Semi-Supervised Medical Image Segmentation Based on Deep Consistent Collaborative Learning
Abstract
1. Introduction
- (1)
- Proposed a semi-supervised segmentation model named DCCLNet based on deep consistent co-training learning. Inspired by the CCT (cross-consistency training) semi-supervised method [10], this model adds different feature perturbations to the output of the backbone network’s CNN encoder, which are then inputted into auxiliary decoders. This encourages consistency between the outputs of the main decoder and the auxiliary decoder, thereby enhancing the robustness of the backbone network CNN.
- (2)
- Added a teacher model to form an MT (mean teacher) architecture [11] with the backbone network. Data with input perturbations are inputted into the teacher model, and a consistency constraint is imposed between the predictions of the teacher model and the backbone network to guide the training of the backbone network further, thereby improving the robustness and accuracy of the backbone network CNN. Moreover, the parameters of the teacher model are obtained from the backbone network, effectively reducing computational complexity.
- (3)
- Utilized the backbone network CNN and ViT to form a co-training architecture, where CNN can better capture local features, and ViT can better capture long-range dependencies. By simultaneously training from the perspectives of two different network architectures and learning pseudo-labels generated from each other’s predictions, the accuracy of the backbone network CNN can be improved.
- (4)
2. Related Works
2.1. Semi-Supervised Medical Image Segmentation
2.2. Consistent Learning
2.3. Co-Training
3. Method
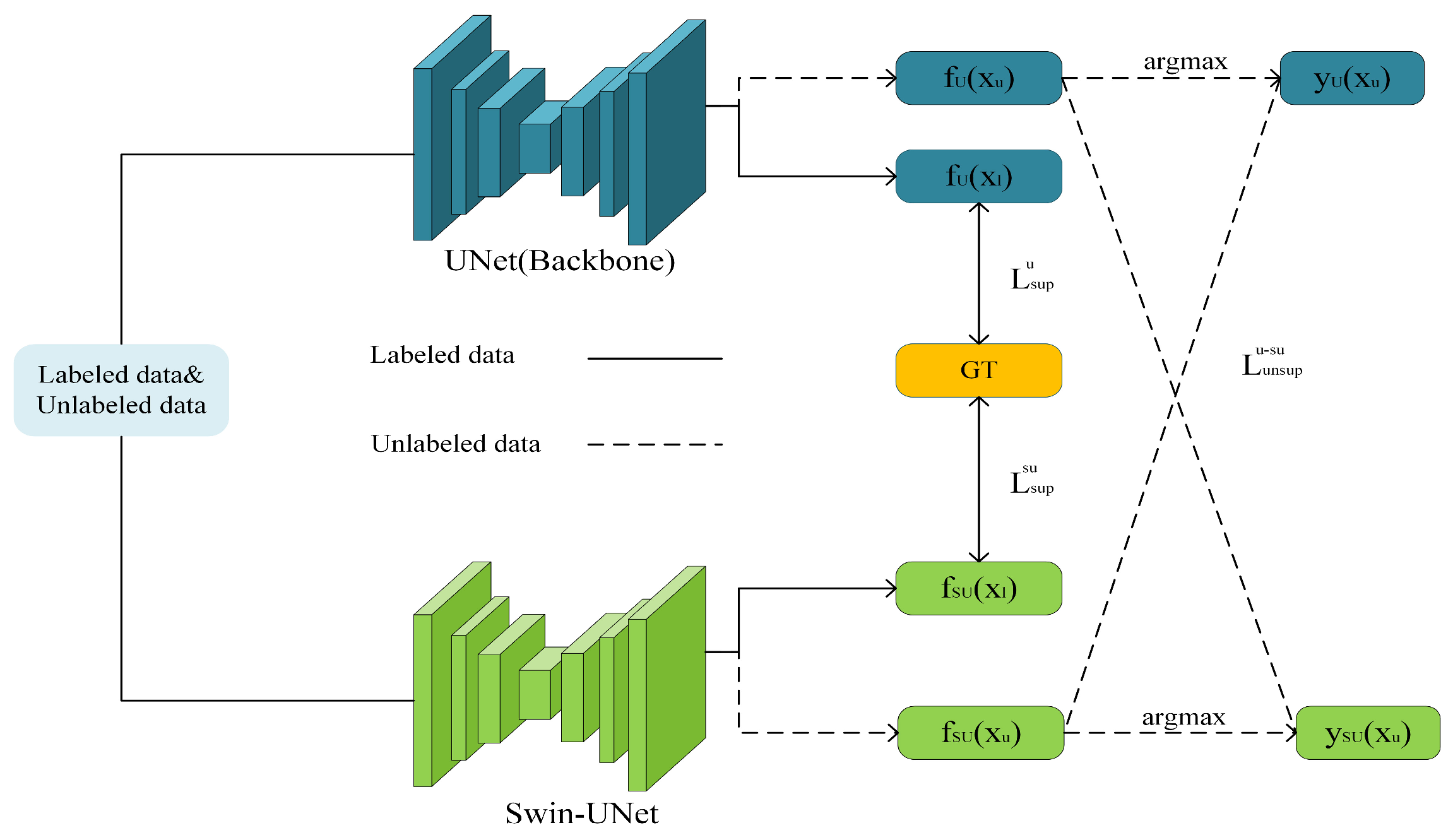
3.1. The Overall Structure of the Model
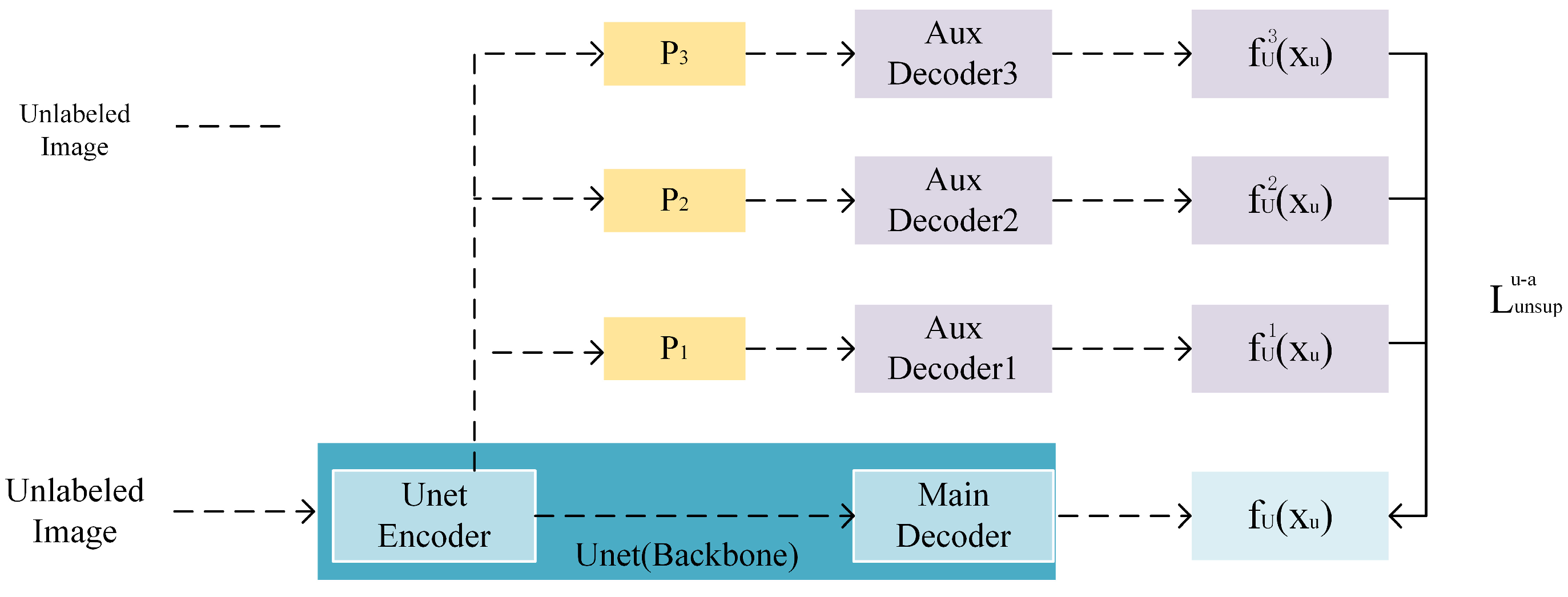
3.2. Auxiliary Decoder Assist
3.2.1. Characteristic Perturbation
3.2.2. Characteristic Perturbation Loss
3.3. Teacher Model Guidance
3.3.1. Teacher Model Parameter Update
3.3.2. Input Disturbance Loss
3.4. CNN and ViT Collaborative Training
3.4.1. Collaborative Training Process
3.4.2. Co-Training Loss
3.5. Overall Loss Function
4. Experiments
4.1. Data Preparation
4.2. Experimental Setup
4.3. Evaluation Index
4.4. Comparative Experimental
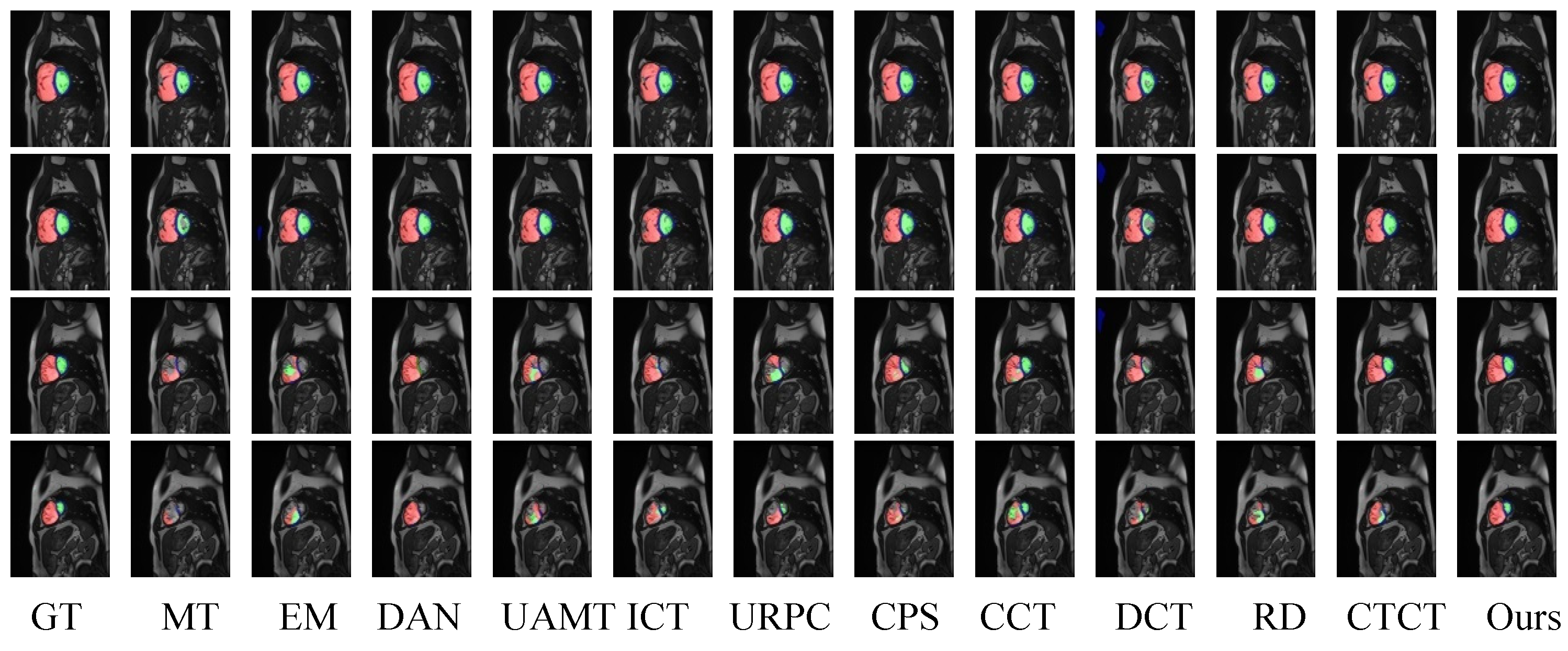
4.4.1. ACDC Dataset Comparative Experimental Analysis
