Optimization Production of an Endo-β-1,4-Xylanase from Streptomyces thermocarboxydus Using Wheat Bran as Sole Carbon Source
Abstract
1. Introduction
2. Results and Discussion
2.1. Production Optimization of Streptomyces thermocarboxydus TKU045 Xylanase Using Wheat Bran Powder as the Sole Carbon Source
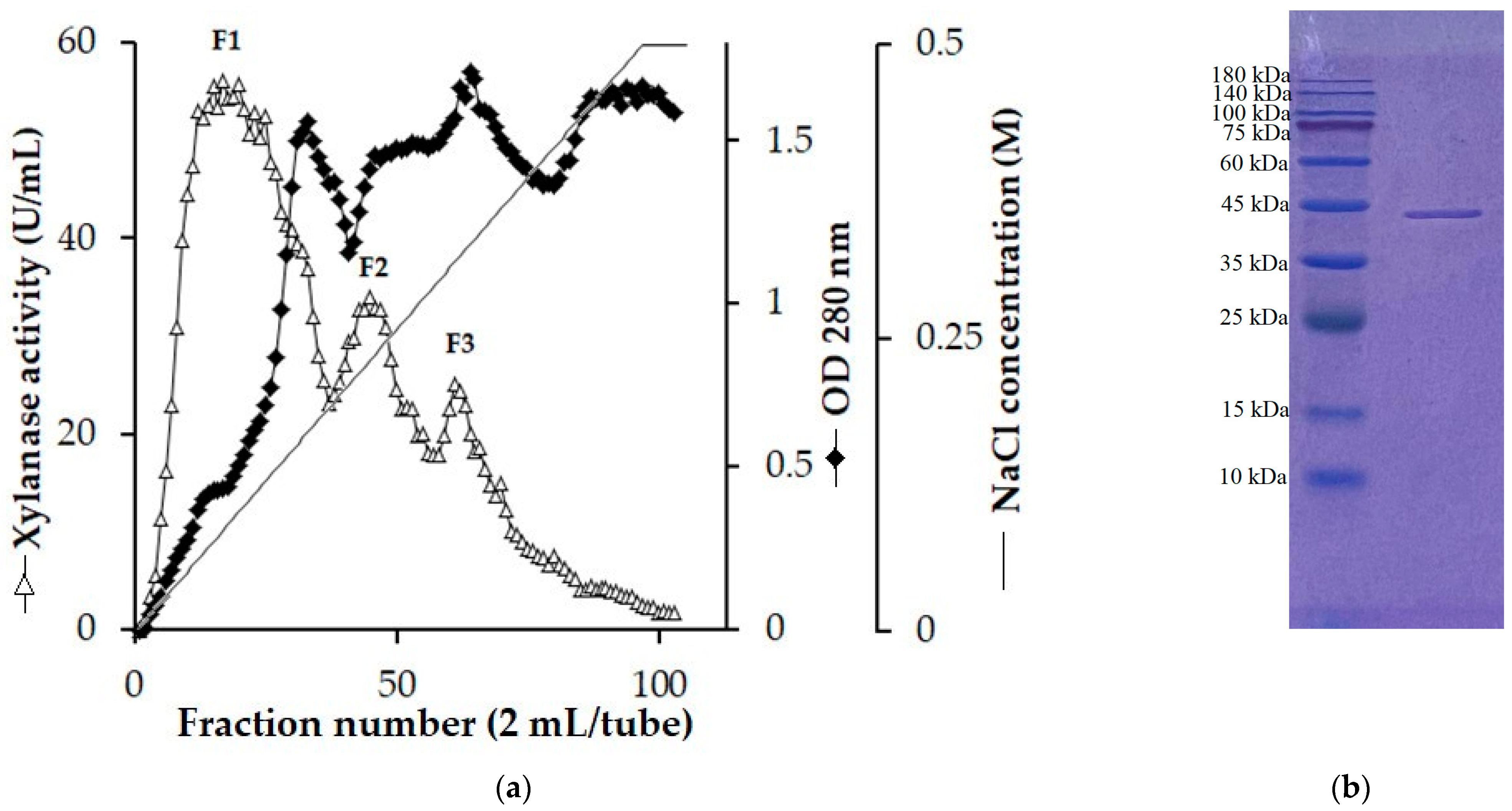
2.2. Enzyme Purification
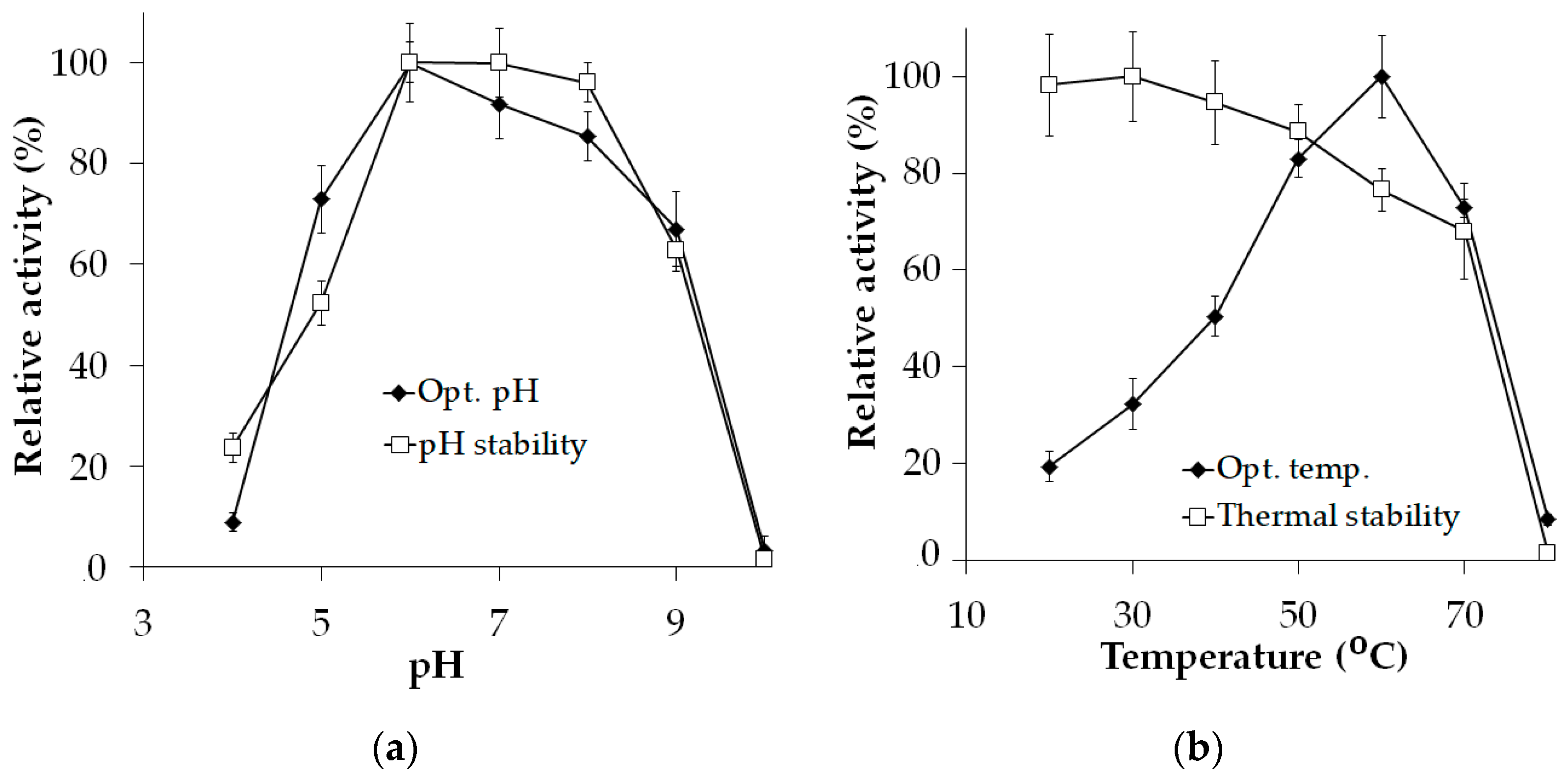
2.3. Biochemical Characterization
2.4. Hydrolysis Pattern and Xylooligosaccharide Production
3. Materials and Methods
3.1. Materials
3.2. Xylanase Assay and Protein Determination
3.3. Optimization of Production
3.4. Enzyme Purification and Identification
3.5. Enzyme Characterization
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Cruz-Davila, J.; Vargas Perez, J.; Sosa del Castillo, D.; Diez, N. Fusarium graminearum as a producer of xylanases with low cellulases when grown on wheat bran. Biotechnol. Rep. 2022, 35, e00738. [Google Scholar]
- Kumari, K.; Goyal, S.; Maan, S. Hyper xylanase production and potential of xylooligosaccharides formation from a novel Bacillus australimaris KS2. Biocatal. Agric. Biotechol. 2023, 54, 102899. [Google Scholar] [CrossRef]
- Rosmine, E.; Sainjan, N.C.; Silvester, R.; Alikkunju, A.; Varghese, S.A. Statistical optimisation of xylanase production by estuarine Streptomyces sp. and its application in clarification of fruit juice. J. Gen. Eng. Biotechnol. 2017, 15, 393–401. [Google Scholar] [CrossRef]
- Güler, F.; Özçelik, F. Screening of xylanase producing Bacillus species and optimization of xylanase process parameters in submerged fermentation. Biocatal. Agric. Biotechol. 2023, 51, 102801. [Google Scholar] [CrossRef]
- Wang, F.; Xu, H.; Wang, M.; Yu, X.; Cui, Y.; Xu, L.; Ma, A.; Ding, Z.; Huo, S.; Zou, B.; et al. Application of immobilized enzymes in juice clarification. Foods 2023, 12, 4258. [Google Scholar] [CrossRef]
- Dhaver, P.; Pletschke, B.; Sithole, B.; Govinden, R. Optimization of xylooligosaccharides production by native and recombinant xylanase hydrolysis of chicken feed substrates. Int. J. Mol. Sci. 2023, 24, 17110. [Google Scholar] [CrossRef]
- Alananbeh, K.M.; Alkfoof, R.; Muhaidat, R.; Massadeh, M. Production of xylanase by Trichoderma species growing on olive mill pomace and barley bran in a packed-bed bioreactor. J. Fungi 2024, 10, 49. [Google Scholar] [CrossRef]
- Ameen, F. Purification and characterization of xylanase produced by Aspergillus fumigatus isolated from the northern border region of Saudi Arabia. Fermentation 2023, 9, 595. [Google Scholar] [CrossRef]
