Comprehensive Evaluation of Multispectral Image Registration Strategies in Heterogenous Agriculture Environment
Abstract
1. Introduction
2. Materials and Methods
2.1. Dataset
2.1.1. Image Acquisition
2.1.2. Annotation of MS Images
2.1.3. Software
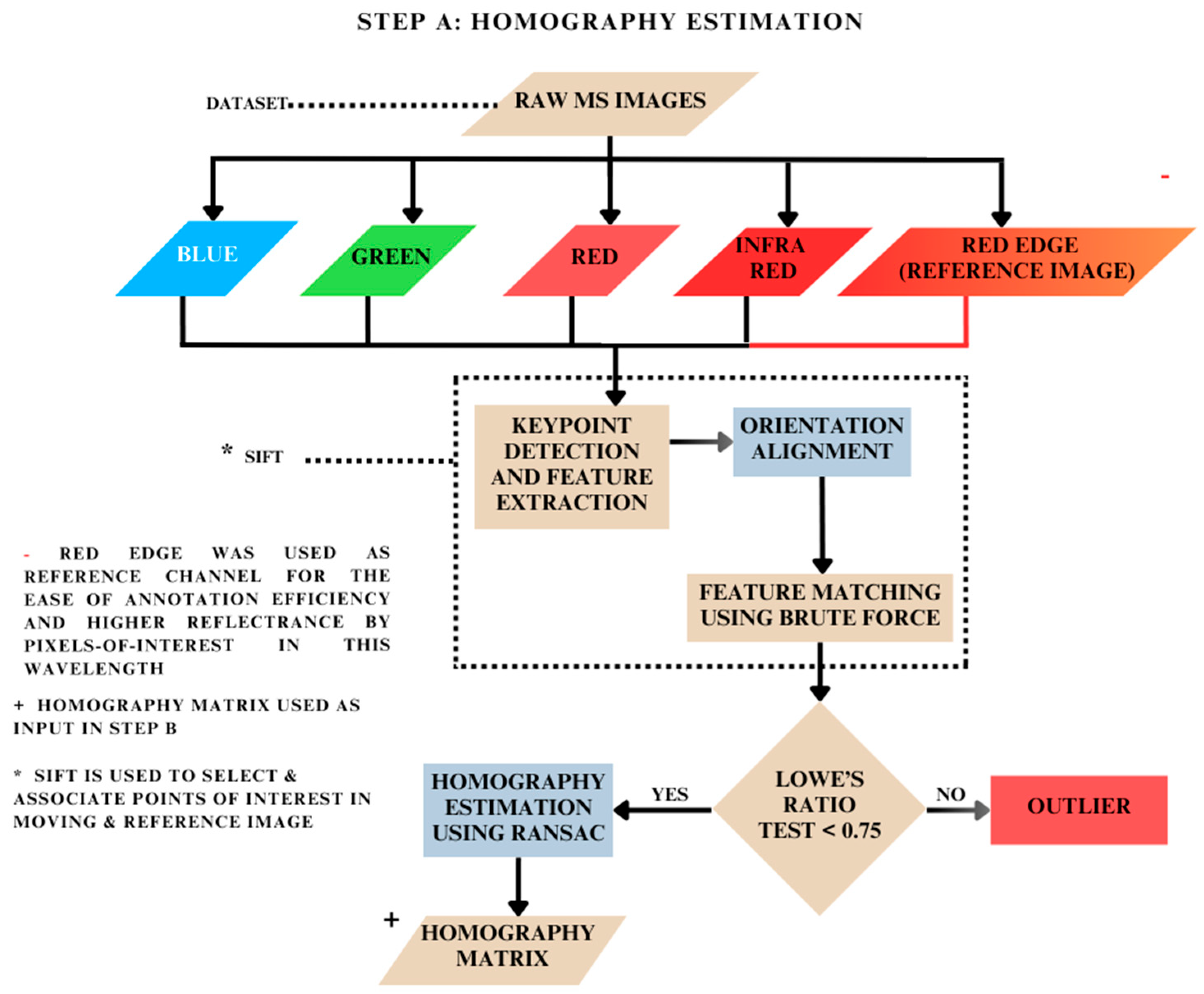
2.2. Homography Matrix Estimation: Step A
2.2.1. Keypoint Detection and Feature Extraction
2.2.2. Feature Matching
2.2.3. Ratio Test and Filtering
2.2.4. Homography Estimation
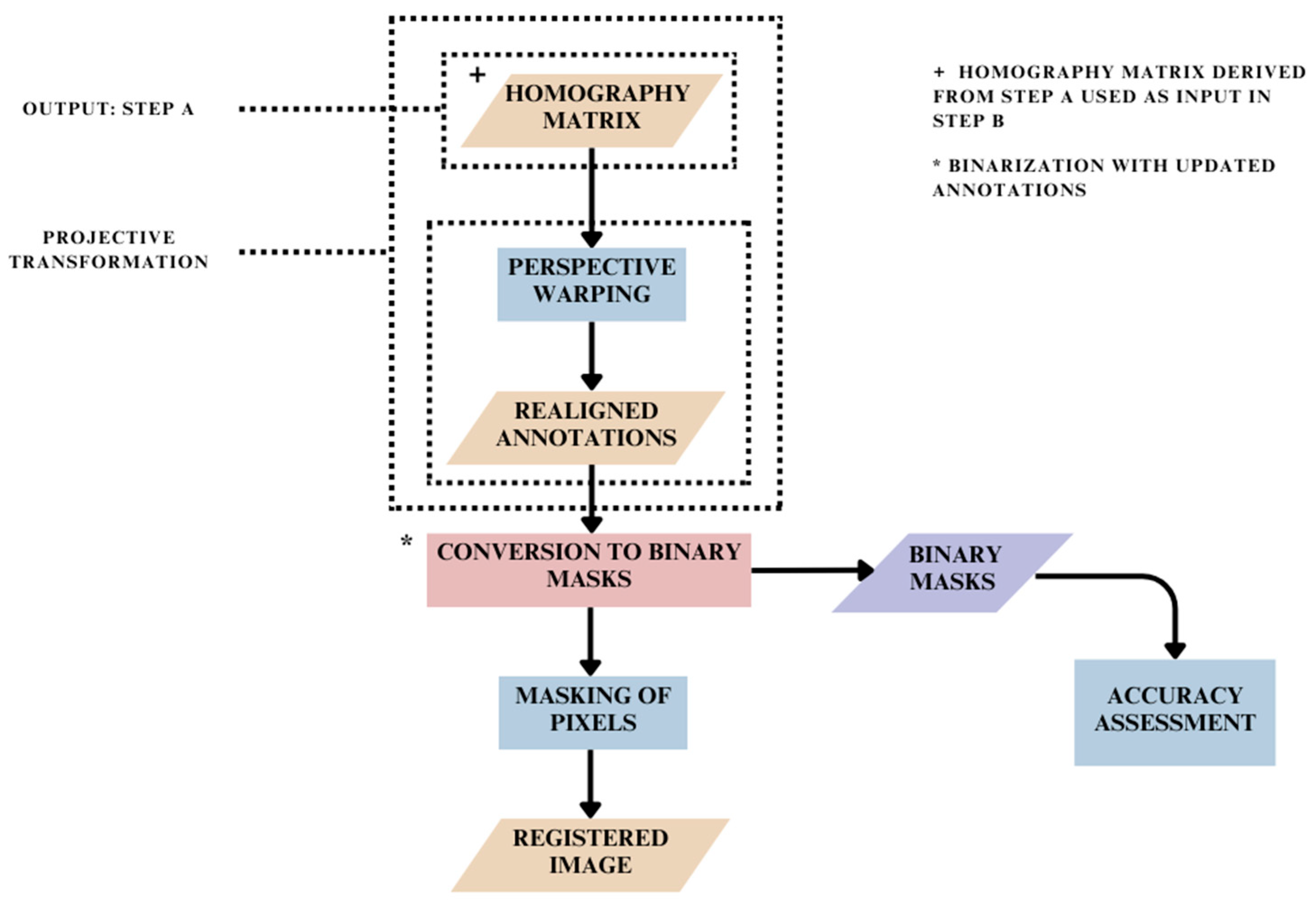
2.3. Spatial Realignment of Pixel Annotations: Step B
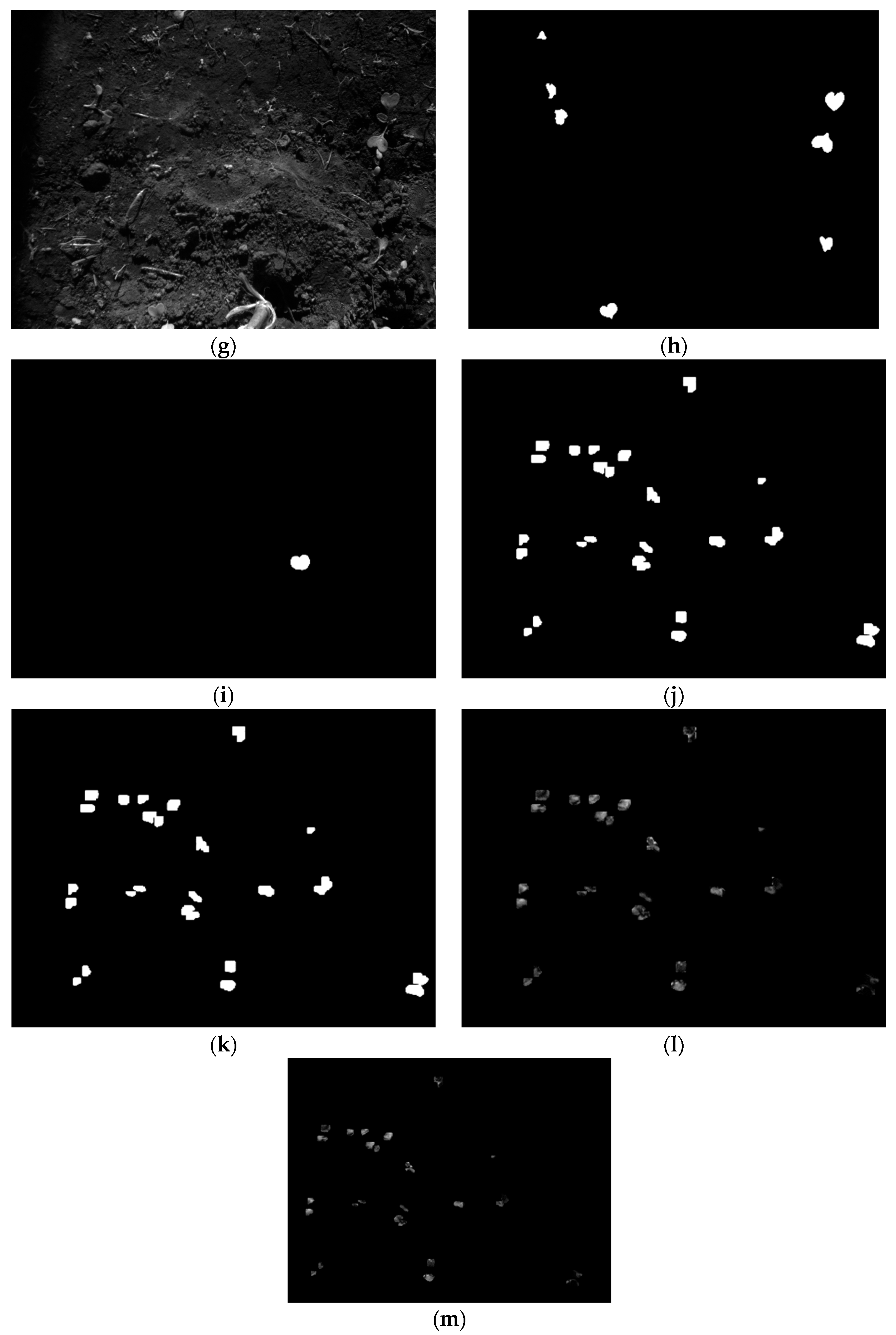
2.4. Registration Based on Binary Mask Images: Step C
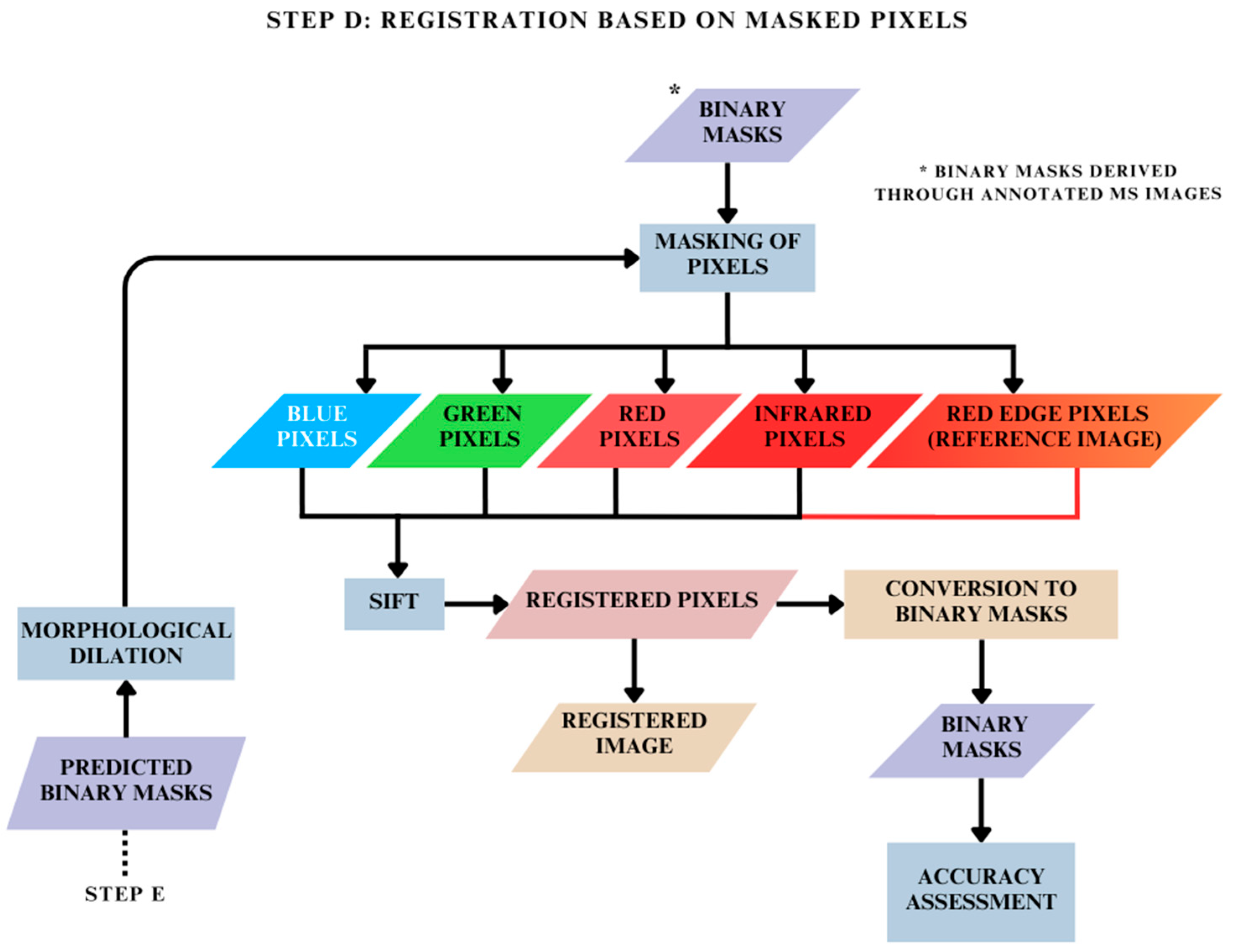
2.5. Registration Based on Masked Pixels: Step D
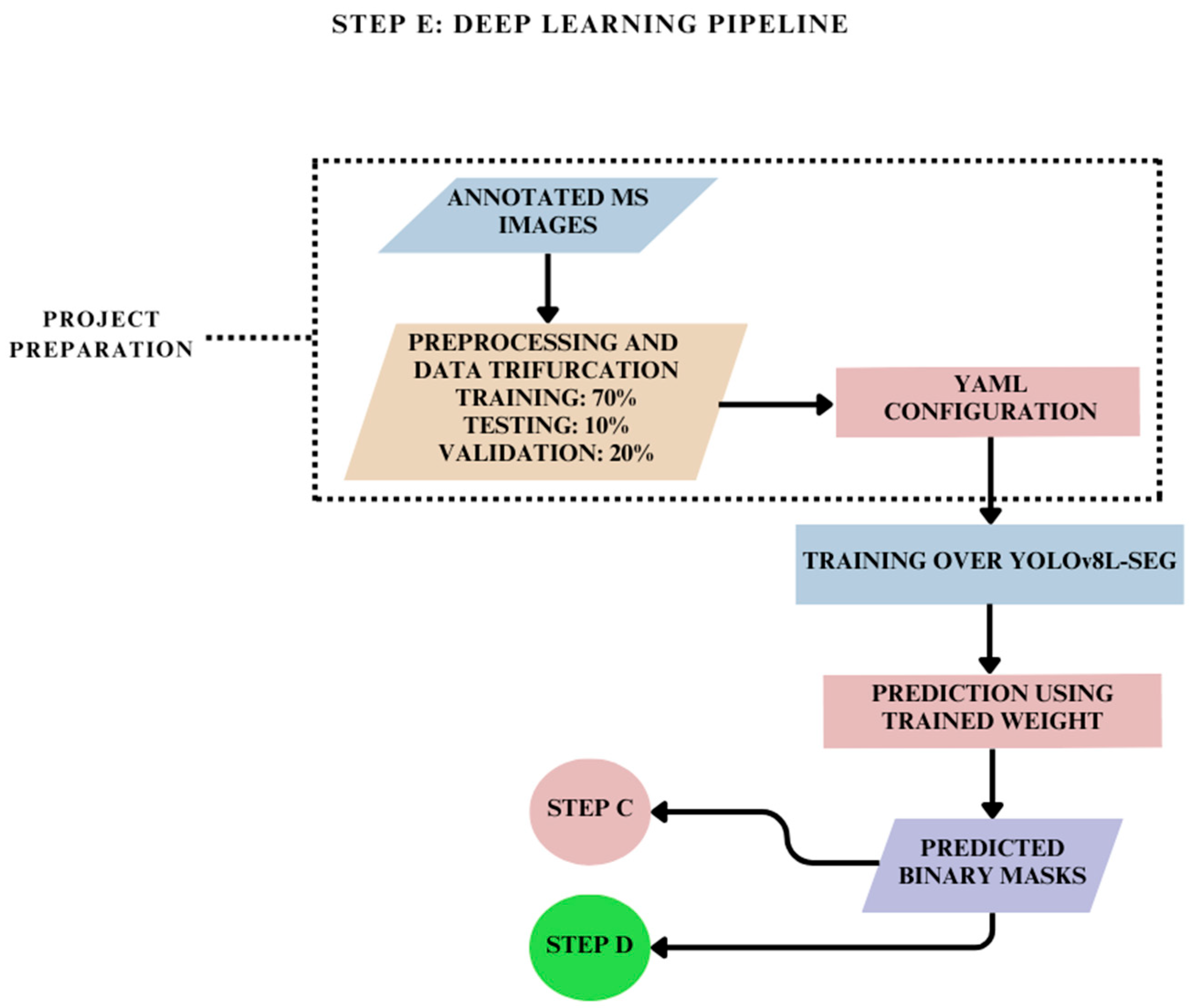
2.6. Deep Learning Pipeline for Training Annotated MS Images: Step E