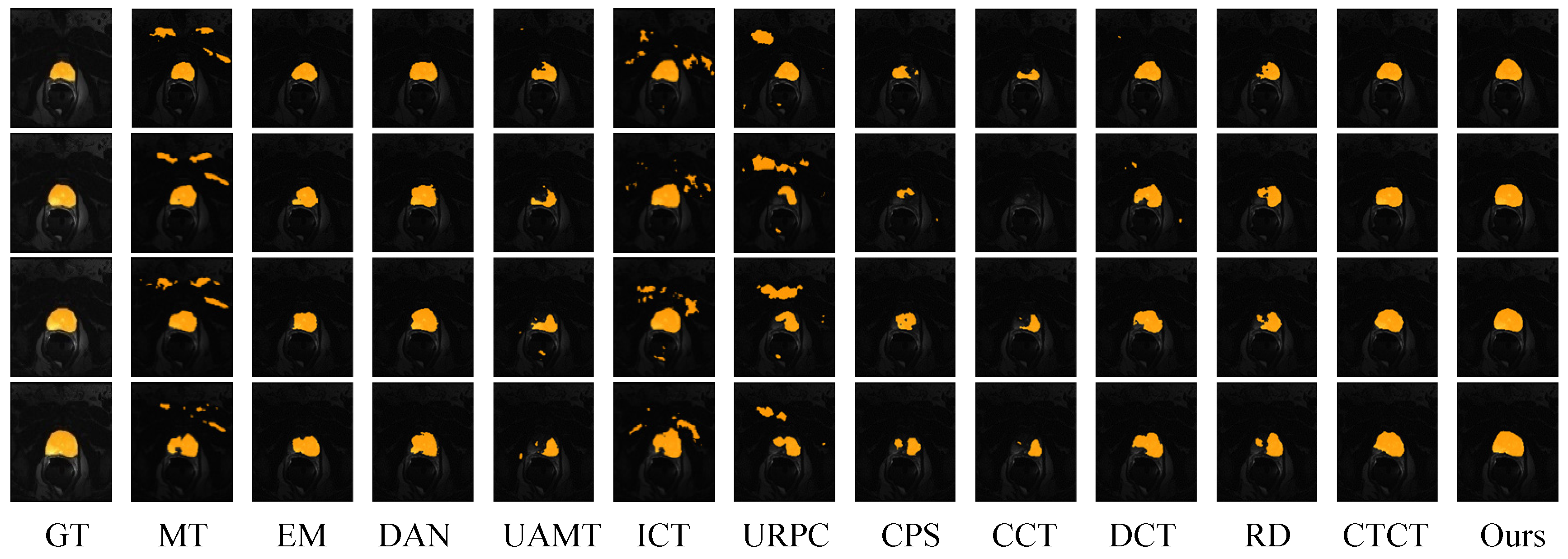
4.4.2. Prostate Dataset Comparative Experimental Analysis
4.5. Ablation Experiment
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, J.; Wei, L.; Wang, L.; Zhou, Q.; Zhu, L.; Qin, J. Boundary-aware transformers for skin lesion segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2021: 24th International Conference, Strasbourg, France, 27 September–1 October 2021; Proceedings, Part I 24. pp. 206–216. [Google Scholar]
- Hatamizadeh, A.; Tang, Y.; Nath, V.; Yang, D.; Myronenko, A.; Landman, B.; Roth, H.R.; Xu, D. Unetr: Transformers for 3D medical image segmentation. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Waikoloa, HI, USA, 3–8 January 2022; pp. 574–584. [Google Scholar]
- Xu, G.; Zhang, X.; He, X.; Wu, X. Levit-unet: Make faster encoders with transformer for medical image segmentation. In Proceedings of the Chinese Conference on Pattern Recognition and Computer Vision (PRCV), Xiamen, China, 13–15 October 2023; pp. 42–53. [Google Scholar]
- Su, J.; Luo, Z.; Lian, S.; Lin, D.; Li, S. Consistency learning with dynamic weighting and class-agnostic regularization for semi-supervised medical image segmentation. Biomed. Signal Process. Control 2024, 90, 105902. [Google Scholar] [CrossRef]
- Wang, P.; Peng, J.; Pedersoli, M.; Zhou, Y.; Zhang, C.; Desrosiers, C. Self-paced and self-consistent co-training for semi-supervised image segmentation. Med. Image Anal. 2021, 73, 102146. [Google Scholar] [CrossRef] [PubMed]
- Chaitanya, K.; Erdil, E.; Karani, N.; Konukoglu, E. Local contrastive loss with pseudo-label based self-training for semi-supervised medical image segmentation. Med. Image Anal. 2023, 87, 102792. [Google Scholar] [CrossRef] [PubMed]
- Peiris, H.; Hayat, M.; Chen, Z.; Egan, G.; Harandi, M. Uncertainty-guided dual-views for semi-supervised volumetric medical image segmentation. Nat. Mach. Intell. 2023, 5, 724–738. [Google Scholar] [CrossRef]
- Yang, L.; Qi, L.; Feng, L.; Zhang, W.; Shi, Y. Revisiting weak-to-strong consistency in semi-supervised semantic segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Vancouver, BC, Canada, 17–24 June 2023; pp. 7236–7246. [Google Scholar]
- Blum, A.; Mitchell, T. Combining labeled and unlabeled data with co-training. In Proceedings of the Eleventh Annual Conference on Computational Learning Theory, Madison, WI, USA, 24–26 July 1998; pp. 92–100. [Google Scholar]
