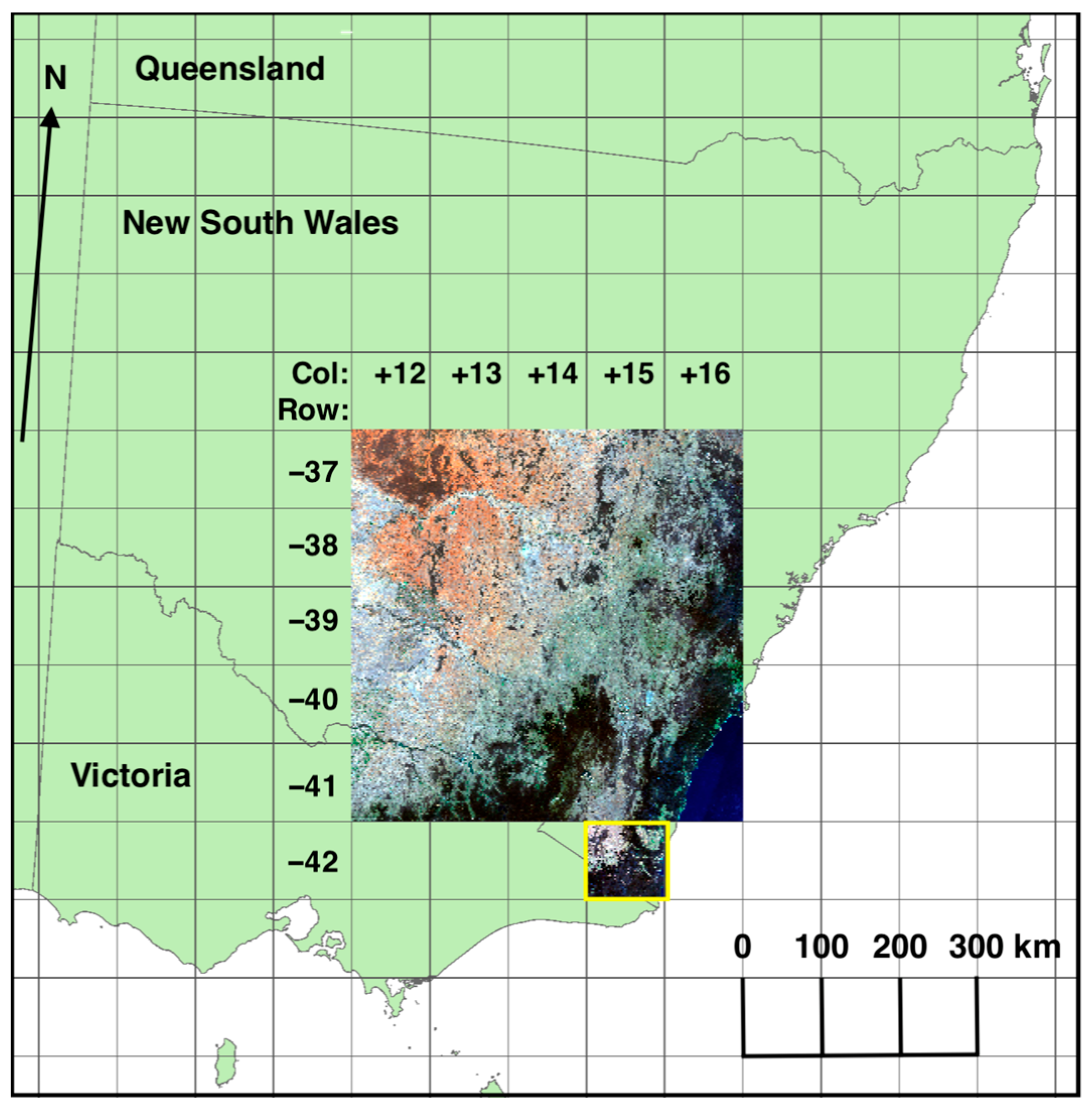
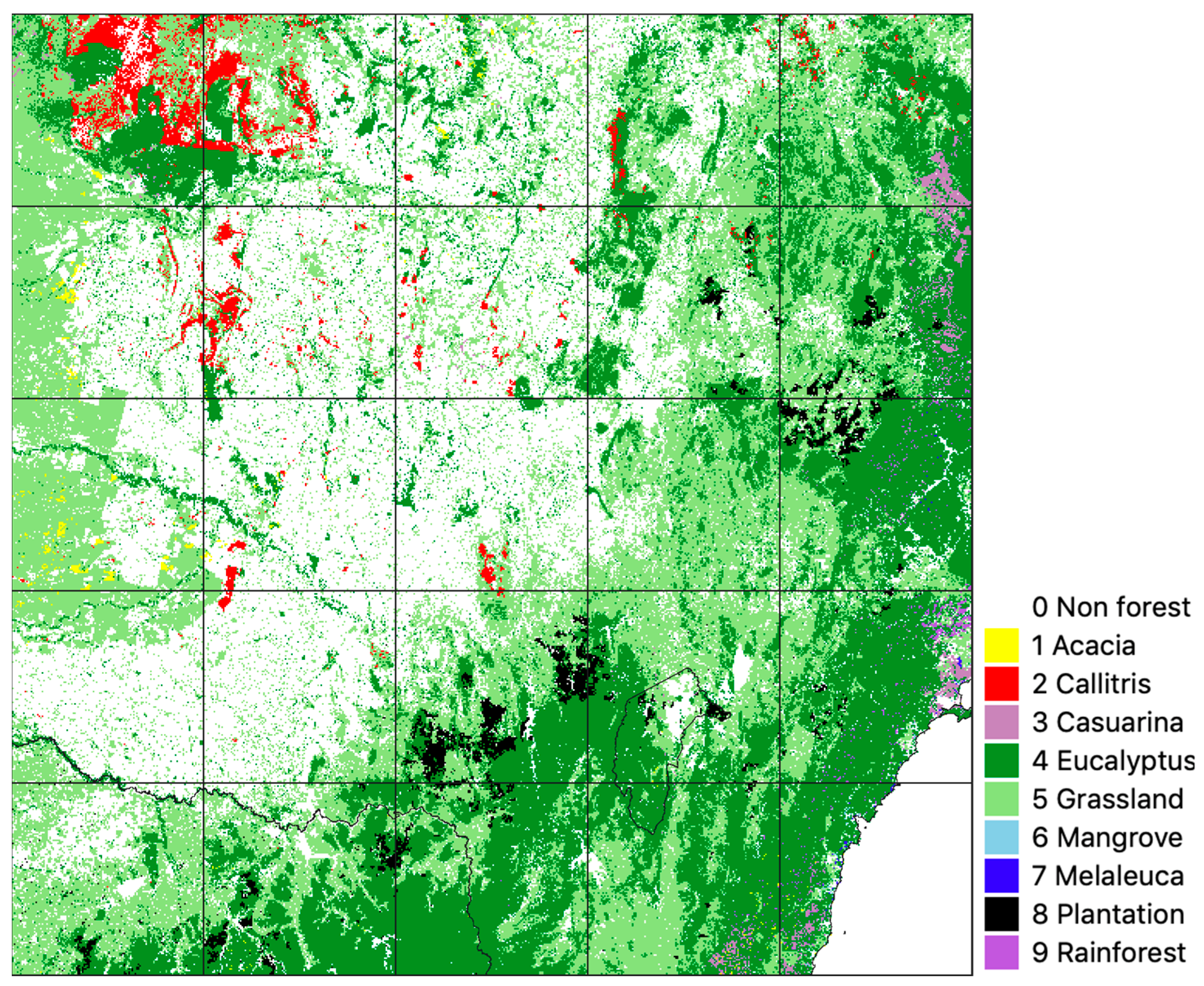
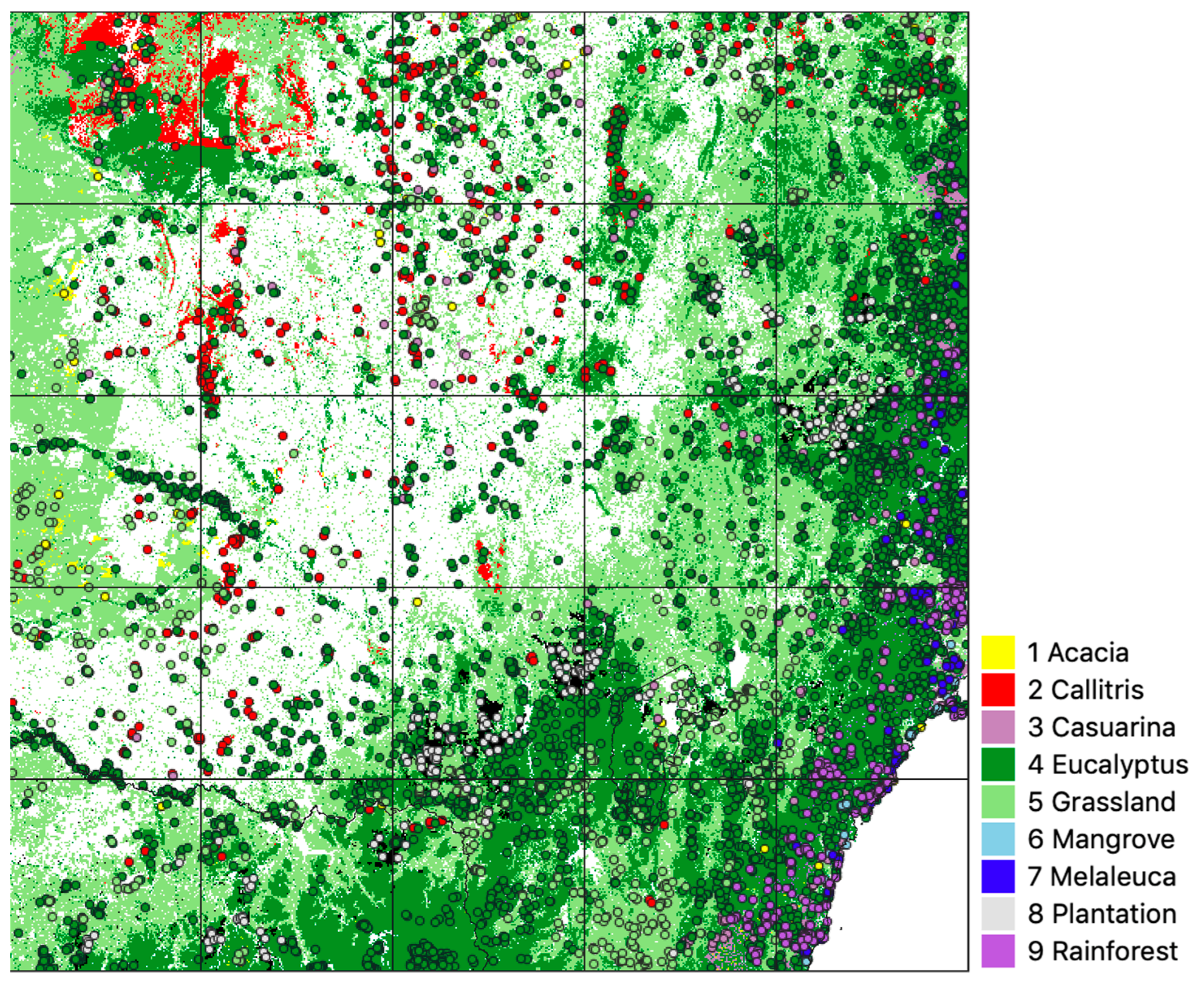
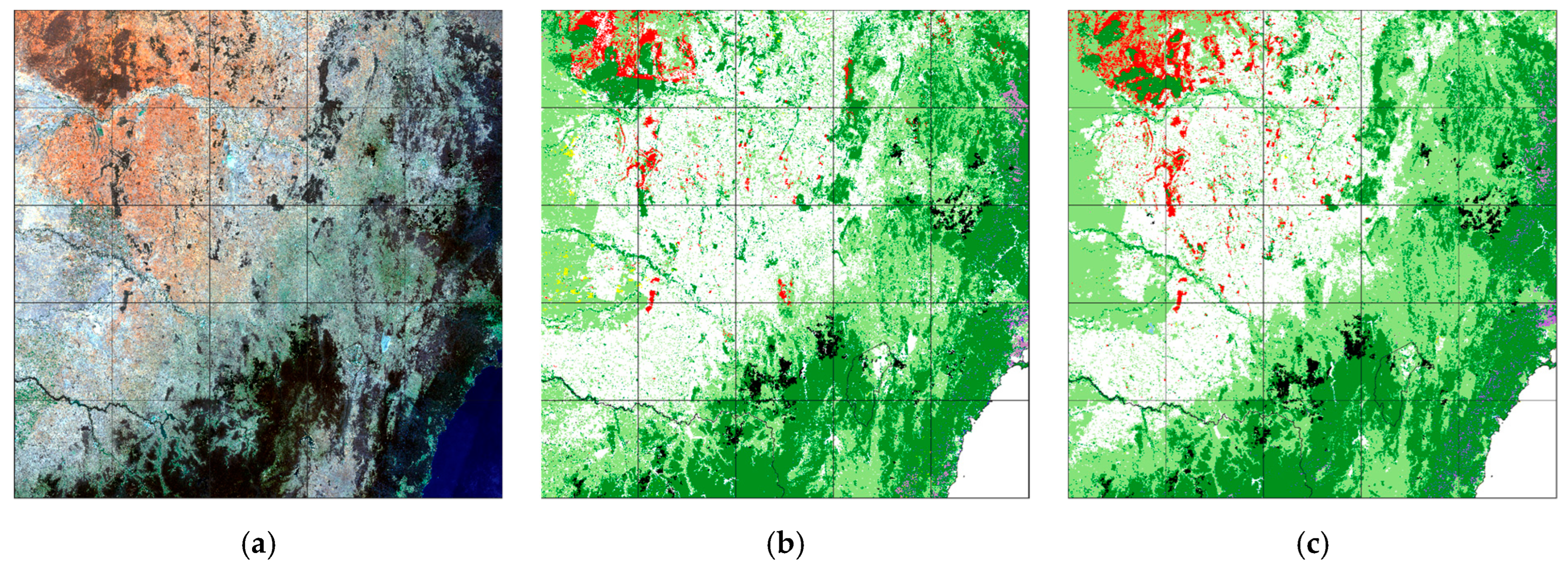

| Class | Source | Comments |
|---|---|---|
| 0 Non-forest | FoA | Based on FoA Non-Forest class. Non-Forest comprises crop and horticulture, water, urban and ocean. This class was reduced in area with the addition of the Grassland class from CLUM |
| 1 Acacia | FoA/NVIS V6 | Class is Acacia IF FoA AND NVIS V6 is Acacia plus coastal Acacia added from NVIS V6 |
| 2 Callitris | FoA | Better definition of Callitris forest vs. Non-Forest in NW of the study area in FoA compared with NVIS V6 |
| 3 Casuarina | FoA | In NE of the study area, a large extent of Casuarina in FoA is labelled Heathland in NVIS V6. Western Casuarina in NVIS V6 is not represented in FoA and does not look like forest in high-resolution satellite imagery. Southern areas of Casuarina are similar in both coverages. Jervis Bay Casuarina in FoA is labelled Eucalypt woodland in NVIS V6 |
| 4 Eucalyptus | FoA | All FoA Eucalypt and Eucalypt Mallee classes were combined. NVIS V6 Eucalypt classes are slightly more extensive in the west of the study area, but FoA appears to be more accurate when compared with high-resolution imagery |