- López-López, A.; Santiago-Hernández, A.; Cayetano-Cruz, M.; García-Huante, Y.; Campos, J.E.; Bustos-Jaimes, I.; Marsch-Moreno, R.; Cano-Ramírez, C.; Benitez-Cardoza, C.G.; Hidalgo-Lara, M.E. TtCel7A: A native thermophilic bifunctional cellulose/xylanase exogluclanase from the thermophilic biomass-degrading fungus Thielavia terrestris Co3Bag1, and its application in enzymatic hydrolysis of agroindustrial derivatives. J. Fungi 2023, 9, 152. [Google Scholar] [CrossRef]
- Hüttner, S.; Granchi, Z.; Nguyen, T.T.; Pelt, S.V.; Larsbrink, J.; Thanh, V.N.; Olsson, L. Genome sequence of Rhizomucor pusillus FCH 5.7, a thermophilic zygomycete involved in plant biomass degradation harbouring putative GH9 endoglucanases. Biotechnol. Rep. 2018, 20, e00279. [Google Scholar] [CrossRef]
- Wu, Q.; Zhang, C.; Zhu, W.; Lu, H.; Li, X.; Yang, Y.; Xu, Y.; Li, W. Improved thermostability, acid tolerance as well as catalytic efficiency of Streptomyces rameus L2001 GH11 xylanase by N-terminal replacement. Enzym. Microb. Technol. 2023, 162, 110143. [Google Scholar] [CrossRef]
- Valenzuela, S.V.; Díaz, P.; Javier Pastor, F.I. Recombinant expression of an alkali stable GH10 xylanase from Paenibacillus barcinonensis. J. Agric. Food Chem. 2010, 58, 4814–4818. [Google Scholar] [CrossRef]
- Veerakumar, S.; Manian, R. Agarase, Amylase and xylanase from Halomonas meridiana: A study on optimization of coproduction for biomass saccharification. Fermentation 2022, 8, 479. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, X.; Lu, C.; Lu, P.; Yin, C.; Ye, Z.; Huang, Z. Identification and characterization of a novel endo-β-1,4-xylanase from Streptomyces sp. T7 and its application in xylo-oligosaccharide production. Molecules 2022, 27, 2516. [Google Scholar] [CrossRef]
- Li, N.; Meng, K.; Wang, Y.; Shi, P.; Luo, H.; Bai, Y.; Yang, P.; Yao, B. Cloning, expression, and characterization of a new xylanase with broad temperature adaptability from Streptomyces sp. S9. Appl. Microbiol. Biotechnol. 2008, 80, 231–240. [Google Scholar] [CrossRef]
- Tran, T.N.; Doan, C.T.; Nguyen, V.B.; Nguyen, A.D.; Wang, S.L. The isolation of chitinase from Streptomyces thermocarboxydus and its application in the preparation of chitin oligomers. Res. Chem. Intermed. 2019, 45, 727–742. [Google Scholar] [CrossRef]
- Tran, T.N.; Doan, C.T.; Nguyen, M.T.; Nguyen, V.B.; Vo, T.P.K.; Nguyen, A.D.; Wang, S.L. An Exochitinase with N-acetyl-β-glucosaminidase-like activity from shrimp head conversion by Streptomyces speibonae and its application in hydrolyzing β-chitin powder to produce N-acetyl-D-glucosamine. Polymers 2019, 11, 1600. [Google Scholar] [CrossRef]
- Govindaraji, P.K.; Vuppu, S. Characterisation of pectin and optimization of pectinase enzyme from novel Streptomyces fumigatiscleroticus VIT-SP4 for drug delivery and concrete crack-healing applications: An eco-friendly approach. Saudi J. Biol. Sci. 2020, 27, 3529–3540. [Google Scholar] [CrossRef]