- Ouali, Y.; Hudelot, C.; Tami, M. Semi-supervised semantic segmentation with cross-consistency training. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 12674–12684. [Google Scholar]
- Tarvainen, A.; Valpola, H. Mean teachers are better role models: Weight-averaged consistency targets improve semi-supervised deep learning results. In Proceedings of the Advances in Neural Information Processing Systems 30: Annual Conference on Neural Information Processing Systems 2017, Long Beach, CA, USA, 4–9 December 2017. [Google Scholar]
- Bernard, O.; Lalande, A.; Zotti, C.; Cervenansky, F.; Yang, X.; Heng, P.-A.; Cetin, I.; Lekadir, K.; Camara, O.; Ballester, M.A.G. Deep learning techniques for automatic MRI cardiac multi-structures segmentation and diagnosis: Is the problem solved? IEEE Trans. Med. Imaging 2018, 37, 2514–2525. [Google Scholar] [CrossRef] [PubMed]
- Litjens, G.; Toth, R.; Van De Ven, W.; Hoeks, C.; Kerkstra, S.; Van Ginneken, B.; Vincent, G.; Guillard, G.; Birbeck, N.; Zhang, J. Evaluation of prostate segmentation algorithms for MRI: The PROMISE12 challenge. Med. Image Anal. 2014, 18, 359–373. [Google Scholar] [CrossRef] [PubMed]
- Bai, W.; Oktay, O.; Sinclair, M.; Suzuki, H.; Rajchl, M.; Tarroni, G.; Glocker, B.; King, A.; Matthews, P.M.; Rueckert, D. Semi-supervised learning for network-based cardiac MR image segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention—MICCAI 2017: 20th International Conference, Quebec City, QC, Canada, 11–13 September 2017; Proceedings, Part II 20. pp. 253–260. [Google Scholar]
- Xu, X.; Sanford, T.; Turkbey, B.; Xu, S.; Wood, B.J.; Yan, P. Shadow-consistent semi-supervised learning for prostate ultrasound segmentation. IEEE Trans. Med. Imaging 2021, 41, 1331–1345. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Yang, K.; Zhang, M.; Chan, L.L.; Ng, T.K.; Ooi, B.C. Semi-supervised unpaired multi-modal learning for label-efficient medical image segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2021: 24th International Conference, Strasbourg, France, 27 September–1 October 2021; Proceedings, Part II 24. pp. 394–404. [Google Scholar]