| 5 Grassland | CLUM | Grassland class based on CLUM, which is much more extensive than Grassland classes in NVIS V6 (19 Tussock Grasslands, 20 Hummock Grasslands and 21 Other Grasslands) |
| 6 Mangrove | FoA/NVIS V6 | Coverages are very similar. NVIS V6 has a slightly larger extent. Limited to coastal areas. Class is Mangrove IF FoA OR NVIS V6 is Mangrove |
| 7 Melaleuca | FoA/NVIS V6 | Coverages are very similar. NVIS V6 is slightly larger extent. Mainly limited to coastal areas. Class is Melaleuca IF FoA OR NVIS V6 is Melaleuca |
| 8 Plantation | FoA | Good definition for softwood plantations. Very small areas on hardwood and mixed species plantation in northern Victoria. Plantation label class includes softwood, hardwood and mixed species plantations |
| 9 Rainforest | FoA | NVIS V6 has no Rainforest in the study area. FoA coverage is largely limited to coastal ranges |
| Dataset Splits | Count | Percent |
|---|---|---|
| Train | 19,906 | 81% |
| Validation | 3736 | 15% |
| Test | 955 | 4% |
| TOTAL | 24,597 | 100% |
| Plot Veg. Type | Criteria for Inclusion | No. |
|---|---|---|
| Acacia | Genus = Acacia and no other major forest species present | 78 |