- Tran, T.N.; Doan, C.T.; Nguyen, V.B.; Nguyen, A.D.; Wang, S.L. Conversion of fishery waste to proteases by Streptomyces speibonae and their application in antioxidant preparation. Fishes 2022, 7, 140. [Google Scholar] [CrossRef]
- Sharma, P.; Sharma, N. RSM approach to pre-treatment of lignocellulosic waste and a statistical methodology for optimizing bioethanol production. Waste Manag. Bull. 2024, 2, 49–66. [Google Scholar] [CrossRef]
- Khangkhachit, W.; Suyotha, W.; Leamdum, C. Production of thermostable xylanase using Streptomyces thermocarboxydus ME742 and application in enzymatic conversion of xylan from oil palm empty fruit bunch to xylooligosaccharides. Biocatal. Agric. Biotechol. 2021, 37, 102180. [Google Scholar] [CrossRef]
- Wu, H.; Cheng, X.; Zhu, Y.; Zeng, W.; Chen, G.; Liang, Z. Purification and characterization of a cellulase-free, thermostable endo-xylanase from Streptomyces griseorubens LH-3 and its use in biobleaching on eucalyptus kraft pulp. J. Biosci. Bioeng. 2018, 125, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Sinjaroonsak, S.; Chaiyaso, T.; H-Kittikun, A. Optimization of cellulase and xylanase productions by Streptomyces thermocoprophilus TC13W using low cost pretreated oil palm empty fruit bunch. Waste Biomass Valori. 2019, 11, 3925–3936. [Google Scholar] [CrossRef]
- Blasi, A.; Verardi, A.; Lopresto, C.G.; Siciliano, S.; Sangiorgio, P. Lignocellulosic agricultural waste valorization to obtain valuable products: An overview. Recycling 2023, 8, 61. [Google Scholar] [CrossRef]
- Ketsakhon, P.; Thammasittirong, A.; Thammasittirong, S.N. Adding value to rice straw waste for high-level xylanase production using a new isolate of Bacillus altitudinis RS3025. Folia Microbiol. 2023, 68, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Gautério, G.V.; da Silva, L.G.G.; Hübner, T.; da Rosa Ribeiro, T.; Kalil, S.J. Maximization of xylanase production by Aureobasidium pullulans using a by-product of rice grain milling as xylan source. Biocatal. Agric. Biotechnol. 2020, 23, 101511. [Google Scholar] [CrossRef]
- Tanwar, E.; Nagar, S.; Kumari, K.; Mallesh, G.; Goyal, S. Enrichment of papaya juice using covalently immobilized xylanase from Bacillus pumilus SV-85S. Biomass Conv. Bioref. 2022, 17, 1–17. [Google Scholar] [CrossRef]
- Alokika; Singh, B. Enhanced production of bacterial xylanase and its utility in saccharification of sugarcane bagasse. Bioprocess. Biosyst. Eng. 2020, 43, 1081–1091. [Google Scholar] [CrossRef] [PubMed]
- Jnawali, P.; Kumar, V.; Tanwar, B.; Hirdyani, H.; Gupta, P. Enzymatic production of xylooligosaccharides from brown coconut husk treated with sodium hydroxide. Waste Biomass Valoriz. 2017, 9, 1757–1766. [Google Scholar] [CrossRef]