- Zhao, Y.-X.; Zhang, Y.-M.; Song, M.; Liu, C.-L. Multi-view semi-supervised 3D whole brain segmentation with a self-ensemble network. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2019: 22nd International Conference, Shenzhen, China, 13–17 October 2019; Proceedings, Part III 22. pp. 256–265. [Google Scholar]
- Wu, H.; Chen, G.; Wen, Z.; Qin, J. Collaborative and adversarial learning of focused and dispersive representations for semi-supervised polyp segmentation. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, QC, Canada, 10–17 October 2021; pp. 3489–3498. [Google Scholar]
- Hou, J.; Ding, X.; Deng, J.D. Semi-supervised semantic segmentation of vessel images using leaking perturbations. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Waikoloa, HI, USA, 3–8 January 2022; pp. 2625–2634. [Google Scholar]
- Wu, J.; Fan, H.; Zhang, X.; Lin, S.; Li, Z. Semi-supervised semantic segmentation via entropy minimization. In Proceedings of the 2021 IEEE International Conference on Multimedia and Expo (ICME), Shenzhen, China, 5–9 July 2021; pp. 1–6. [Google Scholar]
- Wu, H.; Wang, Z.; Song, Y.; Yang, L.; Qin, J. Cross-patch dense contrastive learning for semi-supervised segmentation of cellular nuclei in histopathologic images. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 11666–11675. [Google Scholar]
- Miao, J.; Zhou, S.-P.; Zhou, G.-Q.; Wang, K.-N.; Yang, M.; Zhou, S.; Chen, Y. SC-SSL: Self-correcting Collaborative and Contrastive Co-training Model for Semi-Supervised Medical Image Segmentation. IEEE Trans. Med. Imaging 2023, 43, 1347–1364. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Cao, P.; Yang, H.; Liu, X.; Yang, J.; Zaiane, O.R. Co-training with high-confidence pseudo labels for semi-supervised medical image segmentation. arXiv 2023, arXiv:2301.04465. [Google Scholar]
- Sajjadi, M.; Javanmardi, M.; Tasdizen, T. Regularization with stochastic transformations and perturbations for deep semi-supervised learning. In Proceedings of the Advances in Neural Information Processing Systems 29: Annual Conference on Neural Information Processing Systems 2016, Barcelona, Spain, 5–10 December 2016. [Google Scholar]
- Verma, V.; Kawaguchi, K.; Lamb, A.; Kannala, J.; Solin, A.; Bengio, Y.; Lopez-Paz, D. Interpolation consistency training for semi-supervised learning. Neural Netw. 2022, 145, 90–106. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Cisse, M.; Dauphin, Y.N.; Lopez-Paz, D. mixup: Beyond empirical risk minimization. arXiv 2017, arXiv:1710.09412. [Google Scholar]