2.7. Morphological Dilation of Predicted Masks
2.8. Registration of Predicted Masks
2.9. Registration of Masked Pixels from Predicted Masks
2.10. Accuracy Assessment
2.10.1. Intersection over Union (IoU)
2.10.2. Normalized Coefficient of Correlation (NCC)
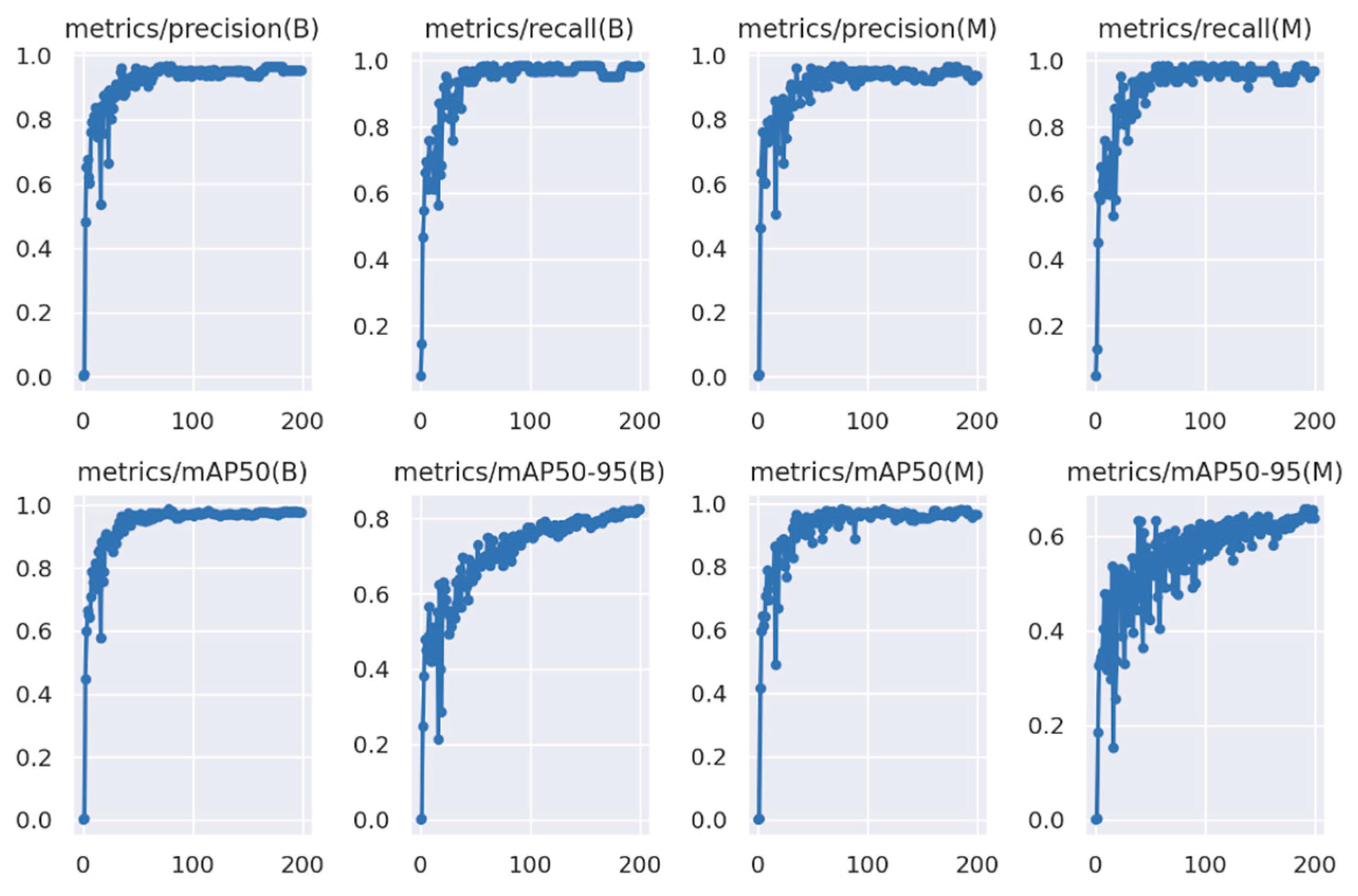
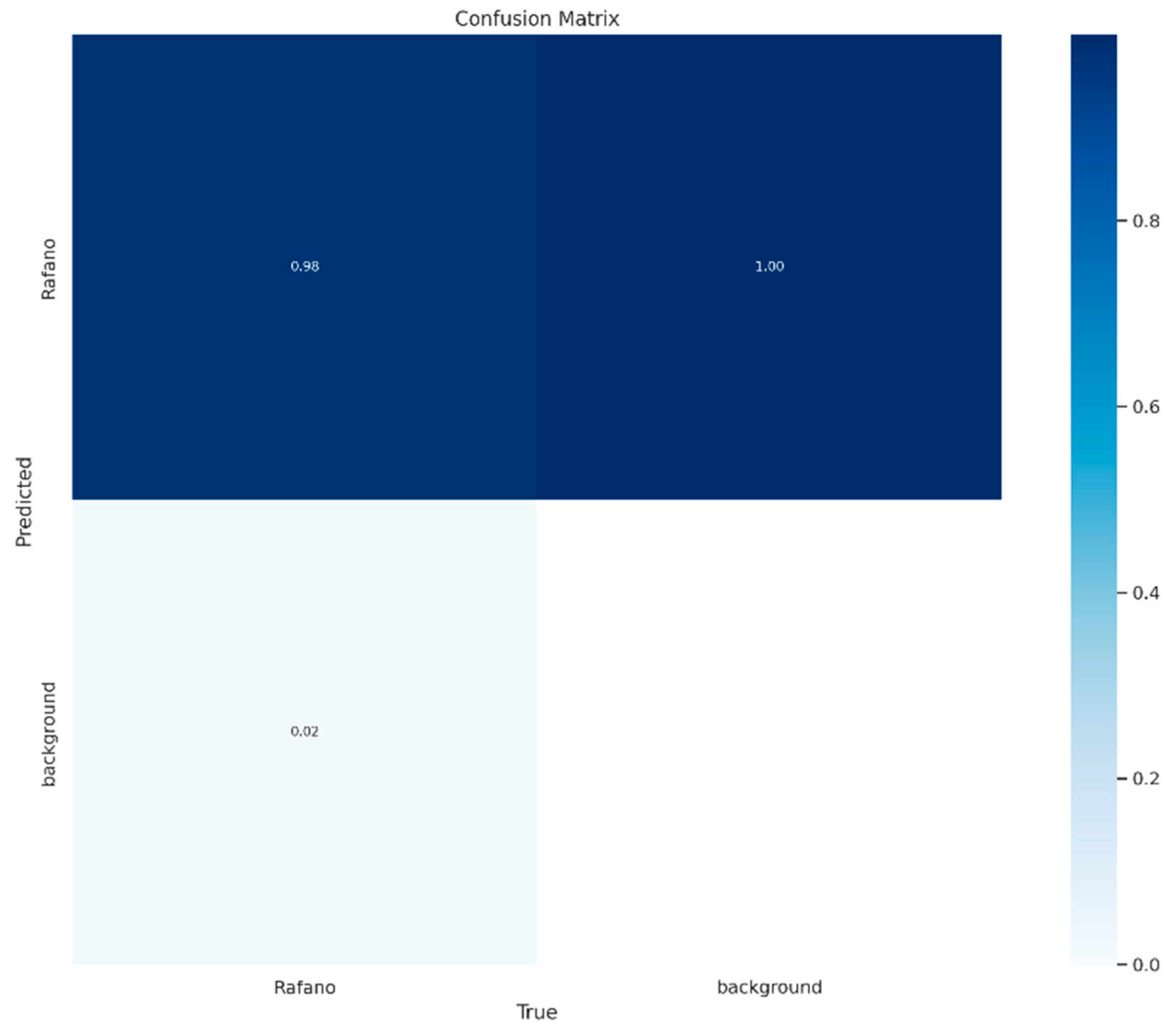
2.10.3. YOLOv8l-Seg Evaluation Metrics
3. Results
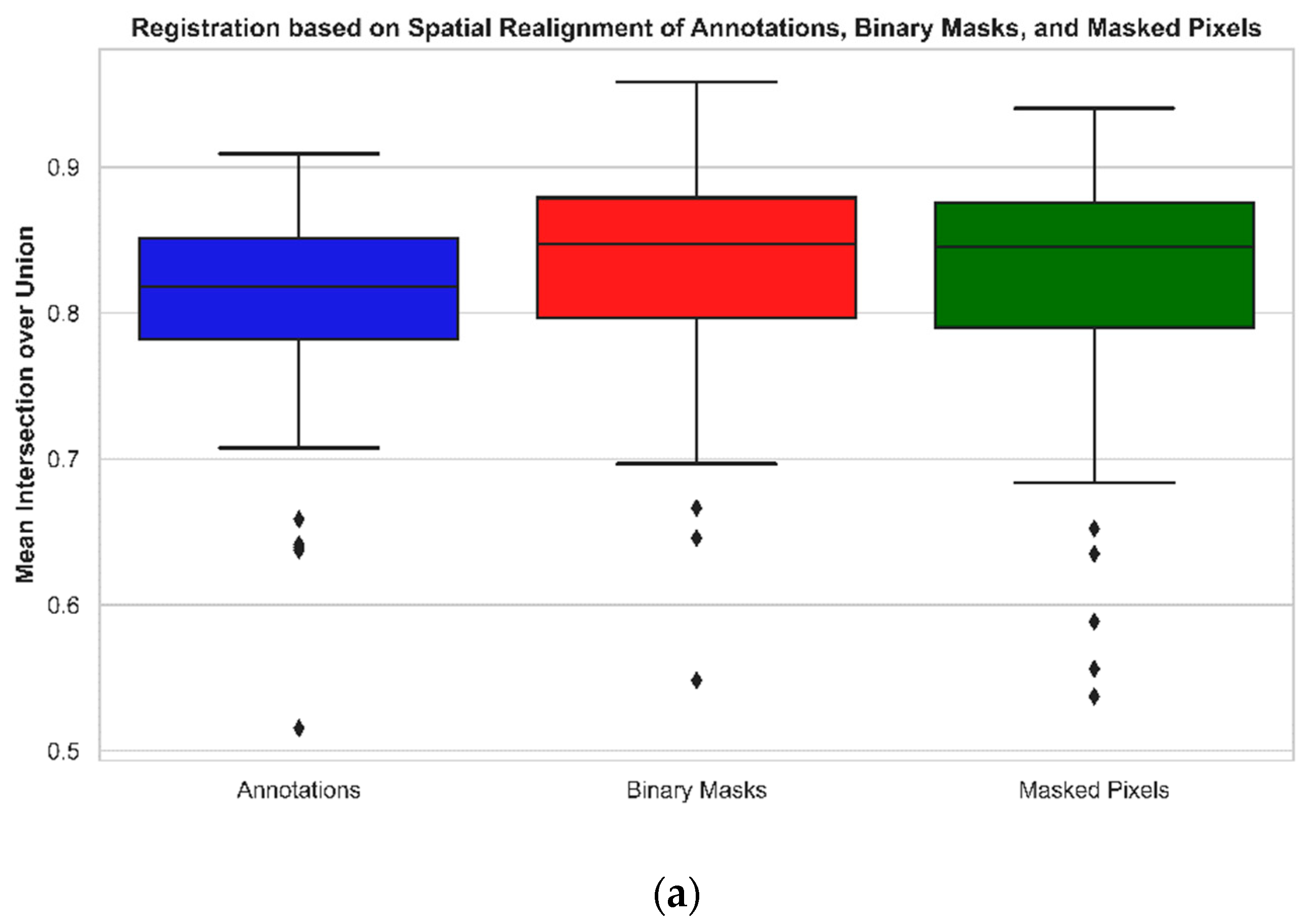
3.1. Evaluation of MS Image Registration Quality
3.2. Evaluation of Registration Quality at Spectral Level
3.3. Evaluation of Registration Quality at Instance Level
3.4. Evaluation of Overall Registration Quality for Predicted Instances
3.5. Evaluation of Registration Quality for Predicted Instances across Individual Spectral Channels
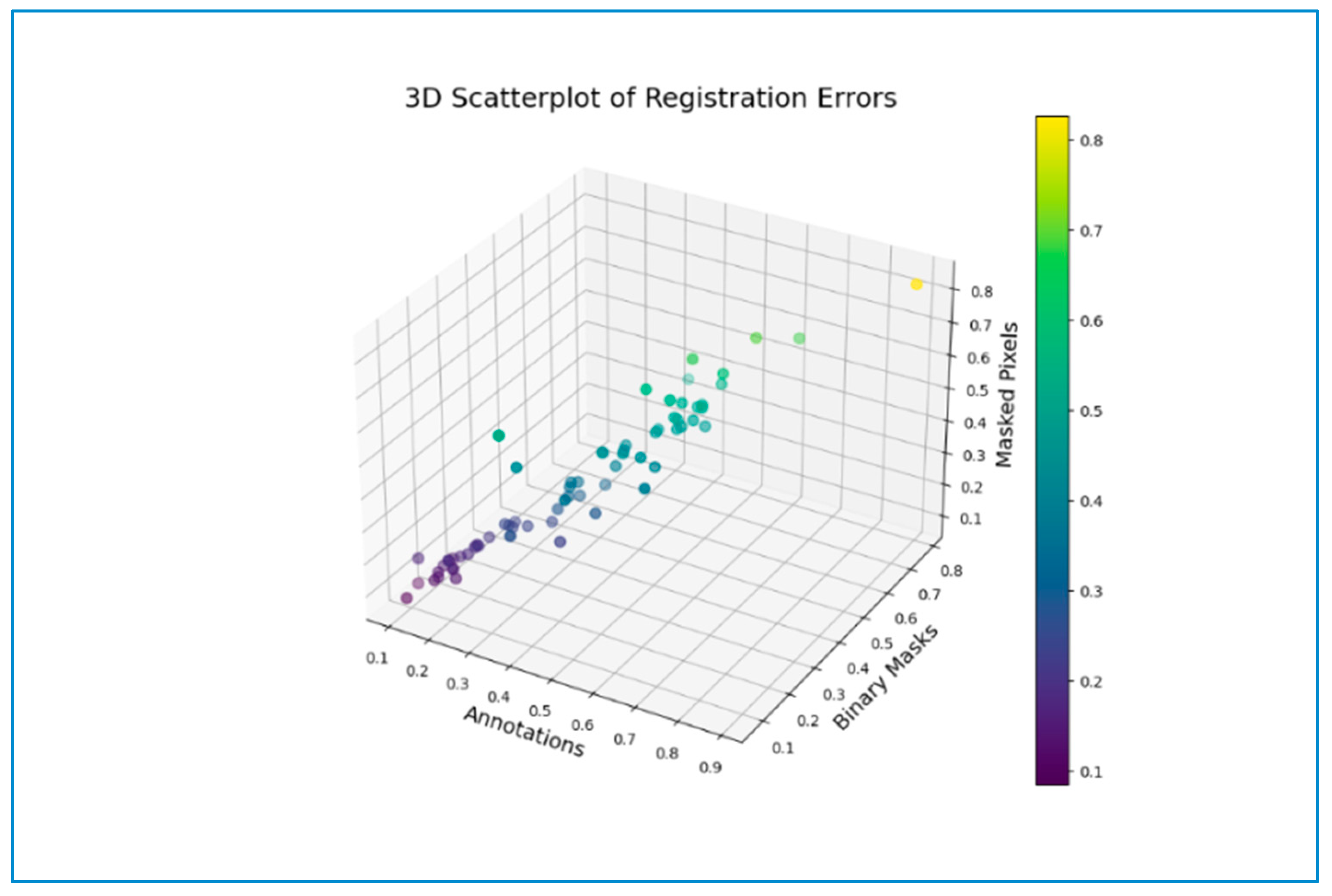