- Zang, H.; Du, X.; Wang, J.; Cheng, Y.; Wang, Y.; Sun, S.; Zhao, X.; Li, D.; Zhang, H.; Li, C. Insight into cold-active xylanase production and xylan degradation pathways in psychrotrophic Acinetobacter sp. HC4 from the cold region of China. Cellulose 2020, 27, 7575–7589. [Google Scholar] [CrossRef]
- Ire, F.S.; Chima, I.J.; Ezebuiro, V. Enhanced xylanase production from UV-mutated Aspergillus niger grown on corn cob and sawdust. Biocatal. Agric. Biotechnol. 2021, 31, 101869. [Google Scholar] [CrossRef]
- Tran, T.N.; Doan, C.T.; Wang, S.L. Conversion of wheat bran to xylanases and dye adsorbent by Streptomyces thermocarboxydus. Polymers 2021, 13, 287. [Google Scholar] [CrossRef] [PubMed]
- Iram, A.; Cekmecelioglu, D.; Demirci, A. Optimization of the fermentation parameters to maximize the production of cellulases and xylanases using DDGS as the main feedstock in stirred tank bioreactors. Biocatal. Agric. Biotechol. 2022, 45, 102514. [Google Scholar]
- Prabha, B.; Ramesh, D.; Sriramajayam, S.; Uma, D. Optimization of pyrolysis process parameters for fuel oil production from the thermal recycling of waste polypropylene grocery bags using the Box–Behnken design. Recycling 2024, 9, 15. [Google Scholar] [CrossRef]
- Lenth, R.V. Response-Surface Methods in R, Using rsm. J. Stat. Softw. 2009, 30, 7–15. [Google Scholar]
- Sanjivkumar, M.; Silambarasan, T.; Balagurunathan, R.; Immanuel, G. Biosynthesis, molecular modeling and statistical optimization of xylanase from a mangrove associated actinobacterium Streptomyces variabilis (MAB3) using Box-Behnken design with its bioconversion efficacy. Int. J. Biol. Macromol. 2018, 118, 195–208. [Google Scholar] [CrossRef] [PubMed]
- Medouni-Haroune, L.; Medouni-Adar, S.; Houfani, A.A.; Bouiche, C.; Azzouz, Z.; Roussos, S.; Desseaux, V.; Madani, K.; Kecha, M. Statistical optimization and partial characterization of xylanases produced by Streptomyces sp. S1M3I using olive pomace as a fermentation substrate. Appl. Biochem. Biotechnol. 2024, 196, 2012–2013. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.Y.; Han, M.K.; Oh, H.W.; Park, D.S.; Kim, S.J.; Lee, S.G.; Shin, D.H.; Son, K.H.; Bae, K.S.; Park, H.Y. Catalytic properties of a GH10 endo-β-1, 4- xylanase from Streptomyces thermocarboxydus HY-15 isolated from the gut of Eisenia fetida. J. Mol. Catal. B Enzym. 2010, 62, 32–39. [Google Scholar] [CrossRef]
- Chi, W.J.; Lim, J.H.; Park, D.Y.; Park, J.S.; Hong, S.K. Production and characterization of a thermostable endo-type β-xylanase produced by a newly-isolated Streptomyces thermocarboxydus subspecies MW8 strain from Jeju Island. Process Biochem. 2013, 48, 1736–1743. [Google Scholar] [CrossRef]