- Huang, H.; Chen, Z.; Chen, C.; Lu, M.; Zou, Y. Complementary consistency semi-supervised learning for 3D left atrial image segmentation. Comput. Biol. Med. 2023, 165, 107368. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Li, J.; Wang, Y.; Meng, Q.; Qin, T.; Chen, W.; Zhang, M.; Liu, T.-Y. R-drop: Regularized dropout for neural networks. Adv. Neural Inf. Process. Syst. 2021, 34, 10890–10905. [Google Scholar]
- Li, Y.; Luo, L.; Lin, H.; Chen, H.; Heng, P.-A. Dual-consistency semi-supervised learning with uncertainty quantification for COVID-19 lesion segmentation from CT images. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2021: 24th International Conference, Strasbourg, France, 27 September–1 October 2021; Proceedings, Part II 24. pp. 199–209. [Google Scholar]
- Chen, X.; Yuan, Y.; Zeng, G.; Wang, J. Semi-supervised semantic segmentation with cross pseudo supervision. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 19–25 June 2021; pp. 2613–2622. [Google Scholar]
- Luo, X.; Hu, M.; Song, T.; Wang, G.; Zhang, S. Semi-supervised medical image segmentation via cross teaching between cnn and transformer. In Proceedings of the International Conference on Medical Imaging with Deep Learning, Zurich, Switzerland, 6–8 July 2022; pp. 820–833. [Google Scholar]
- Peng, J.; Estrada, G.; Pedersoli, M.; Desrosiers, C. Deep co-training for semi-supervised image segmentation. Pattern Recognit. 2020, 107, 107269. [Google Scholar] [CrossRef]
- Wang, Z.; Li, T.; Zheng, J.-Q.; Huang, B. When cnn meet with vit: Towards semi-supervised learning for multi-class medical image semantic segmentation. In Proceedings of the European Conference on Computer Vision, Tel Aviv, Israel, 23–27 October 2022; pp. 424–441. [Google Scholar]
- Huang, S.; Luo, J.; Ou, Y.; Pang, Y.; Nie, X.; Zhang, G. Sd-net: A semi-supervised double-cooperative network for liver segmentation from computed tomography (CT) images. J. Cancer Res. Clin. Oncol. 2024, 150, 79. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015; Proceedings, Part III 18. pp. 234–241. [Google Scholar]