| Callitris | Genus = Callitris | 536 |
| Casuarina | Genus = Allocasuarina or Casuarina | 344 |
| Eucalyptus | Genus = Eucalyptus or Corymbia | 4097 |
| Grassland | Genus = Austrostipa or Rytidosperma or Species = Aotus ericoides or Aristida ramose or Austrostipa bigeniculata or Bothroichloa macra or Carex appressa or Chrysocephalum apiculatum or Cynodon dactylon or Ficinia nodosa or Hemarthria uncinate or Juncus homalocaulis or Lomandra filiformis or Microlaena stipoides or Micromyrtus ciliate or Phragmites australis or Poa labillardierei or Poa poiformis or Poa sieberiana or Pteridium esculentum or Rumex brownie or Themeda australis or Themeda triandra or Typha domingensis or Viola banksia or Zoysia macrantha and no major forest species present | 768 |
| Mangrove | Species = Aegiceras corniculatum or Avicennia marina | 33 |
| Melaleuca | Genus = Melaleuca and Species ≠ Melaleuca uncinata | 76 |
| Plantation | Species = Pinus radiata | 190 |
| Rainforest | Species = Doryphora sassafras or Pittosporum undulatum or Syzygium smithii | 345 |
| TOTAL | 6467 |
| Pixel Count | Label Data | User’s | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CNN | Acacia | Casuarina | Eucalyptus | Grassland | Mangrove | Melaleuca | Plantation | Rainforest | Total | acc. % |