- Boonchuay, P.; Takenaka, S.; Kuntiya, A.; Techapun, C.; Leksawasdi, N.; Seesuriyachan, P.; Chaiyaso, T. Purification, characterization, and molecular cloning of the xylanase from Streptomyces thermovulgaris TISTR1948 and its application to xylooligosaccharide production. J. Mol. Catal. B Enzym. 2016, 129, 61–68. [Google Scholar] [CrossRef]
- He, J.; Su, L.; Sun, X.; Fu, J.; Chen, J.; Wu, J. A novel xylanase from Streptomyces sp. FA1: Purification, characterization, identification, and heterologous expression. Biotechnol. Bioproc. E 2014, 19, 8–17. [Google Scholar] [CrossRef]
- Yan, Q.; Hao, S.; Jiang, Z.; Zhai, Q.; Chen, W. Properties of a xylanase from Streptomyces matensis being suitable for xylooligosaccharides production. J. Mol. Catal. B Enzym. 2009, 58, 72–77. [Google Scholar] [CrossRef]
- Ninawe, S.; Kapoor, M.; Kuhad, R.C. Purification and characterization of extracellular xylanase from Streptomyces cyaneus SN32. Bioresour. Technol. 2007, 99, 1252–1258. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Z.; Shi, P.; Luo, H.; Bai, Y.; Yuan, T.; Yang, P.; Liu, S.; Yao, B. A xylanase with broad pH and temperature adaptability from Streptomyces megasporus DSM 41476, and its potential application in brewing industry. Enzyme Microb. Technol. 2010, 46, 506–512. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Qin, L.; Xu, Y.; Lu, H.; Wu, Q.; Li, W.; Zhang, C.; Li, X. Three molecular modification strategies to improve the thermostability of xylanase XynA from Streptomyces rameus L2001. Foods 2023, 12, 879. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, E.; Zhu, Y.; Teng, C.; Sun, B.; Song, H.; Yang, R. A typical endoxylanase from Streptomyces rameus L2001 and its unique characteristics in xylooligosaccharide production. Carbohydr. Res. 2012, 359, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Li, J.; Teng, C.; Li, X. Cloning, overexpression and characterization of a xylanase gene from a novel Streptomyces rameus L2001 in Pichia pastoris. J. Mol. Catal. B-Enzym. 2016, 131, 85–93. [Google Scholar] [CrossRef]
- Mander, P.; Choi, Y.H.; Pradeep, G.C.; Choi, Y.S.; Hong, J.H.; Cho, S.S.; Yoo, J.C. Biochemical characterization of xylanase produced from Streptomyces sp. CS624 using an agro residue substrate. Process Biochem. 2014, 49, 451–456. [Google Scholar] [CrossRef]
- Manicardi, T.; Baioni e Silva, G.; Longati, A.A.; Paiva, T.D.; Souza, J.P.M.; Pádua, T.F.; Furlan, F.F.; Giordano, R.L.C.; Giordano, R.C.; Milessi, T.S. xylooligosaccharides: A bibliometric analysis and current advances of this bioactive food chemical as a potential product in biorefineries’ portfolios. Foods 2023, 12, 3007. [Google Scholar] [CrossRef]