3.6. Error Map
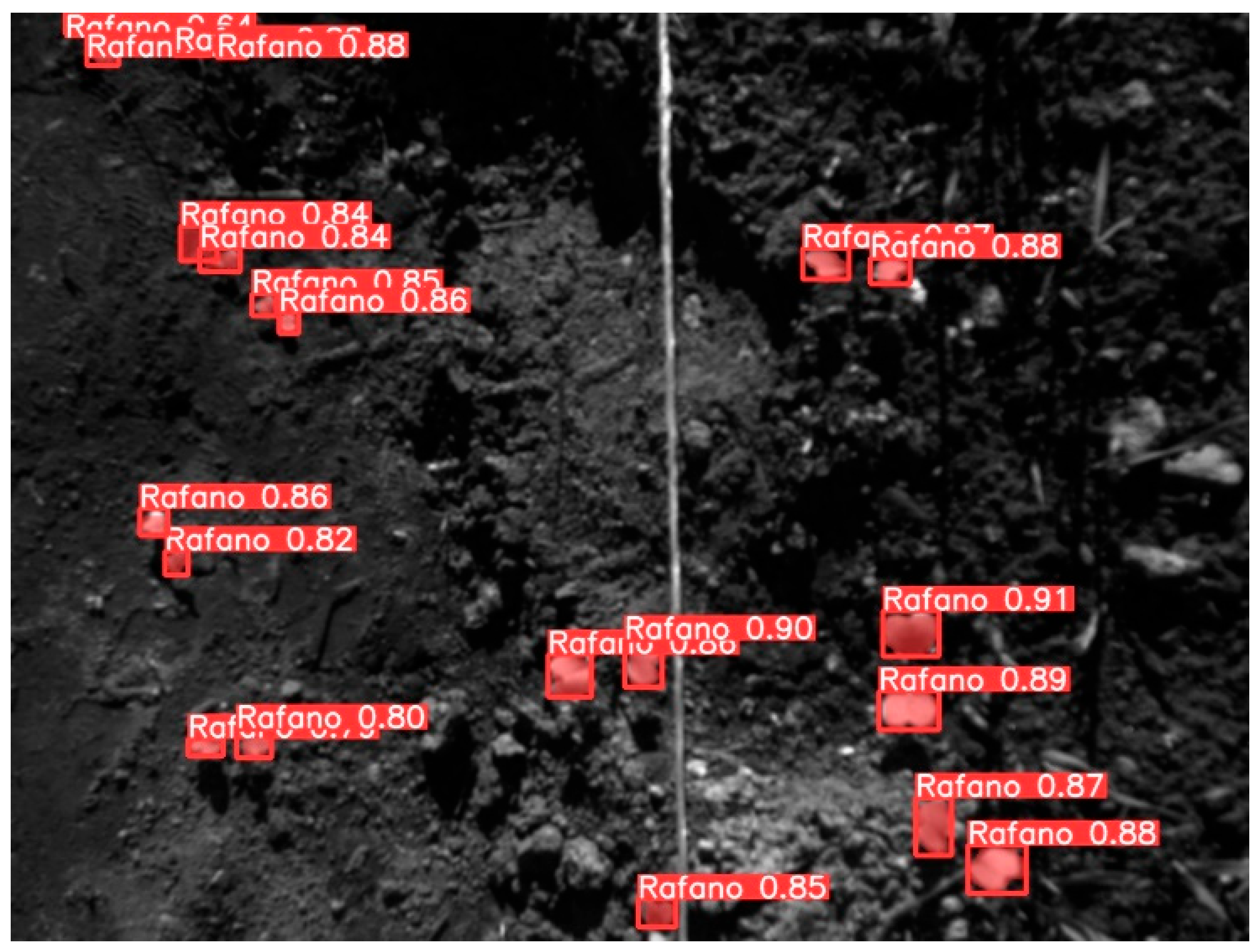
3.7. Performance of YOLOv8l-Seg towards Prediction of Wild Radish Mask Instances
4. Discussion
4.1. Overall Performance of Different Registration Strategies
4.2. Performance of Registration Strategies over Individual Instances across Spectra
4.3. Performance of Registration Methods over Predicted Instances across Different Spectral Channels
4.4. Error Evaluations across Registration Methods
4.5. Automatic Prediction of Wild Radish Instances
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vuletic, J.; Polic, M.; Orsag, M. Introducing Multispectral-Depth (MS-D): Sensor Fusion for Close Range Multispectral Imaging. In Proceedings of the IEEE International Conference on Automation Science and Engineering, Mexico City, Mexico, 20–24 August 2022; pp. 1218–1223. [Google Scholar] [CrossRef]
