1. Introduction
Climate change impacts agriculture through the reduction in crop productivity, becoming a threat to global food security [
1]. This is true particularly in the tropical regions of the world, which are suffering from drought, heat, and floodings. Climate models suggest that global warming will increase the frequency of the Niño Southern Oscillation (ENSO) phenomenon [
2,
3] and tropical precipitation will decrease in certain areas along the margins of the convection zones, which already suffer from water scarcity [
4]. Other expected changes are the increase in carbon dioxide (CO
2) in the air and also the temperature of the environment. Recent evidence shows that temperature has been raised by an average of 0.6 ± 0.2 °C since 1900 [
5]. All these challenges require a novel agricultural model that integrates breeding strategies and different agronomic practices to deal with the main plant stressors which include drought, salinity, elevated CO
2, and temperature (low and high), among others [
1,
5]. Therefore, to develop and select high yielding varieties, adaptation to challenging and specific climate conditions has to be recognized. Generally, plant breeding primarily depends on the presence of substantial genetic variation to address maximum genetic yield potential among crops and exploitation of this variation through effective selection for improvement [
6]. However, adaptation to climate fluctuation and stress conditions is very a critical issue for current breeding programs [
1] and evaluations under a wide range of environmental conditions confirm phenotypic plasticity of promising genotypes, through which agronomic relevance is expressed in the increase in yield productivity. Therefore, it can be considered one of several strategies to tackle climate change [
7].
Agrobiodiversity is key for making production systems more resilient [
8], and its use in evaluations under multi-environmental conditions is an important step in the selection process for ideotypes identification [
1]. Sweet potato (
Ipomoea batatas (L.) Lam) is an herbaceous species from Convolvulaceae family, and
I. batatas is the only species with economic importance as crop [
9]. Sweet potato is a staple crop and is cultivated worldwide using more than 7 million hectares and producing 88 million tons [
10]. It is a versatile species that can adapt to a wide range of environmental conditions [
11]. However, in Colombia, this crop remains neglected and underutilized. Despite its nutritional root quality and broad adaptability, it is still considered an alternative crop [
9,
12], although one which is being introduced into commercial agricultural production systems.
Agronomic evaluation of sweet potato cultivars in multiple locations evidence that the main source of variation is caused by environment effect [
13]. This high plasticity of the sweet potato plant is evidenced also in its adaptation to several conditions, including adaptation at different altitudes, from 0 to 3000 m above sea level (masl) [
14,
15]. Sweet potato is sensitive to environmental variation, as shown by Genotype × Environment (G × E) interaction studies in agronomic traits such as total root yield, commercial root yield, dry matter content, vine length, storage root length, storage root girth, marketable storage root number per plant, weight of above ground biomass, and harvestable index [
16,
17,
18,
19]. Generally, the environmental contribution to total variation in sweet potato yield is greater than that of the genotype [
17] and only some quality traits are relatively more stable [
12,
19]. Only a few cases have identified genotypes with stable performance in different locations (wide adaptability) [
20,
21,
22], while mostly specific adaptability is shown [
23,
24].
Currently, several bioinformatic tools have been established with the primary aim of aiding breeding decisions in early planning and implementation phases through combination of breeding strategies such as phenotypic selection, genomic selection, and speed breeding with genomic information with a high degree of flexibility [
25,
26]. However, plant ideotype is produced in the mind of experienced breeders. Plant ideotype generally integrates a group of desirable traits which can be measured independently in the field. Therefore, selection procedure under current challenging environmental conditions should be systematic and dynamic to better understand the genotype-by-environment interaction in multi-environment trials [
27,
28]. Multi-trait selection was proposed for plant breeding many years ago [
29]. The assignment of subjective economic weight to each trait is performed by the breeder and has been a simple method to analyze. Similar indexes have been proposed for the selection of multiple traits, and an example of its use in defining a plant ideotype was presented. The use of eigenvalues in the analysis helps reduce multicollinearity, and this approach eliminates the need for assigning weights [
30,
31]. However, a selection index (SI) has been a way to simplify the selection process by integrating the information from various desired traits found within a plant ideotype into a single estimated value. Traits such as root yield, dry matter content, plant type, etc., are computed in an SI equation to produce estimated values for specific environmental conditions or a muti-environmental response. In cassava, a SI with weight based on the breeder’s judgement is used; however, the variables should be standardized before in order to avoid a problem due to the range of magnitude among selected variables. The standardization is performed through classical statistical formulas that use total average and standard deviation [
32]. Furthermore, the SI is calculated for selected variables, assigning them optimal weightings based on informed judgment [
33].