- Kim, D.; Yu, J.; Hong, K.; Jung, C.; Kim, H.; Kim, J.; Myung, S. Green production of low-molecular-weight xylooligosaccharides from oil palm empty fruit bunch via integrated enzymatic polymerization and membrane separation for purification. Sep. Purif. Technol. 2022, 293, 121084. [Google Scholar] [CrossRef]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Doan, C.T.; Tran, T.N.; Nguyen, V.B.; Nguyen, A.D.; Wang, S.L. Utilization of seafood processing by-products for production of proteases by Paenibacillus sp. TKU052 and their application in biopeptides’ preparation. Mar. Drugs 2020, 18, 574. [Google Scholar] [CrossRef] [PubMed]
- Doan, C.T.; Tran, T.N.; Tran, T.P.H.; Nguyen, T.T.; Nguyen, H.K.; Tran, T.K.T.; Vu, B.T.; Trinh, T.H.T.; Nguyen, A.D.; Wang, S.L. Chitosanase production from the liquid fermentation of squid pens waste by Paenibacillus elgii. Polymers 2023, 15, 3724. [Google Scholar] [CrossRef]

| Run | Coded Levels | Uncoded Levels | Y (U/mL) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| X1 | X2 | X3 | X4 | X5 | X1 | X2 | X3 | X4 | X5 | ||
| 1 | −1 | −1 | 0 | 0 | 0 | 1 | 1 | 10 | 37 | 2 | 8.494 |
| 2 | +1 | −1 | 0 | 0 | 0 | 3 | 1 | 10 | 37 | 2 | 9.396 |
| 3 | −1 | +1 | 0 | 0 | 0 | 1 | 2 | 10 | 37 | 2 | 1.500 |
| 4 | +1 | +1 | 0 | 0 | 0 | 3 | 2 | 10 | 37 | 2 | 1.651 |
| 5 | 0 | 0 | −1 | −1 | 0 | 2 | 1.5 | 9 | 34 | 2 | 14.488 |
| 6 | 0 | 0 | +1 | −1 | 0 | 2 | 1.5 | 11 | 34 | 2 | 0.043 |
| 7 | 0 | 0 | −1 | +1 | 0 | 2 | 1.5 | 9 | 40 | 2 | 16.373 |
| 8 | 0 | 0 | +1 | +1 | 0 | 2 | 1.5 | 11 | 40 | 2 | 0.355 |
| 9 | 0 | −1 | 0 | 0 | −1 | 2 | 1 | 10 | 37 | 1 | 2.613 |
| 10 | 0 | +1 | 0 | 0 | −1 | 2 | 2 | 10 | 37 | 1 | 0.561 |
| 11 | 0 | −1 | 0 | 0 | +1 | 2 | 1 | 10 | 37 | 3 | 14.571 |
| 12 | 0 | +1 | 0 | 0 | +1 | 2 | 2 | 10 | 37 | 3 | 9.163 |
| 13 | −1 | 0 | −1 | 0 | 0 | 1 | 1.5 | 9 | 37 | 2 | 6.858 |
| 14 | +1 | 0 | −1 | 0 | 0 | 3 | 1.5 | 9 | 37 | 2 | 5.093 |
| 15 | −1 | 0 | +1 | 0 | 0 | 1 | 1.5 | 11 | 37 | 2 | 0.086 |
| 16 | +1 | 0 | +1 | 0 | 0 | 3 | 1.5 | 11 | 37 | 2 | 0.045 |
| 17 | 0 | 0 | 0 | −1 | −1 | 2 | 1.5 | 10 | 34 | 1 | 0.365 |
| 18 | 0 | 0 | 0 | +1 | −1 | 2 | 1.5 | 10 | 40 | 1 | 0.624 |
| 19 | 0 | 0 | 0 | −1 | +1 | 2 | 1.5 | 10 | 34 | 3 | 10.960 |
| 20 | 0 | 0 | 0 | +1 | +1 | 2 | 1.5 | 10 | 40 | 3 | 12.687 |
| 21 | 0 | −1 | −1 | 0 | 0 | 2 | 1 | 9 | 37 | 2 | 18.841 |
| 22 | 0 | +1 | −1 | 0 | 0 | 2 | 2 | 9 | 37 | 2 | 12.625 |
| 23 | 0 | −1 | +1 | 0 | 0 | 2 | 1 | 11 | 37 | 2 | 8.638 |
| 24 | 0 | +1 | +1 | 0 | 0 | 2 | 2 | 11 | 37 | 2 | 0.193 |
| 25 | −1 | 0 | 0 | −1 | 0 | 1 | 1.5 | 10 | 34 | 2 | 8.998 |
| 26 | +1 | 0 | 0 | −1 | 0 | 3 | 1.5 | 10 | 34 | 2 | 6.257 |
| 27 | −1 | 0 | 0 | +1 | 0 | 1 | 1.5 | 10 | 40 | 2 | 9.622 |