- Cao, H.; Wang, Y.; Chen, J.; Jiang, D.; Zhang, X.; Tian, Q.; Wang, M. Swin-unet: Unet-like pure transformer for medical image segmentation. In Proceedings of the European Conference on Computer Vision, Tel Aviv, Israel, 23–27 October 2022; pp. 205–218. [Google Scholar]
- Tompson, J.; Goroshin, R.; Jain, A.; LeCun, Y.; Bregler, C. Efficient object localization using convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Quebec City, QC, Canada, 27–30 September 2015; pp. 648–656. [Google Scholar]
- Laine, S.; Aila, T. Temporal ensembling for semi-supervised learning. arXiv 2016, arXiv:1610.02242. [Google Scholar]
- Vu, T.-H.; Jain, H.; Bucher, M.; Cord, M.; Pérez, P. Advent: Adversarial entropy minimization for domain adaptation in semantic segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 2517–2526. [Google Scholar]
- Liu, J.; Desrosiers, C.; Zhou, Y. Semi-supervised medical image segmentation using cross-model pseudo-supervision with shape awareness and local context constraints. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Singapore, 18–22 September 2022; pp. 140–150. [Google Scholar]
- Luo, X.; Liao, W.; Chen, J.; Song, T.; Chen, Y.; Zhang, S.; Chen, N.; Wang, G.; Zhang, S. Efficient semi-supervised gross target volume of nasopharyngeal carcinoma segmentation via uncertainty rectified pyramid consistency. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2021: 24th International Conference, Strasbourg, France, 27 September–1 October 2021; Proceedings, Part II 24. pp. 318–329. [Google Scholar]


| Labeled Data | Method | RV | Myo | RV | Mean | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DSC | HD95 | ASD | DSC | HD95 | ASD | DSC | HD95 | ASD | DSC | HD95 | ASD | ||
| 5% | MT [11] | 0.425 | 60.1 | 16.8 | 0.586 | 32.4 | 7.2 | 0.625 | 46.3 | 11.5 | 0.545 | 46.3 | 11.5 |
| 3/70 | EM [39] | 0.415 | 40.5 | 11.5 | 0.573 | 20.4 | 4.6 | 0.671 | 25.8 | 6.2 | 0.553 | 29.3 | 7.4 |
| DAN [24] | 0.492 | 44.6 | 17.8 | 0.527 | 37.6 | 9.6 | 0.601 | 38.0 | 8.0 | 0.540 | 40.1 | 11.8 | |
| UAMT [40] | 0.417 | 44.0 | 14.2 | 0.557 | 29.5 | 6.8 | 0.613 | 31.1 | 7.3 | 0.529 | 34.9 | 9.4 | |
| ICT [21] | 0.436 | 29.2 | 11.2 | 0.573 | 21.6 | 5.5 | 0.623 | 25.2 | 7.2 | 0.544 | 25.3 | 8.0 | |
| URPC [41] | 0.387 | 38.9 | 15.5 | 0.441 | 25.7 | 7.2 | 0.545 | 32.9 | 11.7 | 0.458 | 32.5 | 11.5 | |
| CPS [25] | 0.425 | 33.8 | 8.5 | 0.569 | 20.2 | 4.6 | 0.653 | 23.1 | 3.5 | 0.549 | 25.7 | 6.2 | |