Selection analysis complemented by G × A interaction studies allows for the identification of cultivars with high yields for a given region and identification of ideal cultivation conditions. Among the multivariate statistical techniques, AMMI (Additive Main Effects and Multiplicative Interactions) determine the stability of genotypes to adjust their productive capacity to the widest environmental variation [
34]. The G × A interaction is the factor that most interferes in the identification of specific clones for specific environments, which limits the precision in yield estimation [
23,
35].
The aim of this study was to combine multi-trait selection index, stability, and genetic gain analysis to assist phenotypic sweet potato selection from multi-environmental conditions determining their specific or wide adaptability, developing and validating the R software-based script CropInd to facilitate this multi-trait and multi-environmental data analysis.
2. Materials and Methods
This study included the evaluation of several sweet potato genotypes according to two last steps of the breeding cycle to produce new varieties. Therefore, field data collection to evaluate agronomic performance under multi-environmental conditions was conducted and further analysis included development and validation of R software script to support genotype selection.
2.1. Plant Material
Nineteen sweet potato genotypes which are part of a working collection of Agrosavia were evaluated in this study. Although there is evidence of some genotypes being introductions from other countries, all the genotypes were collected in farms of sweet potato producers. The genotypes evaluated in this study were the following: 0113-672COR, 0113-668VAL, 0113-657VAL, 0113-664VAL, 0113-671VAL, 0113-634VAL, 0113-670VAL, 0113-660VAL, 0113-663VAL, 0113-674VAL, 0113-665VAL, 0113-659VAL, 0113-673VAL, 0113-669VAL, 0113-656COR, 0113-662VAL, 0113-658COR, 0113-666VAL and CRIOLLA, a traditional clone, was used as a check.
2.2. Growth Conditions
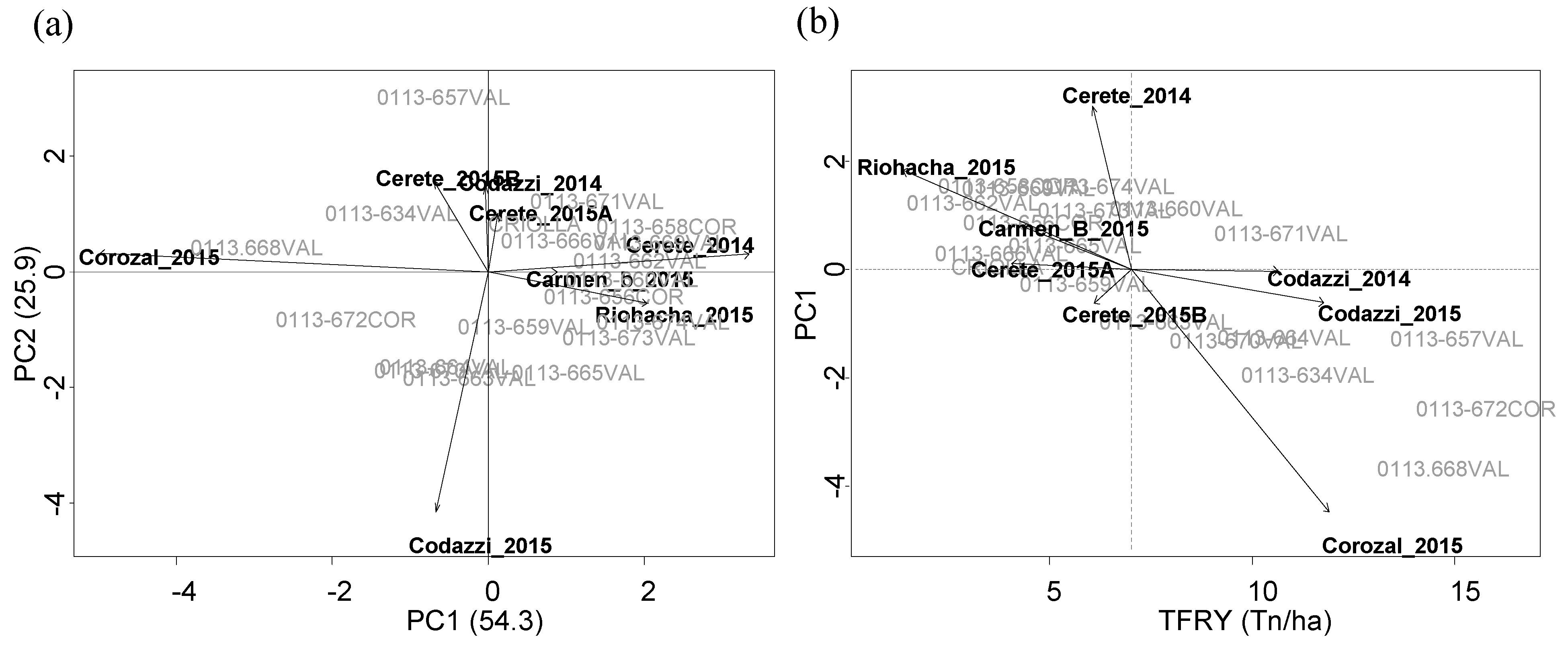
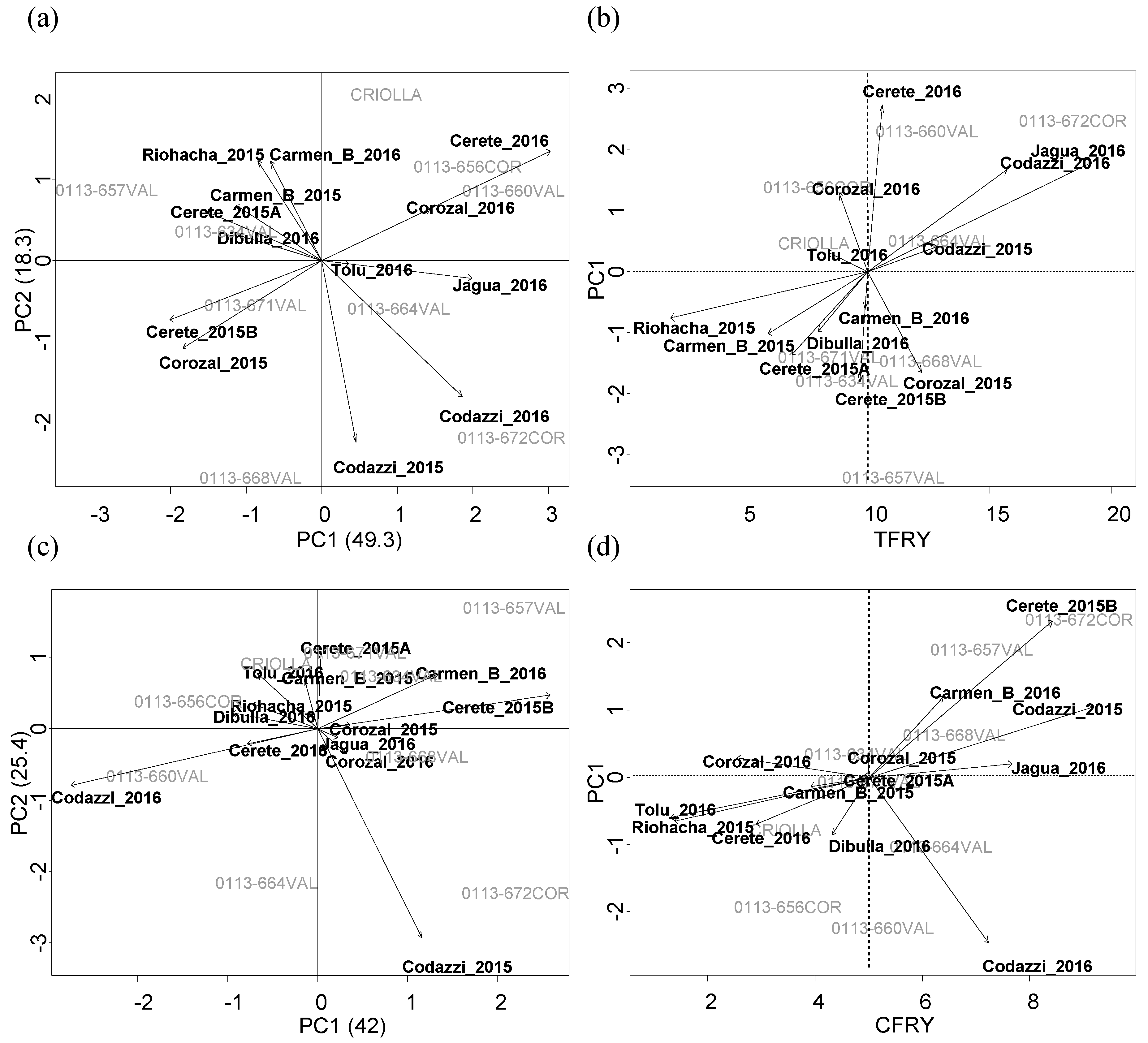
Sweet potato genotypes were established in eight localities in sub-humid region in the Colombian Caribbean coast in crop cycles evaluated during years 2014, 2015, and 2016 (
Table 1). Agroclimatic conditions in this region provide an excellent environment to develop the commercial sweet potato crop [
9]; however, for this, improved varieties were needed. According to breeding process in this population, during 2014 and 2015, a uniform yield test (UYT) was established using the mentioned nineteen genotypes under conditions of Codazzi, Corozal, Cerete, Carmen-B, and Riohacha. Furthermore, genotype selection based in their agronomic performance resulted in nine genotypes, which were evaluated during 2016. This step named the agronomic evaluation test (AET) was established at Jagua, Codazzi, Corozal, Cerete, Carmen-B, Tolu, and Dibulla.