- Jhan, J.P.; Rau, J.Y.; Haala, N.; Cramer, M. Investigation of parallax issues for multi-lens multispectral camera band co-registration. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci.-ISPRS Arch. 2017, 42, 157–163. [Google Scholar] [CrossRef]
- Seidl, K.; Richter, K.; Knobbe, J.; Maas, H.-G. Wide field-of-view all-reflective objectives designed for multispectral image acquisition in photogrammetric applications. In Proceedings of the Complex Systems OCS11, Marseille, Franch, 5–8 September 2011; Volume 8172, p. 817210. [Google Scholar] [CrossRef]
- St-Charles, P.L.; Bilodeau, G.A.; Bergevin, R. Online Mutual Foreground Segmentation for Multispectral Stereo Videos. Int. J. Comput. Vis. 2019, 127, 1044–1062. [Google Scholar] [CrossRef]
- Wu, L.; Shang, Q.; Sun, Y.; Bai, X. A self-adaptive correction method for perspective distortions of image. Front. Comput. Sci. 2019, 13, 588–598. [Google Scholar] [CrossRef]
- Jhan, J.-P.; Rau, J.-Y.; Haala, N. Robust and adaptive band-to-band image transform of UAS miniature multi-lens multispectral camera. ISPRS J. Photogramm. Remote Sens. 2018, 137, 47–60. [Google Scholar] [CrossRef]
- Shahbazi, M.; Cortes, C. Seamless co-registration of images from multi-sensor multispectral cameras. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci.-ISPRS Arch. 2019, 42, 315–322. [Google Scholar] [CrossRef]
- MicaSense Inc. MicaSense RedEdge-MTM Multispectral Camera User Manual. 2017. Available online: https://support.micasense.com/hc/en-us/articles/115003537673-RedEdge-M-User-Manual-PDF-Legacy (accessed on 29 January 2024).