| 28 | +1 | 0 | 0 | +1 | 0 | 3 | 1.5 | 10 | 40 | 2 | 6.374 |
| 29 | 0 | 0 | −1 | 0 | −1 | 2 | 1.5 | 9 | 37 | 1 | 2.645 |
| 30 | 0 | 0 | +1 | 0 | −1 | 2 | 1.5 | 11 | 37 | 1 | 0.000 |
| 31 | 0 | 0 | −1 | 0 | +1 | 2 | 1.5 | 9 | 37 | 3 | 11.229 |
| 32 | 0 | 0 | +1 | 0 | +1 | 2 | 1.5 | 11 | 37 | 3 | 10.778 |
| 33 | −1 | 0 | 0 | 0 | −1 | 1 | 1.5 | 10 | 37 | 1 | 0.526 |
| 34 | +1 | 0 | 0 | 0 | −1 | 3 | 1.5 | 10 | 37 | 1 | 0.680 |
| 35 | −1 | 0 | 0 | 0 | +1 | 1 | 1.5 | 10 | 37 | 3 | 4.983 |
| 36 | +1 | 0 | 0 | 0 | +1 | 3 | 1.5 | 10 | 37 | 3 | 11.855 |
| 37 | 0 | −1 | 0 | −1 | 0 | 2 | 1 | 10 | 34 | 2 | 12.385 |
| 38 | 0 | +1 | 0 | −1 | 0 | 2 | 2 | 10 | 34 | 2 | 8.163 |
| 39 | 0 | −1 | 0 | +1 | 0 | 2 | 1 | 10 | 40 | 2 | 18.121 |
| 40 | 0 | +1 | 0 | +1 | 0 | 2 | 2 | 10 | 40 | 2 | 6.335 |
| 41 | 0 | 0 | 0 | 0 | 0 | 2 | 1.5 | 10 | 37 | 2 | 26.148 |
| 42 | 0 | 0 | 0 | 0 | 0 | 2 | 1.5 | 10 | 37 | 2 | 26.978 |
| 43 | 0 | 0 | 0 | 0 | 0 | 2 | 1.5 | 10 | 37 | 2 | 22.187 |
| 44 | 0 | 0 | 0 | 0 | 0 | 2 | 1.5 | 10 | 37 | 2 | 22.402 |
| 45 | 0 | 0 | 0 | 0 | 0 | 2 | 1.5 | 10 | 37 | 2 | 26.188 |
| 46 | 0 | 0 | 0 | 0 | 0 | 2 | 1.5 | 10 | 37 | 2 | 23.790 |
| Term | Coefficient Estimate | Standard Error Coefficient | t Value | Pr (>|t|) | |
|---|---|---|---|---|---|
| Constant | −2167.200 | 296.800 | −7.302 | <0.00001 | *** |
| X1 | 39.162 | 24.626 | 1.590 | 0.124 | |
| X2 | 138.530 | 49.627 | 2.791 | 0.010 | ** |
| X3 | 180.640 | 28.006 | 6.450 | <0.00001 | *** |
| X4 | 61.012 | 9.894 | 6.167 | <0.00001 | *** |
| X5 | 34.787 | 24.626 | 1.413 | 0.170 | |
| X1X2 | −0.375 | 2.983 | −0.126 | 0.901 | |
| X1X3 | 0.431 | 1.491 | 0.289 | 0.775 | |
| X1X4 | −0.042 | 0.497 | −0.085 | 0.933 | |
| X1X5 | 1.680 | 1.491 | 1.126 | 0.271 | |
| X2X3 | −1.115 | 2.983 | −0.374 | 0.712 | |
| X2X4 | −1.261 | 0.994 | −1.268 | 0.216 | |
| X2X5 | −1.678 | 2.983 | −0.563 | 0.579 | |
| X3X4 | −0.131 | 0.497 | −0.264 | 0.794 | |
| X3X5 | 0.548 | 1.491 | 0.368 | 0.716 | |
| X4X5 | 0.122 | 0.497 | 0.246 | 0.808 | |
| X12 | −11.173 | 1.010 | −11.066 | <0.00001 | *** |
| X22 | −27.745 | 4.038 | −6.870 | <0.00001 | *** |
| X32 | −9.016 | 1.010 | −8.930 | <0.00001 | *** |
| X42 | −0.781 | 0.112 | −6.961 | <0.00001 | *** |
| X52 | −10.187 | 1.010 | −10.091 | <0.00001 | *** |
| Source | Degrees of Freedom | Sum of Squares | Mean Square | F Value | Pr (>F) |
|---|---|---|---|---|---|
| Fo | 5 | 851.01 | 170.20 | 19.133 | <0.00001 |
| TWI | 10 | 32.95 | 3.30 | 0.371 | 0.948 |
| PQ | 5 | 1811.54 | 362.31 | 40.729 | <0.00001 |
| Residuals | 25 | 222.39 | 8.90 | ||
| Lack of fit | 20 | 200.51 | 10.03 | 2.291 | 0.182 |
| Pure error | 5 | 21.88 | 4.38 |
| Step | Total Protein (mg) | Total Activity (U) | Specific Activity (U/mg) | Recovery (%) | Purification (Fold) |