| Acacia | 3396 | 14 | 3531 | 1205 | 179 | 39 | 0 | 15 | 8379 | 40.5 |
| Casuarina | 2235 | 1474 | 49,844 | 10,490 | 3 | 77 | 213 | 160 | 64,496 | 2.3 |
| Eucalyptus | 26,315 | 60,837 | 9,228,705 | 236,205 | 768 | 797 | 106,197 | 85,648 | 9,745,472 | 94.7 |
| Grassland | 3385 | 2588 | 164,483 | 4,189,872 | 82 | 277 | 30,994 | 130 | 4,391,811 | 95.4 |
| Mangrove | 35 | 25 | 2292 | 135 | 1960 | 189 | 15 | 0 | 4651 | 42.1 |
| Melaleuca | 140 | 77 | 4547 | 198 | 336 | 125 | 49 | 0 | 5472 | 2.3 |
| Plantation | 0 | 0 | 55,363 | 9013 | 0 | 0 | 669,958 | 83 | 734,417 | 91.2 |
| Rainforest | 2607 | 4548 | 169,976 | 4659 | 0 | 0 | 233 | 37,502 | 219,525 | 17.1 |
| Total | 38,113 | 69,563 | 9,678,741 | 4,451,777 | 3328 | 1504 | 807,659 | 123,538 | 15,174,223 | |
| Prod.’s acc. % | 8.9 | 2.1 | 95.4 | 94.1 | 58.9 | 8.3 | 83.0 | 30.36 | ||
| F1 % | 14.6 | 2.2 | 95.0 | 94.8 | 49.1 | 3.6 | 86.9 | 21.9 | ||
| Pixel Count | Label Data | User’s | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| RF | Acacia | Casuarina | Eucalyptus | Grassland | Mangrove | Melaleuca | Plantation | Rainforest | Total | acc. % |
| Acacia | 150 | 15 | 2209 | 24,962 | 0 | 0 | 488 | 71 | 27,895 | 0.5 |