- Kwan, C. Methods and challenges using multispectral and hyperspectral images for practical change detection applications. Information 2019, 10, 353. [Google Scholar] [CrossRef]
- Panuju, D.R.; Paull, D.J.; Griffin, A.L. Change detection techniques based on multispectral images for investigating land cover dynamics. Remote Sens. 2020, 12, 1781. [Google Scholar] [CrossRef]
- Parelius, E.J. A Review of Deep-Learning Methods for Change Detection in Multispectral Remote Sensing Images. Remote Sens. 2023, 15, 2092. [Google Scholar] [CrossRef]
- Yuan, K.; Zhuang, X.; Schaefer, G.; Feng, J.; Guan, L.; Fang, H. Deep-Learning-Based Multispectral Satellite Image Segmentation for Water Body Detection. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2021, 14, 7422–7434. [Google Scholar] [CrossRef]
- Lebourgeois, V.; Bégué, A.; Labbé, S.; Mallavan, B.; Prévot, L.; Roux, B. Can commercial digital cameras be used as multispectral sensors? A crop monitoring test. Sensors 2008, 8, 7300–7322. [Google Scholar] [CrossRef]
- Mäkeläinen, A.; Saari, H.; Hippi, I.; Sarkeala, J.; Soukkamäki, J. 2D hyperspectral frame imager camera data in photogrammetric mosaicking. ISPRS-Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2013, XL-1/W2, 263–267. [Google Scholar] [CrossRef]
- Lowe, D.G. Object Recognition from Local Scale-Invariant Features. In Proceedings of the Seventh IEEE International Conference on Computer Vision, Kerkyra, Greece, 20–27 September 1999. [Google Scholar]
- Lowe, D.G. Distinctive Image Features from Scale-Invariant Keypoints. Int. J. Comput. Vis. 2004, 60, 91–110. [Google Scholar] [CrossRef]
- Liu, C.; Xu, J.; Wang, F. A Review of Keypoints’ Detection and Feature Description in Image Registration. Sci. Program. 2021, 2021, 8509164. [Google Scholar] [CrossRef]
- Wang, H.J.; Lee, C.Y.; Lai, J.H.; Chang, Y.C.; Chen, C.M. Image registration method using representative feature detection and iterative coherent spatial mapping for infrared medical images with flat regions. Sci. Rep. 2022, 12, 7932. [Google Scholar] [CrossRef]
- Joshi, K.; Patel, M.I. Recent advances in local feature detector and descriptor: A literature survey. Int. J. Multimed. Inf. Retr. 2020, 9, 231–247. [Google Scholar] [CrossRef]
- Khan Tareen, S.A.; Zahra, S. A Comparative Analysis of SIFT, SURF, KAZE, AKAZE, ORB, and BRISK. In Proceedings of the International Conference on Computing, Mathematics and Engineering Technologies, Sukkur, Pakistan, 3–4 March 2018; Available online: https://ieeexplore.ieee.org/document/8346440 (accessed on 29 January 2024).
- Sharma, S.K.; Jain, K.; Shukla, A.K. A Comparative Analysis of Feature Detectors and Descriptors for Image Stitching. Appl. Sci. 2023, 13, 6015. [Google Scholar] [CrossRef]
- Zhang, D.; Raven, L.A.; Lee, D.J.; Yu, M.; Desai, A. Hardware friendly robust synthetic basis feature descriptor. Electronics 2019, 8, 847. [Google Scholar] [CrossRef]
- Esposito, M.; Crimaldi, M.; Cirillo, V.; Sarghini, F.; Maggio, A. Drone and sensor technology for sustainable weed management: A review. Chem. Biol. Technol. Agric. 2021, 8, 18. [Google Scholar] [CrossRef]
- Fernández, C.I.; Haddadi, A.; Leblon, B.; Wang, J.; Wang, K. Comparison between three registration methods in the case of non-georeferenced close range of multispectral images. Remote Sens. 2021, 13, 396. [Google Scholar] [CrossRef]
- Bressanin, F.N.; Giancotti, P.R.F.; Parreira, M.C.; Alves, P.L.d.C.A. Influence of Raphanus raphanistrum L. Density and Relative Time of Emergence on Bean Crop. J. Agric. Sci. 2013, 5, 199. [Google Scholar] [CrossRef]
- Kebaso, L.; Frimpong, D.; Iqbal, N.; Bajwa, A.A.; Namubiru, H.; Ali, H.H.; Ramiz, Z.; Hashim, S.; Manalil, S.; Chauhan, B.S. Biology, ecology and management of Raphanus raphanistrum L.: A noxious agricultural and environmental weed. Environ. Sci. Pollut. Res. 2020, 27, 17692–17705. [Google Scholar] [CrossRef]
- Eslami, S.V. Ecology of Wild Radish (Raphanus Raphanistrum): Crop-Weed Competition and Seed Dormancy. Ph.D. Thesis, The University of Adelaide, Adelaide, Australia, 2006. Available online: https://digital.library.adelaide.edu.au/dspace/bitstream/2440/59619/2/02whole.pdf (accessed on 29 January 2024).
- Boz, Ö. Economic Threshold for Wild Radish (Raphanus raphanistrum L.) Control in Wheat Fields. Turk. J. Agric. For. 2005, 29, 173–177. Available online: https://journals.tubitak.gov.tr/agriculture/vol29/iss3/3/ (accessed on 29 January 2024).
- Somerville, G.J.; Ashworth, M.B. Adaptations in Wild Radish (Raphanus raphanistrum) flowering time. Part 2: Harvest Weed Seed Control shortens flowering by 12 days. In Weed Science; Cambridge University Press: Cambridge, UK, 2024. [Google Scholar] [CrossRef]
- Walsh, M.J.; Owen, M.J.; Powles, S.B. Frequency and distribution of herbicide resistance in Raphanus raphanistrum populations randomly collected across the Western Australian wheatbelt. Weed Res. 2007, 47, 542–550. [Google Scholar] [CrossRef]
- Foley, J.D.; Fischler, M.A.; Bolles, R.C. Graphics and Image Processing Random Sample Consensus: A Paradigm for Model Fitting with Applications to Image Analysis and Automated Cartography. Commun. ACM 1981, 24, 381–395. [Google Scholar]
- Museyko, O.; Eisa, F.; Hess, A.; Schett, G.; Kalender, W.A.; Engelke, K. Binary segmentation masks can improve intrasubject registration accuracy of bone structures in CT images. Ann. Biomed. Eng. 2010, 38, 2464–2472. [Google Scholar] [CrossRef]
- Bolya, D.; Zhou, C.; Xiao, F.; Lee, Y.J. YOLACT: Real-Time Instance Segmentation. In Proceedings of the IEEE International Conference on Computer Vision, Seoul, Republic of Korea, 27 October–2 November 2019; pp. 9156–9165. [Google Scholar] [CrossRef]
- Szeliski, R. Image alignment and stitching: A tutorial. Found. Trends Comput. Graph. Vis. 2006, 2, 1–104. [Google Scholar] [CrossRef]
- Zitová, B.; Flusser, J. Image registration methods: A survey. Image Vis. Comput. 2003, 21, 977–1000. [Google Scholar] [CrossRef]
- Li, R.; Luo, L.; Zhang, Y. Multiframe Astronomical Image Registration Based on Block Homography Estimation. J. Sens. 2020, 2020, 8849552. [Google Scholar] [CrossRef]
- Mok, T.C.W.; Chung, A.C.S. Affine Medical Image Registration with Coarse-to-Fine Vision Transformer. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 20803–20812. [Google Scholar] [CrossRef]
- Collins, R. Introduction to Computer Vision: Planar Homographies. 2007. Available online: https://www.cse.psu.edu/~rtc12/CSE486/lecture16.pdf (accessed on 29 January 2024).