| CCT [10] | 0.467 | 34.4 | 12.0 | 0.539 | 18.9 | 4.7 | 0.639 | 21.1 | 5.8 | 0.548 | 24.8 | 7.5 | |
| DCT [27] | 0.374 | 40.3 | 12.7 | 0.494 | 22.5 | 6.0 | 0.553 | 25.3 | 7.4 | 0.473 | 20.4 | 1.7 | |
| RD [23] | 0.376 | 36.0 | 11.8 | 0.437 | 23.7 | 5.4 | 0.501 | 26.2 | 6.3 | 0.438 | 28.6 | 7.8 | |
| CTCT [26] | 0.677 | 17.6 | 5.1 | 0.642 | 12.6 | 3.0 | 0.750 | 14.1 | 3.4 | 0.690 | 14.7 | 3.9 | |
| Ours | 0.734 | 14.8 | 3.5 | 0.738 | 15.6 | 3.0 | 0.832 | 9.3 | 2.1 | 0.768 | 13.2 | 2.9 | |
| 10% | MT | 0.791 | 15.5 | 2.7 | 0.764 | 33.3 | 4.8 | 0.832 | 20.0 | 3.9 | 0.796 | 22.9 | 3.8 |
| 7/70 | EM | 0.743 | 3.9 | 1.1 | 0.798 | 7.8 | 1.4 | 0.849 | 11.0 | 2.0 | 0.797 | 7.6 | 1.5 |
| DAN | 0.799 | 8.8 | 1.4 | 0.795 | 6.3 | 1.1 | 0.845 | 11.6 | 2.1 | 0.813 | 8.9 | 1.5 | |
| UAMT | 0.772 | 8.3 | 1.3 | 0.796 | 11.5 | 1.8 | 0.849 | 15.7 | 2.7 | 0.806 | 11.8 | 2.0 | |
| ICT | 0.815 | 5.1 | 1.1 | 0.809 | 10.7 | 1.6 | 0.850 | 16.5 | 2.8 | 0.825 | 10.8 | 1.8 | |
| URPC | 0.817 | 8.7 | 1.9 | 0.812 | 8.3 | 1,4 | 0.886 | 11.7 | 2.3 | 0.838 | 9.6 | 1.9 | |
| CPS | 0.831 | 3.9 | 0.8 | 0.826 | 6.6 | 1.3 | 0.871 | 13.1 | 2.3 | 0.843 | 7.9 | 1.5 | |
| CCT | 0.837 | 5.1 | 0.9 | 0.820 | 6.4 | 1.2 | 0.878 | 11.3 | 1.8 | 0.845 | 7.6 | 1.3 | |
| DCT | 0.757 | 5.9 | 1.3 | 0.762 | 36.1 | 5.8 | 0.855 | 17.8 | 2.6 | 0.792 | 19.9 | 3.2 | |
| RD | 0.814 | 6.6 | 1.3 | 0.810 | 7.4 | 1.2 | 0.869 | 11.0 | 2.0 | 0.831 | 8.4 | 1.5 | |
| CTCT | 0.861 | 5.0 | 1.1 | 0.841 | 6.3 | 1.0 | 0.895 | 13.5 | 1.8 | 0.866 | 8.3 | 1.3 | |
| Ours | 0.888 | 6.8 | 1.3 | 0.861 | 4.8 | 1.0 | 0.921 | 6.5 | 1.4 | 0.890 | 6.0 | 1.2 | |
| Labeled Data | Method | Mean | ||
|---|---|---|---|---|
| DSC | HD95 | ASD | ||
| 10% 4/35 | MT | 0.424 | 94.5 | 25.7 |
| EM | 0.491 | 85.3 | 22.2 | |
| DAN | 0.568 | 96.6 | 23.5 | |
| UAMT | 0.490 | 91.2 | 26.7 | |
| ICT | 0.623 | 65.9 | 16.2 | |
| URPC | 0.317 | 65.1 | 24.5 | |
| CPS | 0.324 | 60.0 | 15.8 | |
| CCT | 0.409 | 57.3 | 21.4 | |
| DCT | 0.410 | 85.6 | 24.3 | |
| RD | 0.432 | 57.6 | 22.3 | |
| CTCT | 0.764 | 25.2 | 7.8 | |
| Ours | 0.792 | 21.2 | 7.3 | |
| 20% 7/35 | MT | 0.635 | 35.1 | 11.6 |
| EM | 0.620 | 41.6 | 13.2 | |
| DAN | 0.695 | 64.9 | 13.5 | |
| UAMT | 0.639 | 30.2 | 10.7 | |
| ICT | 0.734 | 26.5 | 9.2 | |
| URPC | 0.642 | 35.3 | 12.7 | |
| CPS | 0.602 | 47.1 | 13.6 | |