| Casuarina | 61 | 386 | 109,680 | 710 | 0 | 0 | 3535 | 83 | 114,455 | 0.3 |
| Eucalyptus | 36,146 | 67,505 | 9,267,052 | 642,501 | 2489 | 1545 | 392,928 | 108,411 | 10,518,577 | 88.1 |
| Grassland | 2168 | 718 | 86,471 | 3,656,064 | 73 | 100 | 60,653 | 75 | 3,806,322 | 96.1 |
| Mangrove | 1394 | 790 | 41,403 | 5456 | 3197 | 339 | 3165 | 182 | 55,926 | 5.7 |
| Melaleuca | 362 | 238 | 13,231 | 264 | 4 | 11 | 477 | 141 | 14,728 | 0.1 |
| Plantation | 751 | 182 | 151,468 | 158,140 | 13 | 5 | 348,107 | 11,212 | 669,878 | 52.0 |
| Rainforest | 85 | 38 | 14,263 | 94 | 0 | 0 | 1209 | 3515 | 19,204 | 18.3 |
| Total | 41,117 | 69,872 | 9,685,777 | 4,488,191 | 5776 | 2000 | 810,562 | 123,690 | 15,226,985 | |
| Prod.‘s acc. % | 0.4 | 0.6 | 95.7 | 81.5 | 55.3 | 0.6 | 42.9 | 2.8 | ||
| F1 % | 0.4 | 0.4 | 91.7 | 88.2 | 10.4 | 0.1 | 47.0 | 4.9 |
| Class | 2000 (ha) | 2019 (ha) | Change (ha) | Percent |
|---|---|---|---|---|
| Non–forest | 35,545 | 34,966 | −580 | −1.6% |
| Acacia | 751 | 754 | 3 | 0.3% |
| Casuarina | 6604 | 5541 | −1063 | −16.1% |
| Eucalyptus | 611,136 | 619,204 | 8068 | 1.3% |
| Grassland | 301,425 | 283,423 | −18,002 | −6.0% |
| Mangrove | 528 | 358 | −170 | −32.2% |
| Melaleuca | 589 | 415 | −174 | −29.5% |