- Open Source Computer Vision. Basic Concepts of the Homography Explained with Code. Available online: https://docs.opencv.org/4.x/d9/dab/tutorial_homography.html#tutorial_homography_How_the_homography_transformation_can_be_useful (accessed on 18 November 2023).
- Crannell, A.; Frantz, M.; Futamura, F. Factoring a Homography to Analyze Projective Distortion. J. Math. Imaging Vis. 2019, 61, 967–989. [Google Scholar] [CrossRef]
- Mallon, J.; Whelan, P.F. Projective rectification from the fundamental matrix. Image Vis. Comput. 2005, 23, 643–650. [Google Scholar] [CrossRef]
- Santana-Cedrés, D.; Gomez, L.; Alemán-Flores, M.; Salgado, A.; Esclarín, J.; Mazorra, L.; Alvarez, L. Automatic correction of perspective and optical distortions. Comput. Vis. Image Underst. 2017, 161, 1–10. [Google Scholar] [CrossRef]
- Rana, S.; Gerbino, S.; Barretta, D.; Crimaldi, M.; Cirillo, V.; Maggio, A.; Sarghini, F. RafanoSet: Dataset of Manually and Automatically Annotated Raphanus Raphanistrum Weed Images for Object Detection and Segmentation in Heterogenous Agriculture Environment (Version 3) [Data Set]. Zenodo. 2024. Available online: https://zenodo.org/records/10567784 (accessed on 29 January 2024).
- Aljabri, M.; AlAmir, M.; AlGhamdi, M.; Abdel-Mottaleb, M.; Collado-Mesa, F. Towards a better understanding of annotation tools for medical imaging: A survey. Multimed. Tools Appl. 2022, 81, 25877–25911. [Google Scholar] [CrossRef]
- Nousias, G.; Delibasis, K.; Maglogiannis, I. H-RANSAC, an Algorithmic Variant for Homography Image Transform from Featureless Point Sets: Application to Video-Based Football Analytics. 2023. Available online: http://arxiv.org/abs/2310.04912 (accessed on 29 January 2024).
- Zhang, J.; Wang, C.; Liu, S.; Jia, L.; Ye, N.; Wang, J.; Zhou, J.; Sun, J. Content-Aware Unsupervised Deep Homography Estimation. In European Conference on Computer Vision; Springer: Cham, Swizerland, 2020. [Google Scholar] [CrossRef]
- Dhana Lakshmi, M.; Mirunalini, P.; Priyadharsini, R.; Mirnalinee, T.T. Review of feature extraction and matching methods for drone image stitching. In Lecture Notes in Computational Vision and Biomechanics; Springer: Cham, Swizerland, 2019; Volume 30, pp. 595–602. [Google Scholar] [CrossRef]
- Jakubović, A.J.; Velagić, J.V. Image Feature Matching and Object Detection Using Brute-Force Matchers. In Proceedings of the 2018 International Symposium ELMAR, Zadar, Croatia, 16–19 September 2018. [Google Scholar] [CrossRef]
- Kaplan, A.; Avraham, T.; Lidenbaum, M. Interpreting the Ratio Criterion for Matching SIFT Descriptors. In Lecture Notes in Computer Science; Leibe, B., Matas, J., Sebe, N., Welling, M., Eds.; Springer International Publishing: Berlin/Heidelberg, Germany, 2016; Volume 9909, pp. 697–712. [Google Scholar] [CrossRef]
- Hartley, R.; Zisserman, A. Multiple View Geometry in Computer Vision; Cambridge University Press: Cambridge, UK, 2003. [Google Scholar]
- Ultralytics. Instance Segmentation. Ultralytics YOLOv8 Docs. 2023. Available online: https://docs.ultralytics.com/tasks/segment/ (accessed on 29 January 2024).
- Marques, O. Morphological Image Processing. In Practical Image and Video Processing Using MATLAB; Wiley: Hoboken, NJ, USA, 2011; pp. 299–334. [Google Scholar] [CrossRef]
- Rahman, M.A.; Wang, Y. Optimizing Intersection-Over-Union in Deep Neural Networks for Image Segmentation; Bebis, G., Boyle, R., Parvin, B., Koracin, D., Porikli, F., Skaff, S., Entezari, A., Min, J., Iwai, D., Sadagic, A., et al., Eds.; Springer International Publishing: Berlin/Heidelberg, Germany, 2016; Volume 10072. [Google Scholar] [CrossRef]
- Rezatofighi, H.; Tsoi, N.; Gwak, J.; Sadeghian, A.; Reid, I.; Savarese, S. Generalized Intersection over Union: A Metric and A Loss for Bounding Box Regression. arXiv 2019. Available online: http://arxiv.org/abs/1902.09630 (accessed on 29 January 2024).
- Nanni, L.; Lumini, A.; Loreggia, A.; Formaggio, A.; Cuza, D. An Empirical Study on Ensemble of Segmentation Approaches. Signals 2022, 3, 341–358. [Google Scholar] [CrossRef]
- Sarvaiya, J.N.; Patnaik, S.; Bombaywala, S. Image registration by template matching using normalized cross-correlation. In Proceedings of the ACT 2009—International Conference on Advances in Computing, Control and Telecommunication Technologies, Bangalore, India, 28–29 December 2009; pp. 819–822. [Google Scholar] [CrossRef]
- Feng, Z.; Qingming, H.; Wen, G. Image matching by normalized cross-correlation. In Proceedings of the IEEE International Conference on Acoustics, Speech and Signal Processing, ICASSP 2006, Toulouse, France, 14–19 May 2006. [Google Scholar] [CrossRef]
- Heo, Y.S.; Lee, K.M.; Lee, S.U. Robust Stereo Matching Using Adaptive Normalized Cross-Correlation. IEEE Trans. Pattern Anal. Mach. Intell. 2011, 33, 807–822. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.Y.; Maire, M.; Belongie, S.; Bourdev, L.; Girshick, R.; Hays, J.; Perona, P.; Ramanan, D.; Zitnick, C.L.; Dollár, P. Microsoft COCO: Common Objects in Context. In Computer Vision–ECCV 2014; Fleet, D., Pajdla, T., Schiele, B., Tuytelaars, T., Eds.; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2014; Volume 8693. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, D.; Liu, Y. Box-supervised dynamical instance segmentation for in-field cotton. Comput. Electron. Agric. 2023, 215, 108390. [Google Scholar] [CrossRef]