|---|---|---|---|---|---|
| Cultural supernatant | 4858.7 | 26,111.0 | 5.4 | 100 | 1.0 |
| Ethanol precipitation | 2439.1 | 25,078.6 | 10.3 | 96 | 1.9 |
| High Q column | 57.7 | 10,857.8 | 188.1 | 42 | 35.0 |
| DEAE sepharose column | 3.2 | 3882.330 | 1212.7 | 15 | 225.7 |
| Matched Peptide Sequence | Identified Protein and Coverage Rate | Strain |
|---|---|---|
| 90IDATEPQR97 144QAMIDHINGVMAHYK158 161 IVQWDVVNEA FADGSSGAR179 208LCYNDYNVENWTWAK222 247GVPIDCVGFQSHFNSGSPYNSNFR260 418VQIYSCWGGDNQK430 | Endo-1,4-beta-xylanase 19% | Streptomyces lividans |
| Chemical | Relative Activity (%) |
|---|---|
| Control | 100.000 ± 10.057 |
| Zn2+ | 2.676 ± 4.837 |
| Fe2+ | 51.140 ± 9.080 |
| Fe3+ | 0.793 ± 6.868 |
| Cu2+ | 0.000 ± 3.156 |
| Mg2+ | 142.616 ± 10.016 |
| Mn2+ | 219.029 ± 18.431 |
| Ba2+ | 168.682 ± 18.554 |
| Ca2+ | 170.565 ± 11.894 |
| 2-mercaptoethanol | 166.501 ± 8.121 |
| Tween 20 | 164.420 ± 5.252 |
| Tween 40 | 161.943 ± 27.468 |
| Triton X-100 | 149.653 ± 13.872 |
| SDS | 3.964 ± 5.152 |
| EDTA | 105.198 ± 4.454 |
| Chemical | Relative Activity (%) |
|---|---|
| Birchwood xylan * | 100.000 ± 6.585 |
| Beechwood xylan | 93.433 ± 6.935 |
| Oatspelt xylan | 95.646 ± 5.945 |
| Starch | ND |
| Pectin | ND |
| Cellulose | ND |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tran, T.N.; Doan, C.T.; Dinh, T.K.L.; Duong, T.H.N.; Phan, T.T.U.; Le, T.T.L.; Tran, T.D.; Hoang, P.H.Q.; Nguyen, A.D.; Wang, S.-L. Optimization Production of an Endo-β-1,4-Xylanase from Streptomyces thermocarboxydus Using Wheat Bran as Sole Carbon Source. Recycling 2024, 9, 50. https://doi.org/10.3390/recycling9030050
Tran TN, Doan CT, Dinh TKL, Duong THN, Phan TTU, Le TTL, Tran TD, Hoang PHQ, Nguyen AD, Wang S-L. Optimization Production of an Endo-β-1,4-Xylanase from Streptomyces thermocarboxydus Using Wheat Bran as Sole Carbon Source. Recycling. 2024; 9(3):50. https://doi.org/10.3390/recycling9030050
Chicago/Turabian StyleTran, Thi Ngoc, Chien Thang Doan, Thi Kieu Loan Dinh, Thi Hai Ninh Duong, Thi Thuc Uyen Phan, Thi Thuy Loan Le, Trung Dung Tran, Pham Hung Quang Hoang, Anh Dzung Nguyen, and San-Lang Wang. 2024. "Optimization Production of an Endo-β-1,4-Xylanase from Streptomyces thermocarboxydus Using Wheat Bran as Sole Carbon Source" Recycling 9, no. 3: 50. https://doi.org/10.3390/recycling9030050
APA StyleTran, T. N., Doan, C. T., Dinh, T. K. L., Duong, T. H. N., Phan, T. T. U., Le, T. T. L., Tran, T. D., Hoang, P. H. Q., Nguyen, A. D., & Wang, S.-L. (2024). Optimization Production of an Endo-β-1,4-Xylanase from Streptomyces thermocarboxydus Using Wheat Bran as Sole Carbon Source. Recycling, 9(3), 50. https://doi.org/10.3390/recycling9030050