| Plantation | 33,252 | 45,828 | 12,576 | 37.8% |
| Rainforest | 10,170 | 9511 | −660 | −6.5% |
| Total | 1,000,000 | 1,000,000 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boston, T.; Van Dijk, A.; Thackway, R. U-Net Convolutional Neural Network for Mapping Natural Vegetation and Forest Types from Landsat Imagery in Southeastern Australia. J. Imaging 2024, 10, 143. https://doi.org/10.3390/jimaging10060143
Boston T, Van Dijk A, Thackway R. U-Net Convolutional Neural Network for Mapping Natural Vegetation and Forest Types from Landsat Imagery in Southeastern Australia. Journal of Imaging. 2024; 10(6):143. https://doi.org/10.3390/jimaging10060143
Chicago/Turabian StyleBoston, Tony, Albert Van Dijk, and Richard Thackway. 2024. "U-Net Convolutional Neural Network for Mapping Natural Vegetation and Forest Types from Landsat Imagery in Southeastern Australia" Journal of Imaging 10, no. 6: 143. https://doi.org/10.3390/jimaging10060143
APA StyleBoston, T., Van Dijk, A., & Thackway, R. (2024). U-Net Convolutional Neural Network for Mapping Natural Vegetation and Forest Types from Landsat Imagery in Southeastern Australia. Journal of Imaging, 10(6), 143. https://doi.org/10.3390/jimaging10060143