- Nie, L.; Li, B.; Jiao, F.; Shao, J.Y.; Yang, T.; Liu, Z. ASPP-YOLOv5: A study on constructing pig facial expression recognition for heat stress. Comput. Electron. Agric. 2023, 214, 108346. [Google Scholar] [CrossRef]
- Kulkarni, A.; Chong, D.; Batarseh, F.A. 2020. Foundations of data imbalance and solutions for a data democracy. In Data Demoicracy; Elsevier: Amsterdam, The Netherlands, 2020; pp. 83–106. [Google Scholar] [CrossRef]
- Zeng, Y.; Hao, D.; Huete, A.; Dechant, B.; Berry, J.; Chen, J.M.; Joiner, J.; Frankenberg, C.; Bond-Lamberty, B.; Ryu, Y.; et al. Optical vegetation indices for monitoring terrestrial ecosystems globally. Nat. Rev. Earth Environ. 2022, 3, 477–493. [Google Scholar] [CrossRef]
- Maria, K.; René, G.; Cecilie, H.; Yi, P.; Per, M.; Lis, W.; Mogens, H.G. Comparing predictive ability of laser-induced breakdown spectroscopy to visible near-infrared spectroscopy for soil property determination. Biosyst. Eng. 2017, 156, 157–172. [Google Scholar] [CrossRef]
- Li, X.; Gao, J.; Jin, S.; Jiang, C.; Zhao, M.; Lu, M. Towards robust registration of heterogeneous multispectral UAV imagery: A two-stage approach for cotton leaf lesion grading. Comput. Electron. Agric. 2023, 212, 108153. [Google Scholar] [CrossRef]

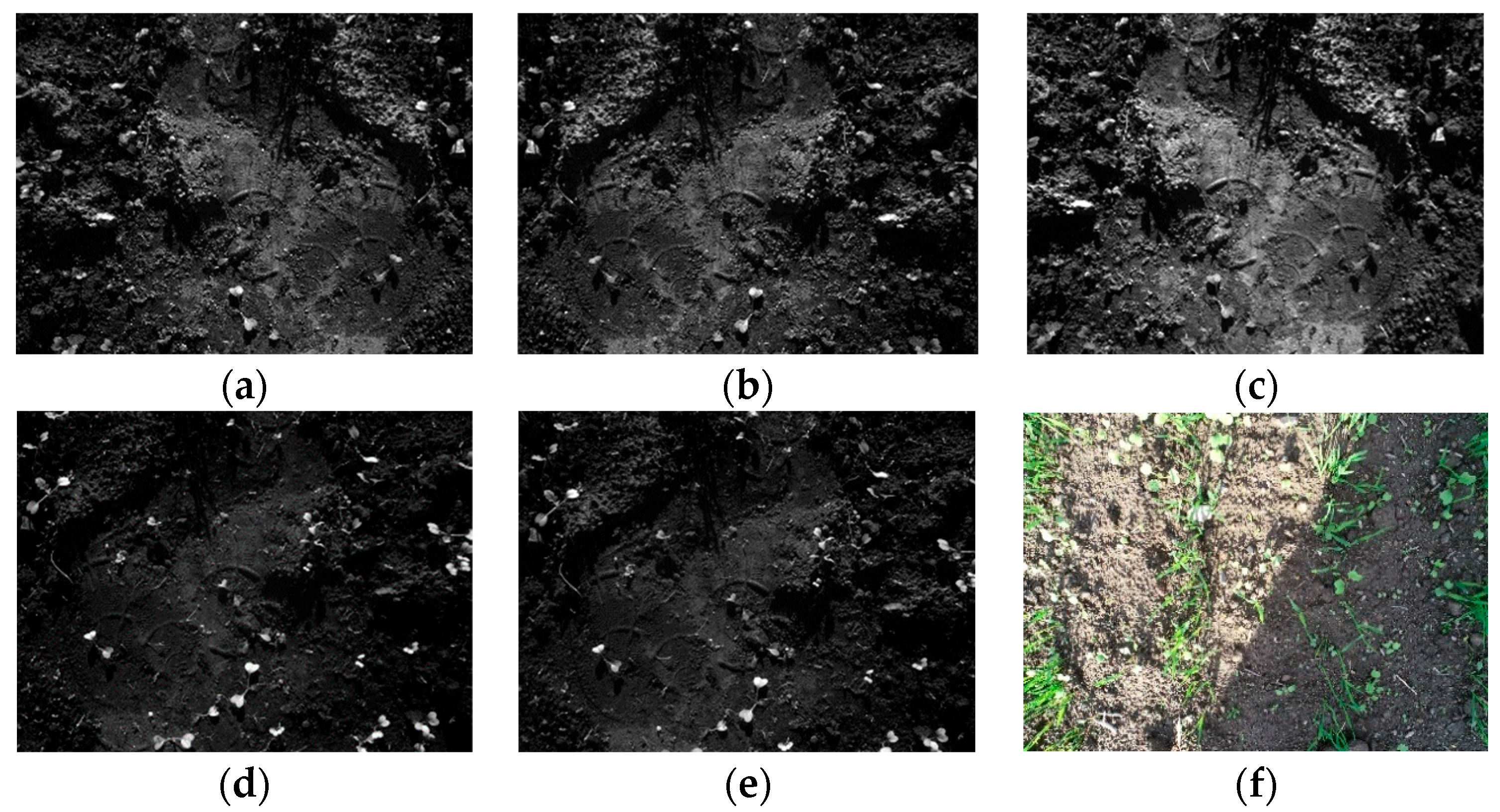





| Spatial resolution | 0.07 cm/pixel at 1 m altitude |
| Frame rate | 1 image/second |
| Wavelength (nm) | Red (668 nm), green (560 nm), blue (475 nm), near-IR (840 nm), RedEdge (717 nm) |
| Method | Intersection over Union | Normalized Coefficient of Correlation |
|---|---|---|
| Registration based on spatial realignment of annotations | 0.8055 | 0.8900 |
| Registration applied on binary masks | 0.8314 | 0.9059 |
| Registration applied on masked pixels | 0.8183 | 0.8971 |
| Cumulative Performance | Spatial Realignment of Annotations | Registration Based on Binary Masks | Registration Based on Masked Pixels | |||||
|---|---|---|---|---|---|---|---|---|
| Spectral Channels | IoU | NCC | IoU | NCC | IoU | NCC | IoU | NCC |
| Blue | 0.8206 | 0.8996 | 0.8091 | 0.8929 | 0.8364 | 0.9094 | 0.8162 | 0.8965 |
| Green | 0.8408 | 0.9128 | 0.8266 | 0.9043 | 0.8476 | 0.9155 | 0.8481 | 0.9173 |
| Red | 0.7509 | 0.853 | 0.7415 | 0.8474 | 0.7690 | 0.8654 | 0.7421 | 0.8462 |
| Near-infrared | 0.8614 | 0.9252 | 0.8449 | 0.9155 | 0.8726 | 0.9170 | 0.8667 | 0.9284 |
| Spectral Channels | Registration Based on Spatial Realignment of Annotations | Registration Based on Binary Masks | Registration Based on Masked Pixels | |||
|---|---|---|---|---|---|---|
| IoU | NCC | IoU | NCC | IoU | NCC | |
| Blue | 0.8962 | 0.945 | 0.8961 | 0.9453 | 0.8081 | 0.892 |
| Green | 0.8969 | 0.9457 | 0.9339 | 0.9658 | 0.7539 | 0.7539 |
| Red | 0.8894 | 0.9413 | 0.8872 | 0.9282 | 0.9159 | 0.9573 |
| Near-infrared | 0.8249 | 0.9046 | 0.9326 | 0.9652 | 0.8871 | 0.9407 |
| Cumulative average | 0.8768 | 0.9341 | 0.9124 | 0.9511 | 0.8412 | 0.8859 |
| Method | IoU | NCC |
|---|---|---|
| Registration based on predicted binary masks | 0.7188 | 0.8099 |
| Registration based on predicted and masked pixels | 0.7185 | 0.8016 |
| Spectral Channels | Registration Based on Predicted Binary Masks | Registration Based on Predicted and Masked Pixels | ||
|---|---|---|---|---|
| IoU | NCC | IoU | NCC | |
| Blue | 0.7481 | 0.8525 | 0.7171 | 0.8313 |
| Green | 0.7223 | 0.8357 | 0.7378 | 0.8462 |
| Red | 0.6723 | 0.8009 | 0.6751 | 0.8019 |
| Near-infrared | 0.7353 | 0.8456 | 0.7144 | 0.8298 |
| Cumulative mean | 0.7195 | 0.8336 | 0.7111 | 0.8273 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rana, S.; Gerbino, S.; Crimaldi, M.; Cirillo, V.; Carillo, P.; Sarghini, F.; Maggio, A. Comprehensive Evaluation of Multispectral Image Registration Strategies in Heterogenous Agriculture Environment. J. Imaging 2024, 10, 61. https://doi.org/10.3390/jimaging10030061
Rana S, Gerbino S, Crimaldi M, Cirillo V, Carillo P, Sarghini F, Maggio A. Comprehensive Evaluation of Multispectral Image Registration Strategies in Heterogenous Agriculture Environment. Journal of Imaging. 2024; 10(3):61. https://doi.org/10.3390/jimaging10030061
Chicago/Turabian StyleRana, Shubham, Salvatore Gerbino, Mariano Crimaldi, Valerio Cirillo, Petronia Carillo, Fabrizio Sarghini, and Albino Maggio. 2024. "Comprehensive Evaluation of Multispectral Image Registration Strategies in Heterogenous Agriculture Environment" Journal of Imaging 10, no. 3: 61. https://doi.org/10.3390/jimaging10030061
APA StyleRana, S., Gerbino, S., Crimaldi, M., Cirillo, V., Carillo, P., Sarghini, F., & Maggio, A. (2024). Comprehensive Evaluation of Multispectral Image Registration Strategies in Heterogenous Agriculture Environment. Journal of Imaging, 10(3), 61. https://doi.org/10.3390/jimaging10030061