2.3. Experimental Design
Genotypes were established under experimental conditions of two steps in a conventional breeding scheme [
35]: a uniform yield test (UYT, also named multi-local trials) and an agronomic evaluation test (AET, following regulations in Colombia for new varieties development). Experimental units using the UYT were established under randomized complete block design (RCBD) with three replications. Each plot consisted of four rows, each measuring five meters in length, with a planting density of 25,000 plants per hectare (spaced at 1 m between rows and 0.4 m within rows). This resulted in a total of 48 plants per plot. Using the AET, experimental units followed same experimental design (RCBD) with four replications, with each plot with five rows, each measuring five meters in length and with the same previous density, which resulted in a total number of 60 plants per plot. At 120 days after planting, survival percentage was estimated from established versus initially planted plants. Forage yield was estimated from a sample taken in a 1 m
2 square. Furthermore, yield components (number and weight) were determined for plant from five plants per experimental unit. Total and commercial roots (first category) from experimental unit were weighted to further estimate total and commercial yield, respectively.
2.4. Phenotypic Selection Index and CropInd Implementation
Breeders select genotypes according to their superiority in desired traits regarding average population performance. To consider multiple traits in the selection process, a selection index (SI) is routinely used in some crops. The selection index in this study applied to sweet potato was based in traits such as total fresh root yield (TFRY), high commercial fresh root yield (CFRY), and good establishment represented by survival percentage (SP).
In order to simplify the multi-traits analysis across selection process, an R software-based script CropInd was developed. CropInd required a data frame, where the first three columns of excel file should be ordered as environment (it could combine location and year in a single column), genotype, and repetition. The following columns included any traits collected in the field, and in this case, were TFRY, CFRY, and SP (in the file more traits can be added). Selected desired traits that were used for SI together with their respective assigned judgment weight were added in the first two rows of the excel file.
The breeder’s selection index was established based on the breeder’s expertise. Specific traits were chosen and assigned optimal weights based on informed judgment [
33], demonstrated as follows:
The values in the selection index formula depend on their contribution to improve the characteristics of a breed genotype; thus, in traits in which meaning is related to detriment of genotype, the assign value should be subtracted, then the negative weight should be added, such root cracking in sweet potato, which is a desired target. Desired targets are represented by a lower number (1 desired and 5 undesired character). In general, a SI is defined as a linear function of two or more phenotypes weighted according to their relevance, economic importance, or selection objectives. The SI can be written as:
where,
is the phenotypic value of the
trait to select and
corresponds to their assigned weight.
Initially,
CropInd calculates standardized values for each trait as follows:
where,
is the standardized phenotypic value,
corresponds to the individual phenotipic value,
is the phenotypic mean in the sample, and
is the standard deviation from entire sample
(General Selection Index). SI for specific environment is calculated through standardization as follows:
where,
is the phenotypic value,
corresponds to the individual phenotypic value,
is the phenotypic mean in specific environment, and
is the standard deviation from specific environment Further, averaged SI for each genotype is estimated from single values in replicates.
Subsequently, the phenotypic values for each individual are transformed into individual selection indices for the entire evaluated population, as well as for each environment specified by the user.
To improve the user-friendliness during analysis performance, a start file named Start CropInd was designed. To perform the CropInd analysis in an R session, the following instructions can be followed:
source(“CropInd.R”) # Load CropInd functions
data = read.table(“File.txt”, header=T, na.string=“.”, skip=2) # Read data
IS =read.table(“File.txt”,na.string=“.”, nrow=2) # Read Selection Index Weights
IdxRun(data, IS) # Perform Selection Index estimation.
The first row of
File.txt must provide information about desired traits and the second row must provide the weights based on trait importance or selection criteria. The third row contains the column names of data, and subsequently rows provide the sample information and traits values to evaluate (
Supplementary File S1). There is no limit in the number of traits to evaluate, as well no limits in environments or genotypes. The user can include experimental replicates (column Replicates in
File.txt) with a number to differentiate each. In case of absence of replicates, user must include this column using a number one for each sample in this column.
2.5. AMMI and Data Analysis
A combined analysis of variance was used to test genotype and environment effects and magnitude of G × E interaction. AMMI analysis was used to determine main or additive genotype and environmental effects, and multiplicative effects for G × E interaction. Tukey’s multiple comparison test was used for mean comparison between genotypes and locations (α = 0.01). Statistical Analysis System (SAS, version 9.4) y R (version 3.3.0) were used to perform the analysis.