| CCT | 0.572 | 87.7 | 21.9 | |
| DCT | 0.659 | 36.1 | 12.1 | |
| RD | 0.633 | 39.6 | 12.8 | |
| CTCT | 0.783 | 26.9 | 8.4 | |
| Ours | 0.812 | 19.3 | 6.4 | |
| Method | RV | Myo | LV | Mean | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DSC | HD95 | ASD | DSC | HD95 | ASD | DSC | HD95 | ASD | DSC | HD95 | ASD | |
| UNet | 0.673 | 16.3 | 3.6 | 0.785 | 10.0 | 1.7 | 0.874 | 13.8 | 2.6 | 0.778 | 13.8 | 2.6 |
| UNet + Aux | 0.791 | 15.5 | 2.7 | 0.764 | 33.3 | 4.8 | 0.832 | 20.0 | 3.9 | 0.796 | 22.9 | 3.8 |
| UNet + Tea | 0.837 | 5.1 | 1.9 | 0.820 | 6.4 | 1.2 | 0.878 | 11.3 | 1.8 | 0.845 | 7.6 | 1.3 |
| UNet + ViT | 0.861 | 5.0 | 1.1 | 0.841 | 6.3 | 1.0 | 0.895 | 13.5 | 1.8 | 0.866 | 8.3 | 1.3 |
| UNet + Tea + Aux | 0.801 | 10.1 | 2.0 | 0.808 | 10.2 | 1.7 | 0.871 | 21.3 | 3.2 | 0.826 | 13.9 | 2.3 |
| UNet + Tea + ViT | 0.870 | 7.4 | 1.5 | 0.857 | 7.1 | 1.2 | 0.913 | 11.7 | 2.0 | 0.880 | 8.7 | 1.6 |
| UNet + Aux + ViT | 0.880 | 6.8 | 1.4 | 0.860 | 4.1 | 1.0 | 0.910 | 9.4 | 1.6 | 0.884 | 6.8 | 1.3 |
| DCCLNet | 0.888 | 6.8 | 1.3 | 0.861 | 4.8 | 1.0 | 0.921 | 6.5 | 1.4 | 0.890 | 6.0 | 1.2 |
| Method | Mean | ||
|---|---|---|---|
| DSC | HD95 | ASD | |
| UNet | 0.563 | 95.3 | 24.6 |
| UNet + Aux | 0.572 | 87.7 | 21.9 |
| UNet + Tea | 0.635 | 35.1 | 11.6 |
| UNet + ViT | 0.783 | 26.9 | 8.4 |
| UNet + Tea + Aux | 0.695 | 33.2 | 10.1 |
| UNet + Tea + ViT | 0.794 | 24.6 | 7.9 |
| UNet + Aux + ViT | 0.807 | 21.0 | 7.2 |
| DCCLNet | 0.812 | 19.3 | 6.4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, X.; Wang, W. Semi-Supervised Medical Image Segmentation Based on Deep Consistent Collaborative Learning. J. Imaging 2024, 10, 118. https://doi.org/10.3390/jimaging10050118
Zhao X, Wang W. Semi-Supervised Medical Image Segmentation Based on Deep Consistent Collaborative Learning. Journal of Imaging. 2024; 10(5):118. https://doi.org/10.3390/jimaging10050118
Chicago/Turabian StyleZhao, Xin, and Wenqi Wang. 2024. "Semi-Supervised Medical Image Segmentation Based on Deep Consistent Collaborative Learning" Journal of Imaging 10, no. 5: 118. https://doi.org/10.3390/jimaging10050118
APA StyleZhao, X., & Wang, W. (2024). Semi-Supervised Medical Image Segmentation Based on Deep Consistent Collaborative Learning. Journal of Imaging, 10(5), 118. https://doi.org/10.3390/jimaging10050118






